Cervical Cancer Stages, Human Papillomavirus Integration, and Malignant Genetic Mutations: Integrative Analysis of Datasets from Four Different Cohorts
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets Used in This Study
- Datasets generated by The Cancer Genome Atlas Research (TCGA) under the “Integrated genomic and molecular characterization of cervical cancer [17] (https://www.cancer.gov/tcga, accessed on 7 October 2023) and the Cancer Genome Characterization Initiative (https://ocg.cancer.gov/programs/cgci, accessed on 7 October 2023)” were accessed via the supplemental material of a Nature Communications paper [33]. The number of cervical cancer patients for the TCGA cohort, N = 228.
- Datasets from the MOCC (Chinese) cohort were obtained from the GitHub repository of the Cell Genomics paper “Multi-omics characterization of silent and productive HPV integration in cervical cancer” [34] (https://github.com/FlyPythons/MOCC/tree/v1.0.0/data, accessed on 7 October 2023). The number of cervical cancer patients, N = 106.
- Datasets for the Ugandan cohort were obtained from an online repository of the Nature Genetics paper “Analysis of Ugandan cervical carcinomas identifies HPV clade–specific epigenome and transcriptome landscapes” [35] (https://www.nature.com/articles/s41588-020-0673-7#Sec46, accessed on 7 October 2023). The number of cervical cancer patients, N = 212.
- Datasets on cervical cancer (risk factors) were collected at the ‘Hospital Universitario de Caracas’ in Caracas, Venezuela, and the datasets focus on the prediction of indicators or diagnosis of cervical cancer. These datasets comprise demographic information, habits, and historic medical records of 858 subjects [36] (https://archive.ics.uci.edu/dataset/383/cervical+cancer+risk+factors, accessed on 7 or 13 October 2023; DOI: 10.24432/C5Z310). The latter is licensed under a Creative Commons Attribution 4.0 International license (CC BY 4.0). Several patients decided not to answer some of the questions because of privacy concerns (missing values).
2.2. Statistical and Machine Learning Methods, Gene Networks, and Visualization
3. Results
3.1. Occurrence Frequencies of Cervical Cancer Stages across Three Different Cohorts
3.2. Clinical Features Conserved across Cohorts
3.3. Correlation of Stages of Cervical Cancer with Clinical and Demographic Features
3.4. Biological Processes and Molecular Pathways Significantly Associated with Genes Mutated in Cervical Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Disclaimer
References
- Munoz, N.; Bosch, F.X.; Castellsague, X.; Diaz, M.; de Sanjose, S.; Hammouda, D.; Shah, K.V.; Meijer, C.J. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int. J. Cancer 2004, 111, 278–285. [Google Scholar] [CrossRef]
- Kim, K.S.; Park, S.A.; Ko, K.N.; Yi, S.; Cho, Y.J. Current status of human papillomavirus vaccines. Clin. Exp. Vaccine Res. 2014, 3, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Bogani, G.; Sopracordevole, F.; Ciavattini, A.; Vizza, E.; Vercellini, P.; Ghezzi, F.; Scambia, G.; Di Donato, V.; Giannini, A.; Raspagliesi, F. HPV persistence after cervical surgical excision of high-grade cervical lesions. Cancer Cytopathol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bogani, G.; Sopracordevole, F.; Ciavattini, A.; Vizza, E.; Vercellini, P.; Giannini, A.; Ghezzi, F.; Scambia, G.; Raspagliesi, F.; Di Donato, V.; et al. Duration of human papillomavirus persistence and its relationship with recurrent cervical dysplasia. Eur. J. Cancer Prev. 2023, 32, 525–532. [Google Scholar] [CrossRef]
- Pirtea, L.; Grigoras, D.; Matusz, P.; Pirtea, M.; Moleriu, L.; Tudor, A.; Ilina, R.; Secosan, C.; Horhat, F.; Mazilu, O. Age and HPV type as risk factors for HPV persistence after loop excision in patients with high grade cervical lesions: An observational study. BMC Surg. 2016, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Chotewutmontri, S.; Wolf, S.; Klos, U.; Schmitz, M.; Durst, M.; Schwarz, E. Multiplex Identification of Human Papillomavirus 16 DNA Integration Sites in Cervical Carcinomas. PLoS ONE 2013, 8, e66693. [Google Scholar] [CrossRef]
- Koneva, L.A.; Zhang, Y.; Virani, S.; Hall, P.B.; McHugh, J.B.; Chepeha, D.B.; Wolf, G.T.; Carey, T.E.; Rozek, L.S.; Sartor, M.A. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol. Cancer Res. 2018, 16, 90–102. [Google Scholar] [CrossRef]
- Tjalma, W.A.; Depuydt, C.E. Don’t forget HPV-45 in cervical cancer screening. Am. J. Clin. Pathol. 2012, 137, 161–162; author reply 162–163. [Google Scholar] [CrossRef]
- Arbyn, M.; Tommasino, M.; Depuydt, C.; Dillner, J. Are 20 human papillomavirus types causing cervical cancer? J. Pathol. 2014, 234, 431–435. [Google Scholar] [CrossRef]
- Bachtiary, B.; Obermair, A.; Dreier, B.; Birner, P.; Breitenecker, G.; Knocke, T.H.; Selzer, E.; Potter, R. Impact of multiple HPV infection on response to treatment and survival in patients receiving radical radiotherapy for cervical cancer. Int. J. Cancer 2002, 102, 237–243. [Google Scholar] [CrossRef]
- Xi, L.F.; Toure, P.; Critchlow, C.W.; Hawes, S.E.; Dembele, B.; Sow, P.S.; Kiviat, N.B. Prevalence of specific types of human papillomavirus and cervical squamous intraepithelial lesions in consecutive, previously unscreened, West-African women over 35 years of age. Int. J. Cancer 2003, 103, 803–809. [Google Scholar] [CrossRef]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef]
- Munger, K.; Jones, D.L. Human papillomavirus carcinogenesis: An identity crisis in the retinoblastoma tumor suppressor pathway. J. Virol. 2015, 89, 4708–4711. [Google Scholar] [CrossRef]
- Bahrami, A.; Hasanzadeh, M.; Shahidsales, S.; Farazestanian, M.; Hassanian, S.M.; Moetamani Ahmadi, M.; Maftouh, M.; Gharib, M.; Yousefi, Z.; Kadkhodayan, S.; et al. Genetic susceptibility in cervical cancer: From bench to bedside. J. Cell. Physiol. 2018, 233, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Vinokurova, S.; Wentzensen, N.; Kraus, I.; Klaes, R.; Driesch, C.; Melsheimer, P.; Kisseljov, F.; Durst, M.; Schneider, A.; von Knebel Doeberitz, M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008, 68, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhu, W. ERBB3 mediates the PI3K/AKT/mTOR pathway to alter the epithelial-mesenchymal transition in cervical cancer and predict immunity filtration outcome. Exp. Ther. Med. 2023, 25, 146. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, Y.; Zhang, Y.; Ge, Y.; Yin, Y.; Zhu, H. A New HPV score System Predicts the Survival of Patients With Cervical Cancers. Front. Genet. 2021, 12, 747090. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Bai, R.; Liu, Y.; Wang, K.; Wang, Y.; Yang, J.; Cai, H.; Yang, P. Multi-region sequencing depicts intratumor heterogeneity and clonal evolution in cervical cancer. Med. Oncol. 2023, 40, 78. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lei, W.; Chao, X.; Song, X.; Bi, Y.; Wu, H.; Wu, M.; Li, L. Genomic alterations caused by HPV integration in a cohort of Chinese endocervical adenocarcinomas. Cancer Gene Ther. 2021, 28, 1353–1364. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Allen-Hoffmann, B.L.; Lambert, P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995, 69, 2989–2997. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, I.; Marimon, L.; Barnadas, E.; Saco, A.; Rodriguez-Carunchio, L.; Fuste, P.; Marti, C.; Rodriguez-Trujillo, A.; Torne, A.; Del Pino, M.; et al. HPV-negative tumors of the uterine cervix. Mod. Pathol. 2019, 32, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Shiraishi, K.; Kato, T. Molecular Pathology of Human Papilloma Virus-Negative Cervical Cancers. Cancers 2021, 13, 6351. [Google Scholar] [CrossRef]
- Matsuo, K.; Mabuchi, S.; Okazawa, M.; Kawano, M.; Kuroda, H.; Kamiura, S.; Kimura, T. Clinical implication of surgically treated early-stage cervical cancer with multiple high-risk factors. J. Gynecol. Oncol. 2015, 26, 3–11. [Google Scholar] [CrossRef][Green Version]
- Lim, S.; Lee, S.H.; Lee, K.B.; Park, C.Y. The influence of number of high risk factors on clinical outcomes in patients with early-stage cervical cancer after radical hysterectomy and adjuvant chemoradiation. Obstet. Gynecol. Sci. 2016, 59, 184–191. [Google Scholar] [CrossRef]
- Odongua, N.; Chae, Y.M.; Kim, M.R.; Yun, J.E.; Jee, S.H. Associations between smoking, screening, and death caused by cervical cancer in Korean women. Yonsei Med. J. 2007, 48, 192–200. [Google Scholar] [CrossRef]
- Mutyaba, T.; Mmiro, F.A.; Weiderpass, E. Knowledge, attitudes and practices on cervical cancer screening among the medical workers of Mulago Hospital, Uganda. BMC Med. Educ. 2006, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Vignat, J.; Lorenzoni, V.; Eslahi, M.; Ginsburg, O.; Lauby-Secretan, B.; Arbyn, M.; Basu, P.; Bray, F.; Vaccarella, S. Global estimates of incidence and mortality of cervical cancer in 2020: A baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob. Health 2023, 11, e197–e206. [Google Scholar] [CrossRef]
- Zhao, X.L.; Hu, S.Y.; Hu, J.W.; Wang, H.H.; Wen, T.M.; Feng, Y.S.; Qiao, Y.L.; Zhao, F.H.; Zhang, Y. Tackling barriers to scale up human papillomavirus vaccination in China: Progress and the way forward. Infect. Dis. Poverty 2023, 12, 86. [Google Scholar] [CrossRef]
- Nakayita, R.M.; Benyumiza, D.; Nekesa, C.; Misuk, I.; Kyeswa, J.; Nalubuuka, A.; Murungi, T.; Udho, S.; Kumakech, E. Factors associated with uptake of human papilloma virus vaccine among school girls aged 9-14 years in Lira City northern Uganda: A cross-sectional study. BMC Womens Health 2023, 23, 362. [Google Scholar] [CrossRef]
- Nogueira-Rodrigues, A.; Flores, M.G.; Macedo Neto, A.O.; Braga, L.A.C.; Vieira, C.M.; de Sousa-Lima, R.M.; de Andrade, D.A.P.; Machado, K.K.; Guimaraes, A.P.G. HPV vaccination in Latin America: Coverage status, implementation challenges and strategies to overcome it. Front. Oncol. 2022, 12, 984449. [Google Scholar] [CrossRef]
- Chakravarthy, A.; Reddin, I.; Henderson, S.; Dong, C.; Kirkwood, N.; Jeyakumar, M.; Rodriguez, D.R.; Martinez, N.G.; McDermott, J.; Su, X.; et al. Integrated analysis of cervical squamous cell carcinoma cohorts from three continents reveals conserved subtypes of prognostic significance. Nat. Commun. 2022, 13, 5818. [Google Scholar] [CrossRef]
- Fan, J.; Fu, Y.; Peng, W.; Li, X.; Shen, Y.; Guo, E.; Lu, F.; Zhou, S.; Liu, S.; Yang, B.; et al. Multi-omics characterization of silent and productive HPV integration in cervical cancer. Cell Genom. 2023, 3, 100211. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.; Porter, V.L.; Zong, Z.; Bowlby, R.; Titmuss, E.; Namirembe, C.; Griner, N.B.; Petrello, H.; Bowen, J.; Chan, S.K.; et al. Analysis of Ugandan cervical carcinomas identifies human papillomavirus clade-specific epigenome and transcriptome landscapes. Nat. Genet. 2020, 52, 800–810. [Google Scholar] [CrossRef]
- Fernandes, K.; Cardoso, J.; Fernandes, J. Cervical cancer (Risk Factors). UC Irvine Mach. Learn. Repos. 2017. [Google Scholar] [CrossRef]
- Chongsuvivatwong, V. Epidemiological Data Display Package; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Garnier, S.; Ross, N.; Rudis, B.; Filipovic-Pierucci, A.; Galili, T.; Timelyportfolio; O’Callaghan, A.; Greenwell, B.; Sievert, C.; Harris, D.J.; et al. Viridis(Lite)—Colorblind-Friendly Color Maps for R. virAidis Package; Version 0.6.4; R Core Team: Vienna, Austria, 2023. [Google Scholar] [CrossRef]
- Deng, H.; Runger, G. Gene Selection with Guided Regularized Random Forest. Pattern Recognit. 2013, 46, 3483–3489. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V. R Package ‘Corrplot’: Visualization of a Correlation Matrix; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Han, Z.; Zhi, Y.; Ruan, Y.; Cao, G.; Wang, G.; Xu, X.; Mu, J.; Kang, J.; Dai, F.; et al. Long-read sequencing reveals oncogenic mechanism of HPV-human fusion transcripts in cervical cancer. Transl. Res. 2023, 253, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Wang, Y.; Liu, B.; Rao, X.; Cao, C.; Peng, F.; Zhi, W.; Wu, P.; Peng, T.; Wei, Y.; et al. Multi-omics data reveals novel impacts of human papillomavirus integration on the epigenomic and transcriptomic signatures of cervical tumorigenesis. J. Med. Virol. 2023, 95, e28789. [Google Scholar] [CrossRef]
- Iden, M.; Tsaih, S.W.; Huang, Y.W.; Liu, P.; Xiao, M.; Flister, M.J.; Rader, J.S. Multi-omics mapping of human papillomavirus integration sites illuminates novel cervical cancer target genes. Br. J. Cancer 2021, 125, 1408–1419. [Google Scholar] [CrossRef] [PubMed]


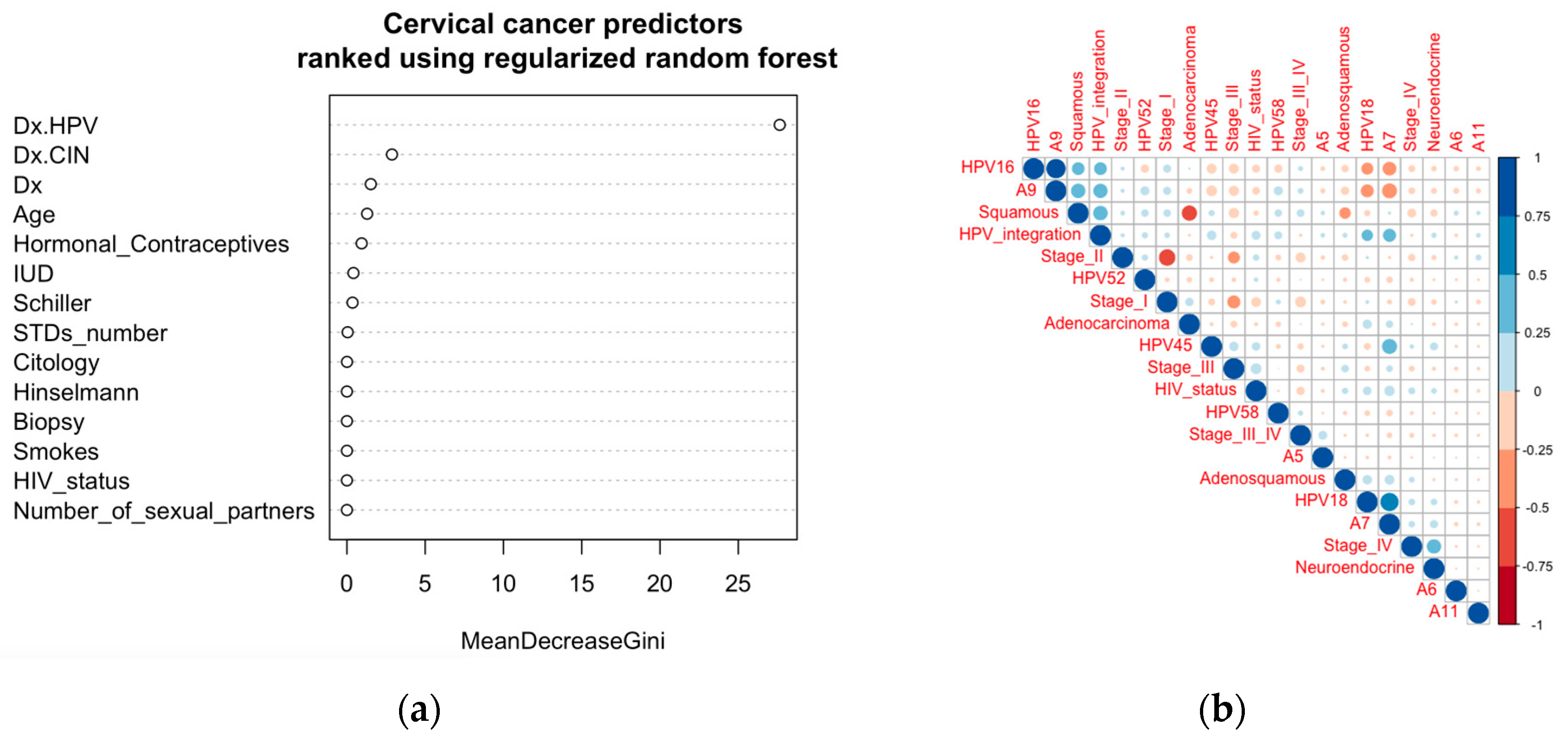
| Cervical Cancer Stage | I | II | III | III/IV | IV | NA | |||
|---|---|---|---|---|---|---|---|---|---|
| 225 | 178 | 83 | 42 | 10 | 8 | ||||
| Histology | Adenocarcinoma | Adenosquamous | Neuroendocrine | Squamous | Undifferentiated | NA | |||
| 51 | 16 | 2 | 387 | 1 | 89 | ||||
| Cancer Grade | G1 | G2 | G3 | G4 | NA | ||||
| 21 | 176 | 113 | 3 | 233 | |||||
| HPV Status | Negative | Positive | NA | ||||||
| 11 | 391 | 144 | |||||||
| HPV Type | HPV16 | HPV18 | HPV26 | HPV30 | HPV31 | HPV33 | HPV34 | HPV35 | |
| 220 | 62 | 1 | 2 | 11 | 9 | 1 | 2 | ||
| HPV39 | HPV45 | HPV51 | HPV52 | HPV56 | HPV58 | HPV59 | HPV6 | ||
| 3 | 29 | 1 | 13 | 2 | 14 | 6 | 1 | ||
| HPV66 | HPV68 | HPV69 | HPV70 | HPV73 | HPV82 | HPV9 | Negative | NA | |
| 2 | 3 | 2 | 1 | 2 | 3 | 1 | 11 | 144 | |
| HPV Clade | A5 | A6 | A7 | A9 | A11 | Negative | Other | NA | |
| 1 | 2 | 104 | 269 | 1 | 11 | 14 | 144 | ||
| HPV Integration | Non-integrated | Productive integrated | Silent integrated | NA | |||||
| 54 | 315 | 25 | 152 | ||||||
| HIV Status | Negative | Positive | NA | ||||||
| 122 | 89 | 335 | |||||||
| Age Range | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | 71–80 | 81–90 | NA | |
| 29 | 111 | 174 | 145 | 59 | 22 | 5 | 1 | ||
| Age Stat | Min. | 1st Qu. | Median | Mean | 3rd Qu. | Max. | NA | ||
| 21 | 40.24 | 48 | 49.01 | 56.71 | 89 | 1 | |||
| Race | Alaska Native | Asian | Black | White | NA | ||||
| 8 | 125 | 231 | 163 | 19 | |||||
| Cohort (Region) | TCGA (USA) | MOCC (China) | HTMCP (Uganda) | ||||||
| 228 | 106 | 212 |
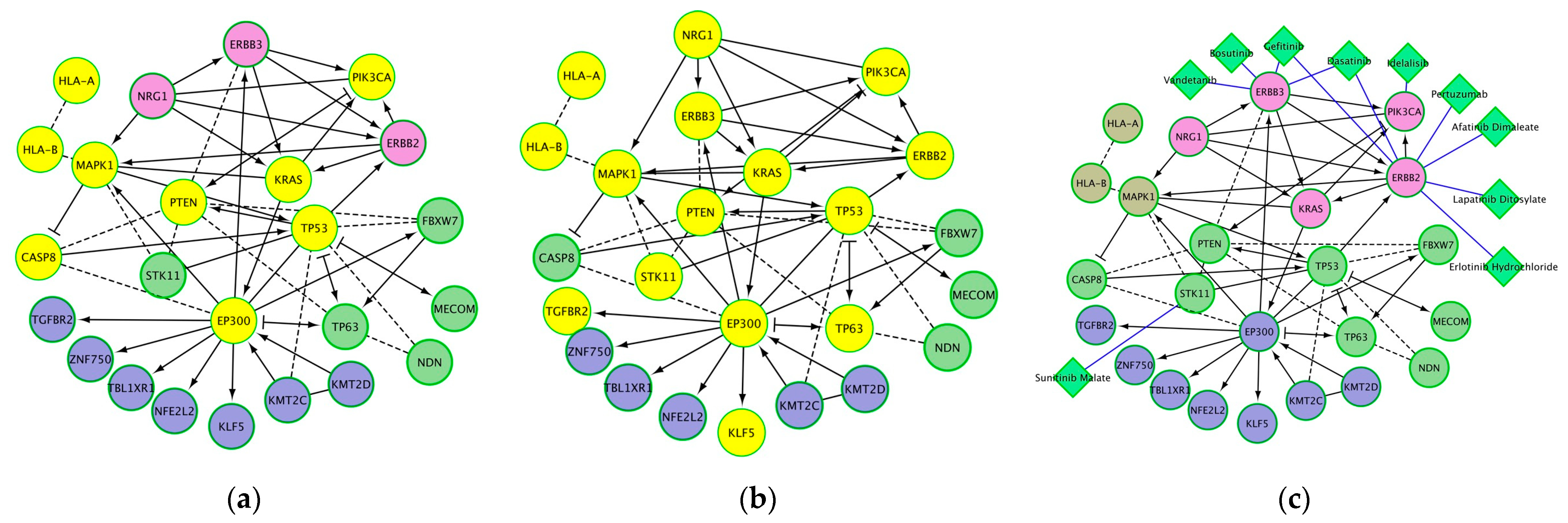
| Significantly Enriched Pathway | Proteins from Network | False Discovery Rate | Nodes |
|---|---|---|---|
| EGFR tyrosine kinase inhibitor resistance | 7 | 1.01 × 10−7 | PTEN, ERBB3, ERBB2, MAPK1, NRG1, PIK3CA, KRAS |
| Human T-cell leukemia virus 1 infection | 9 | 1.01 × 10−7 | PTEN, MAPK1, EP300, HLA-B, HLA-A, TGFBR2, PIK3CA, KRAS, TP53 |
| Cellular senescence | 8 | 1.31 × 10−7 | PTEN, MAPK1, HLA-B, HLA-A, TGFBR2, PIK3CA, KRAS, TP53 |
| Prostate cancer | 7 | 1.31 × 10−7 | PTEN, ERBB2, MAPK1, EP300, PIK3CA, KRAS, TP53 |
| p53 pathway feedback loops 2 | 5 | 1.74 × 10−7 | PTEN, TP63, PIK3CA, KRAS, TP53 |
| Endometrial cancer | 6 | 1.88 × 10−7 | PTEN, ERBB2, MAPK1, PIK3CA, KRAS, TP53 |
| Kaposi sarcoma-associated herpesvirus infection | 8 | 2.26 × 10−7 | CASP8, MAPK1, EP300, HLA-B, HLA-A, PIK3CA, KRAS, TP53 |
| Pathways in cancer | 11 | 2.35 × 10−7 | PTEN, CASP8, MECOM, ERBB2, MAPK1, EP300, TGFBR2, PIK3CA, KRAS, TP53, NFE2L2 |
| Viral carcinogenesis | 8 | 3.15 × 10−7 | CASP8, MAPK1, EP300, HLA-B, HLA-A, PIK3CA, KRAS, TP53 |
| Central carbon metabolism in cancer | 6 | 3.15 × 10−7 | PTEN, ERBB2, MAPK1, PIK3CA, KRAS, TP53 |
| MicroRNAs in cancer | 9 | 3.43 × 10−7 | PTEN, ERBB3, ERBB2, MAPK1, EP300, TP63, PIK3CA, KRAS, TP53 |
| FoxO signaling pathway | 7 | 3.43 × 10−7 | PTEN, STK11, MAPK1, EP300, TGFBR2, PIK3CA, KRAS |
| Chronic myeloid leukemia | 6 | 3.94 × 10−7 | MECOM, MAPK1, TGFBR2, PIK3CA, KRAS, TP53 |
| Human papillomavirus infection | 9 | 4.65 × 10−7 | PTEN, CASP8, MAPK1, EP300, HLA-B, HLA-A, PIK3CA, KRAS, TP53 |
| ErbB2/ErbB3 signaling events | 5 | 4.65 × 10−7 | ERBB3, ERBB2, MAPK1, NRG1, KRAS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammed, F.A.; Tune, K.K.; Jett, M.; Muhie, S. Cervical Cancer Stages, Human Papillomavirus Integration, and Malignant Genetic Mutations: Integrative Analysis of Datasets from Four Different Cohorts. Cancers 2023, 15, 5595. https://doi.org/10.3390/cancers15235595
Mohammed FA, Tune KK, Jett M, Muhie S. Cervical Cancer Stages, Human Papillomavirus Integration, and Malignant Genetic Mutations: Integrative Analysis of Datasets from Four Different Cohorts. Cancers. 2023; 15(23):5595. https://doi.org/10.3390/cancers15235595
Chicago/Turabian StyleMohammed, Foziya Ahmed, Kula Kekeba Tune, Marti Jett, and Seid Muhie. 2023. "Cervical Cancer Stages, Human Papillomavirus Integration, and Malignant Genetic Mutations: Integrative Analysis of Datasets from Four Different Cohorts" Cancers 15, no. 23: 5595. https://doi.org/10.3390/cancers15235595
APA StyleMohammed, F. A., Tune, K. K., Jett, M., & Muhie, S. (2023). Cervical Cancer Stages, Human Papillomavirus Integration, and Malignant Genetic Mutations: Integrative Analysis of Datasets from Four Different Cohorts. Cancers, 15(23), 5595. https://doi.org/10.3390/cancers15235595







