The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Antibodies
2.3. Cell Viability and Apoptosis Measurements
2.4. Cell Modification, CRISPR/spCas9-Mediated Gene Knockout
2.5. Competitive Cell Proliferation Assays
2.6. Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC)
2.7. Mass Spectrometry and Data Analyses
2.8. Cell Lysis, SDS-PAGE, and Western Blot
2.9. Biomarker Analysis
3. Results
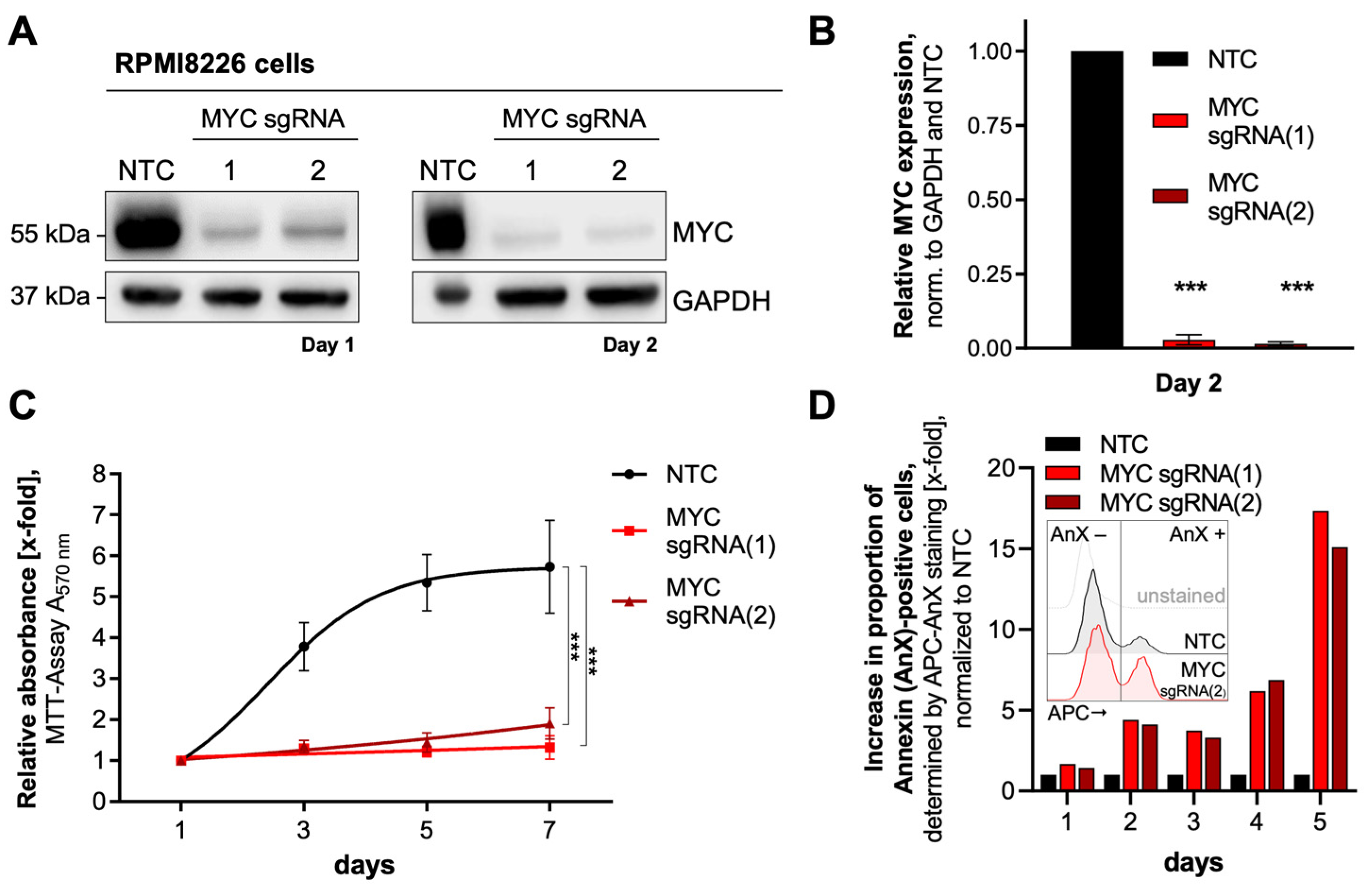
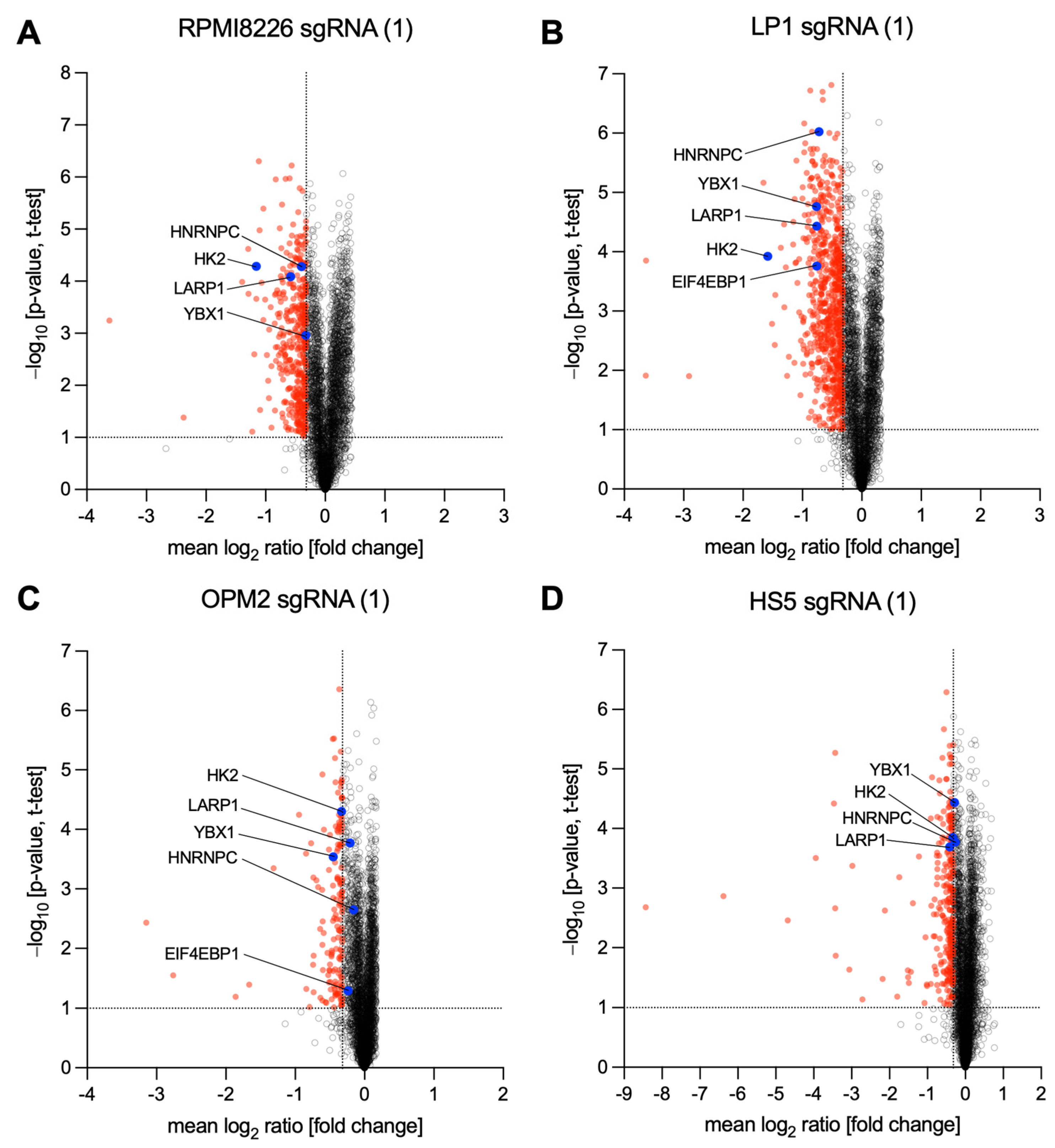
3.1. Identification of Potential MYC Target Proteins by SILAC and Mass Spectrometry
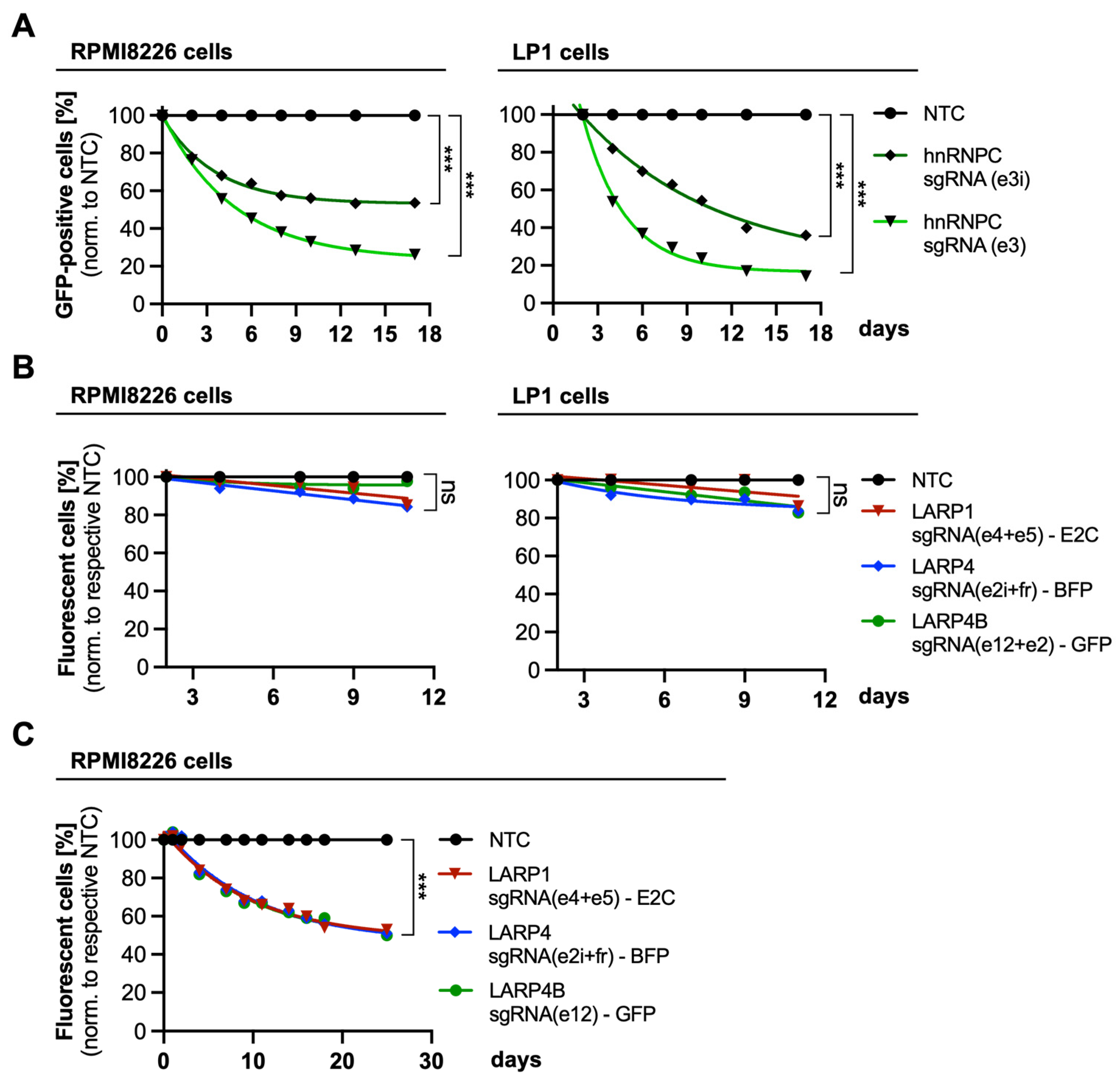
3.2. Investigation of MM-Specific MYC-Driven Pathways
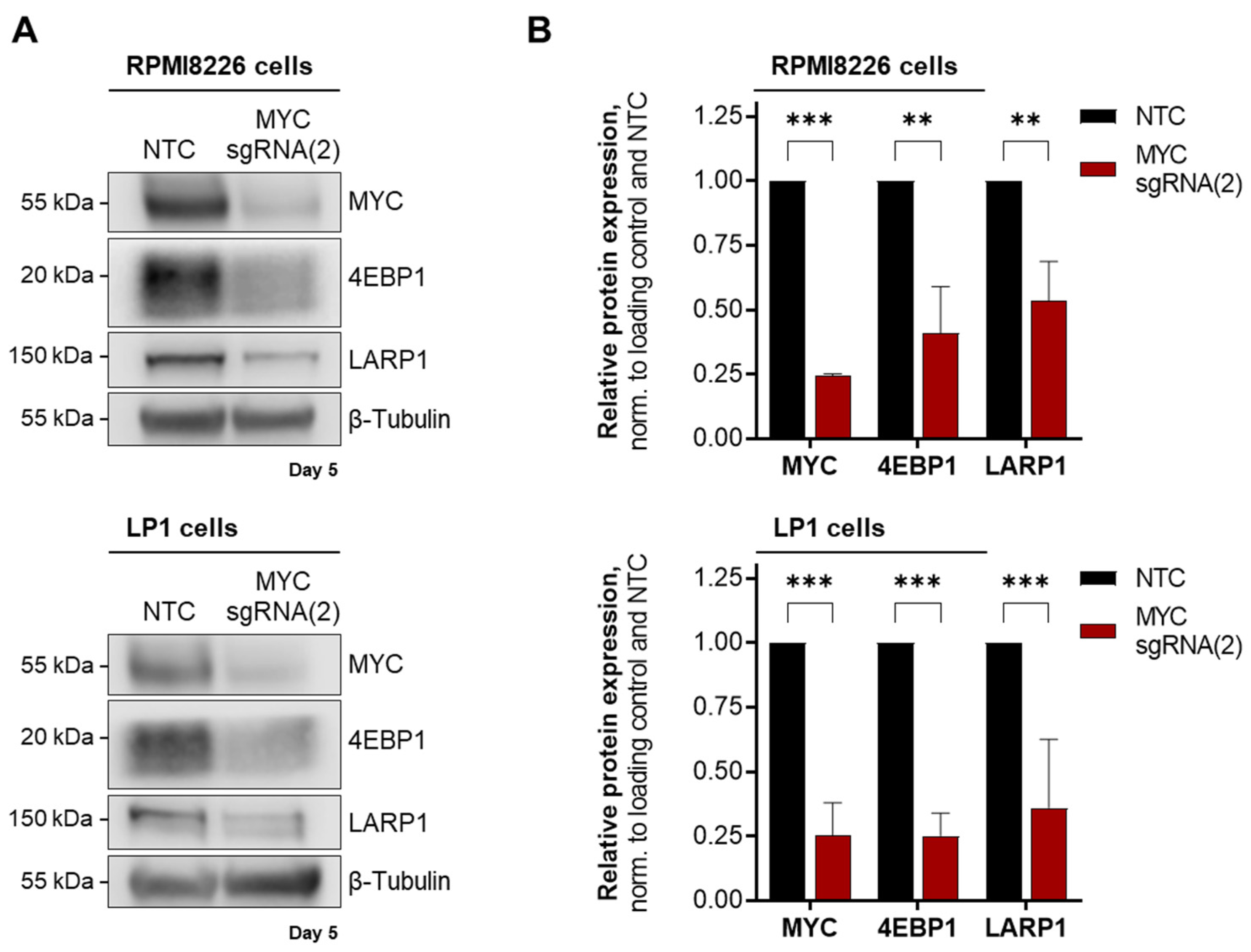
3.3. Deregulation of MYC-Driven Translation in MM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kyle, R.A.; Rajkumar, S.V. Multiple myeloma. N. Engl. J. Med. 2004, 351, 1860–1873. [Google Scholar] [CrossRef]
- Anderson, K.C. Progress and Paradigms in Multiple Myeloma. Clin. Cancer Res. 2016, 22, 5419–5427. [Google Scholar] [CrossRef]
- Hansen, D.K.; Sidana, S.; Peres, L.C.; Colin Leitzinger, C.; Shune, L.; Shrewsbury, A.; Gonzalez, R.; Sborov, D.W.; Wagner, C.; Dima, D.; et al. Idecabtagene Vicleucel for Relapsed/Refractory Multiple Myeloma: Real-World Experience from the Myeloma CAR T Consortium. J. Clin. Oncol. 2023, 41, 2087–2097. [Google Scholar] [CrossRef]
- Chng, W.J.; Huang, G.F.; Chung, T.H.; Ng, S.B.; Gonzalez-Paz, N.; Troska-Price, T.; Mulligan, G.; Chesi, M.; Bergsagel, P.L.; Fonseca, R. Clinical and biological implications of MYC activation: A common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011, 25, 1026–1035. [Google Scholar] [CrossRef]
- Sabò, A.; Kress, T.R.; Pelizzola, M.; De Pretis, S.; Gorski, M.M.; Tesi, A.; Morelli, M.J.; Bora, P.; Doni, M.; Verrecchia, A.; et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014, 511, 488–492. [Google Scholar] [CrossRef]
- Kress, T.R.; Sabò, A.; Amati, B. MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 2015, 15, 593–607. [Google Scholar] [CrossRef]
- Berg, T.; Cohen, S.B.; Desharnais, J.; Sonderegger, C.; Maslyar, D.J.; Goldberg, J.; Boger, D.L.; Vogt, P.K. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. USA 2002, 99, 3830–3835. [Google Scholar] [CrossRef]
- Kiessling, A.; Sperl, B.; Hollis, A.; Eick, D.; Berg, T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem. Biol. 2006, 13, 745–751. [Google Scholar] [CrossRef]
- Madden, S.K.; de Araujo, A.D.; Gerhardt, M.; Fairlie, D.P.; Mason, J.M. Taking the Myc out of cancer: Toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 2021, 20, 3. [Google Scholar] [CrossRef]
- Garralda, E.; Moreno, V.; Alonso, G.; Corral, E.; Hernandez-Guerrero, T.; Ramon, J.; Doger de Spéville, B.; Martinez, E.; Soucek, L.; Niewel, M.; et al. Dose escalation study of OMO-103, a first in class Pan-MYC-Inhibitor in patients (pts) with advanced solid tumors. Eur. J. Cancer 2022, 174, S5–S6. [Google Scholar] [CrossRef]
- Massó-Vallés, D.; Soucek, L. Blocking Myc to Treat Cancer: Reflecting on Two Decades of Omomyc. Cells 2020, 9, 883. [Google Scholar] [CrossRef]
- Ruggero, D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009, 69, 8839. [Google Scholar] [CrossRef]
- Manier, S.; Huynh, D.; Shen, Y.J.; Zhou, J.; Yusufzai, T.; Salem, K.Z.; Ebright, R.Y.; Shi, J.; Park, J.; Glavey, S.V.; et al. Inhibiting the oncogenic translation program is an effective therapeutic strategy in multiple myeloma. Sci. Transl. Med. 2017, 9, eaal2668. [Google Scholar] [CrossRef]
- Dib, A.; Gabrea, A.; Glebov, O.K.; Bergsagel, P.L.; Kuehl, W.M. Characterization of MYC Translocations in Multiple Myeloma Cell Lines. J. Natl. Cancer Inst. Monogr. 2008, 8, 25. [Google Scholar] [CrossRef]
- Shou, Y.; Martelli, M.L.; Gabrea, A.; Qi, Y.; Brents, L.A.; Roschke, A.; Dewald, G.; Kirsch, I.R.; Bergsagel, P.L.; Kuehl, W.M. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc. Natl. Acad. Sci. USA 2000, 97, 228–233. [Google Scholar] [CrossRef]
- Tiscornia, G.; Singer, O.; Verma, I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006, 1, 241–245. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO::TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A 160 software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Giusti, K. Company Profile: Multiple Myeloma Research Foundation. Per. Med. 2012, 9, 333–336. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Chng, W.J.; Kumar, S.; VanWier, S.; Ahmann, G.; Price-Troska, T.; Henderson, K.; Chung, T.H.; Kim, S.; Mulligan, G.; Bryant, B.; et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007, 67, 2982–2989. [Google Scholar] [CrossRef]
- Zeller, K.I.; Jegga, A.G.; Aronow, B.J.; O’Donnell, K.A.; Dang, C.V. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003, 4, R69. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Schmidt, E.V. The role of c-myc in regulation of translation initiation. Oncogene 2004, 23, 3217–3221. [Google Scholar] [CrossRef]
- Aoki, K.; Adachi, S.; Homoto, M.; Kusano, H.; Koike, K.; Natsume, T. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 2013, 587, 2173–2178. [Google Scholar] [CrossRef]
- Tcherkezian, J.; Cargnello, M.; Romeo, Y.; Huttlin, E.L.; Lavoie, G.; Gygi, S.P.; Roux, P.P. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014, 28, 357. [Google Scholar] [CrossRef]
- Pause, A.; Belsham, G.J.; Gingras, A.C.; Donzé, O.; Lin, T.A.; Lawrence, J.C.; Sonenberg, N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 1994, 371, 762–767. [Google Scholar] [CrossRef]
- Dang, C.V.; Reddy, E.P.; Shokat, K.M.; Soucek, L. Drugging the “undruggable” cancer targets. Nat. Rev. Cancer 2017, 17, 502–508. [Google Scholar] [CrossRef]
- Morcelle, C.; Menoyo, S.; Morón-Duran, F.D.; Tauler, A.; Kozma, S.C.; Thomas, G.; Gentilella, A. Oncogenic MYC Induces the Impaired Ribosome Biogenesis Checkpoint and Stabilizes p53 Independent of Increased Ribosome Content. Cancer Res. 2019, 79, 4348–4359. [Google Scholar] [CrossRef]
- Balakumaran, B.S.; Porrello, A.; Hsu, D.S.; Glover, W.; Foye, A.; Leung, J.Y.; Sullivan, B.A.; Hahn, W.C.; Loda, M.; Febbo, P.G. MYC activity mitigates response to rapamycin in prostate cancer through 4EBP1-mediated inhibition of autophagy. Cancer Res. 2009, 69, 7803. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2016, 16, 288–304. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731. [Google Scholar] [CrossRef]
- Pourdehnad, M.; Truitt, M.L.; Siddiqi, I.N.; Ducker, G.S.; Shokat, K.M.; Ruggero, D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl. Acad. Sci. USA 2013, 110, 11988–11993. [Google Scholar] [CrossRef]
- Dowling, R.J.O.; Topisirovic, I.; Alain, T.; Bidinosti, M.; Fonseca, B.D.; Petroulakis, E.; Wang, X.; Larsson, O.; Selvaraj, A.; Liu, Y.; et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010, 328, 1172–1176. [Google Scholar] [CrossRef]
- Fonseca, B.D.; Zakaria, C.; Jia, J.J.; Graber, T.E.; Svitkin, Y.; Tahmasebi, S.; Healy, D.; Hoang, H.D.; Jensen, J.M.; Diao, I.T.; et al. La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1). J. Biol. Chem. 2015, 290, 15996–16020. [Google Scholar] [CrossRef]
- Levy, S.; Avni, D.; Hariharan, N.; Perry, R.P.; Meyuhas, O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc. Natl. Acad. Sci. USA 1991, 88, 3319–3323. [Google Scholar] [CrossRef]
- Hsieh, A.C.; Liu, Y.; Edlind, M.P.; Ingolia, N.T.; Janes, M.R.; Sher, A.; Shi, E.Y.; Stumpf, C.R.; Christensen, C.; Bonham, M.J.; et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012, 485, 55–61. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef]
- Philippe, L.; Vasseur, J.J.; Debart, F.; Thoreen, C.C. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 2018, 46, 1457–1469. [Google Scholar] [CrossRef]
- Meyuhas, O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000, 267, 6321–6330. [Google Scholar] [CrossRef]
- Philippe, L.; van den Elzen, A.M.G.; Watson, M.J.; Thoreen, C.C. Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5′ TOP motifs. Proc. Natl. Acad. Sci. USA 2020, 117, 5319–5328. [Google Scholar] [CrossRef]
- Desi, N.; Tong, Q.Y.; Teh, V.; Chan, J.J.; Zhang, B.; Tabatabaeian, H.; Tan, H.Q.; Kapeli, K.; Jin, W.; Lim, C.Y.; et al. Global analysis of RNA-binding proteins identifies a positive feedback loop between LARP1 and MYC that promotes tumorigenesis. Cell. Mol. Life Sci. 2022, 79, 147. [Google Scholar] [CrossRef]
- Bommert, K.S.; Effenberger, M.; Leich, E.; Küspert, M.; Murphy, D.; Langer, C.; Moll, R.; Janz, S.; Mottok, A.; Weissbach, S.; et al. The feed-forward loop between YB-1 and MYC is essential for multiple myeloma cell survival. Leukemia 2013, 27, 441–450. [Google Scholar] [CrossRef]
- Choi, Y.D.; Grabowski, P.J.; Sharp, P.A.; Dreyfuss, G. Heterogeneous nuclear ribonucleoproteins: Role in RNA splicing. Science 1986, 231, 1534–1539. [Google Scholar] [CrossRef]
- Kim, J.H.; Paek, K.Y.; Choi, K.; Kim, T.-D.; Hahm, B.; Kim, K.-T.; Jang, S.K. Heterogeneous Nuclear Ribonucleoprotein C Modulates Translation of c-myc mRNA in a Cell Cycle Phase-Dependent Manner. Mol. Cell. Biol. 2003, 23, 708. [Google Scholar] [CrossRef]
- Küspert, M.; Murakawa, Y.; Schäffler, K.; Vanselow, J.T.; Wolf, E.; Juranek, S.; Schlosser, A.; Landthaler, M.; Fischer, U. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA 2015, 21, 1294. [Google Scholar] [CrossRef]
- Schäffler, K.; Schulz, K.; Hirmer, A.; Wiesner, J.; Grimm, M.; Sickmann, A.; Fischer, U. A stimulatory role for the La-related protein 4B in translation. RNA 2010, 16, 1488–1499. [Google Scholar] [CrossRef]
- Yang, R.; Gaidamakov, S.A.; Xie, J.; Lee, J.; Martino, L.; Kozlov, G.; Crawford, A.K.; Russo, A.N.; Conte, M.R.; Gehring, K.; et al. La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol. Cell. Biol. 2011, 31, 542–556. [Google Scholar] [CrossRef]
- Attar-Schneider, O.; Drucker, L.; Zismanov, V.; Tartakover-Matalon, S.; Lishner, M. Targeting eIF4GI translation initiation factor affords an attractive therapeutic strategy in multiple myeloma. Cell. Signal. 2014, 26, 1878–1887. [Google Scholar] [CrossRef]
- Li, S.; Fu, J.; Lu, C.; Mapara, M.Y.; Raza, S.; Hengst, U.; Lentzsch, S. Elevated Translation Initiation Factor eIF4E Is an Attractive Therapeutic Target in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 711–719. [Google Scholar] [CrossRef]
- Wang, W.; Xu, S.W.; Zhu, X.Y.; Guo, Q.Y.; Zhu, M.; Mao, X.L.; Chen, Y.H.; Li, S.W.; Luo, W. Da Identification and Validation of a Novel RNA-Binding Protein-Related Gene-Based Prognostic Model for Multiple Myeloma. Front. Genet. 2021, 12, 665173. [Google Scholar] [CrossRef]



| Cell Lines | Chr | A | N | Abnormality |
|---|---|---|---|---|
| RPMI8226 | 60 | 1 | 3 | MYC insertion on t(16;22)(q23;q11):der(16) |
| LP1 | 80 | 5 | 1 | t(8;14) variant: der(8) with VH |
| OPM2 | 67 | 2 | 2 | CH insertion on t(1;8)(q12?;q24):der(8) |
| sgRNA ID | 5′-3′ Sequences | Fluorescent Marker |
|---|---|---|
| MYC (sgRNA(1)) | sense: CACCGgacgctgtgcccgcgggcg antisense: AAACcgcccgcgggcacagcgtcC | GFP |
| MYC (sgRNA(2)) | sense: CACCGacgttgcggtcacaccctt antisense: AAACaagggtgtgaccgcaacgtC | GFP |
| hnRNPC (sgRNA(e3)) | sense: CACCGgagtagaggggacggagaa antisense: AAACttctccgtcccctctactcC | GFP |
| hnRNPC (sgRNA(e3i)) | sense: CACCGgttccggacctgagtagag antisense: AAACctctactcaggtccggaacC | GFP |
| LARP1 (sgRNA(e4)) | sense: CACCGaaatcagatgaatcagggg antisense: AAACcccctgattcatctgatttC | E2Crimson (E2C) |
| LARP1 (sgRNA(e5)) | sense: CACCGtatgtgtctaggctccggt antisense: AAACaccggagcctagacacataC | E2Crimson (E2C) |
| LARP4 (sgRNA(e2i)) | sense: CACCGatatttaagacttaccctc antisense: AAACgagggtaagtcttaaatatC | BFP |
| LARP4 (sgRNA(fr)) | sense: CACCGcatcaggtgctcatcctga antisense: AAACtcaggatgagcacctgatgC | BFP |
| LARP4B (sgRNA(e12)) | sense: CACCGcgattcctagtgcagagag antisense: AAACctctctgcactaggaatcgC | GFP |
| LARP4B (sgRNA(e2)) | sense: CACCGgatggaacttgtctgagaa antisense: AAACttctcagacaagttccatcC | GFP |
| NTC | sense: CACCGttccgggctaacaagtcct antisense: AAACaggacttgttagcccggaaC | GFP, BFP, or E2C |
| Putative MYC Target Proteins | |
|---|---|
| MM-derived cell lines (RPMI8226, LP1, and OPM2) | PABPC1, EIF5B, EIF4G1, EIF4EBP1, EIF4B, EIF4A1, EIF3K, EIF3I, EIF3G, EIF3F, EIF3E, EIF1AX + (RPS9, RPS7, RPS6, RPS3, RPS29, RPS27, RPS21, RPS20, RPS18, RPS17, RPS15, RPS14, RPS12, RPS11, RPS10, RPL9, RPL5, RPL38, RPL36AL, RPL23) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seibert, M.; Koschade, S.E.; Stolp, V.; Häupl, B.; Wempe, F.; Serve, H.; Kurrle, N.; Schnütgen, F.; von Metzler, I. The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival. Cancers 2023, 15, 5508. https://doi.org/10.3390/cancers15235508
Seibert M, Koschade SE, Stolp V, Häupl B, Wempe F, Serve H, Kurrle N, Schnütgen F, von Metzler I. The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival. Cancers. 2023; 15(23):5508. https://doi.org/10.3390/cancers15235508
Chicago/Turabian StyleSeibert, Marcel, Sebastian E. Koschade, Verena Stolp, Björn Häupl, Frank Wempe, Hubert Serve, Nina Kurrle, Frank Schnütgen, and Ivana von Metzler. 2023. "The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival" Cancers 15, no. 23: 5508. https://doi.org/10.3390/cancers15235508
APA StyleSeibert, M., Koschade, S. E., Stolp, V., Häupl, B., Wempe, F., Serve, H., Kurrle, N., Schnütgen, F., & von Metzler, I. (2023). The MYC-Regulated RNA-Binding Proteins hnRNPC and LARP1 Are Drivers of Multiple Myeloma Cell Growth and Disease Progression and Negatively Predict Patient Survival. Cancers, 15(23), 5508. https://doi.org/10.3390/cancers15235508







