Unveiling Disrupted Lipid Metabolism in Benign Prostate Hyperplasia, Prostate Cancer, and Metastatic Patients: Insights from a Colombian Nested Case–Control Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
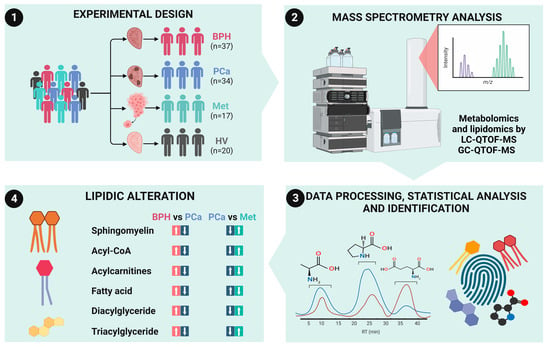
2.1. Study Participants
2.2. Metabolomics Analysis by RP-LC-QTOF-MS
2.3. Metabolomics Analysis by GC-QTOF-MS
2.4. Lipidomic Analysis by RP-LC-QTOF-MS
2.5. Quality Assurance (QA) and Quality Control Samples (QC)
2.6. Data Processing and Analysis
2.7. Metabolites Identification
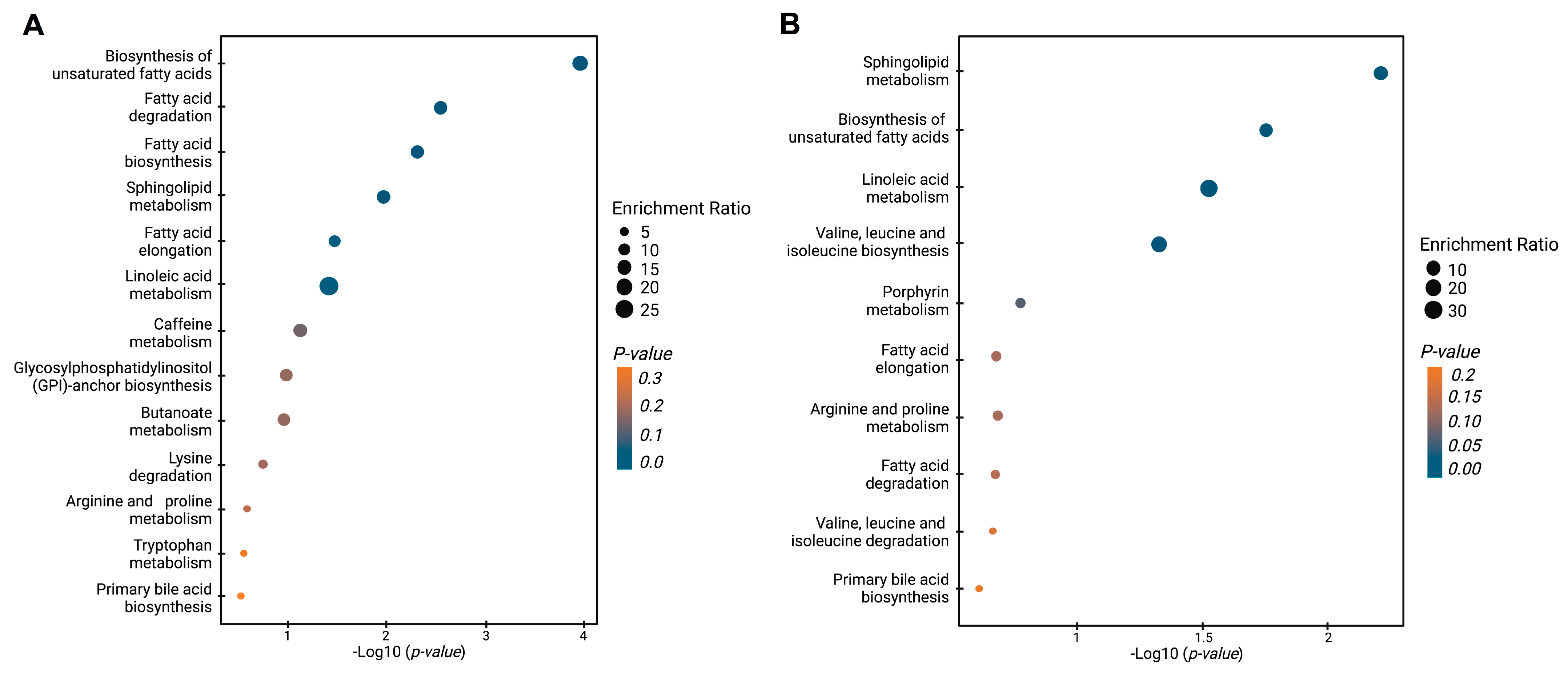
2.8. Altered Metabolite Pathway Mapping and Identification of Potential Diagnostic Biomarkers for PCA Patients
3. Results
3.1. Description of the Cohorts
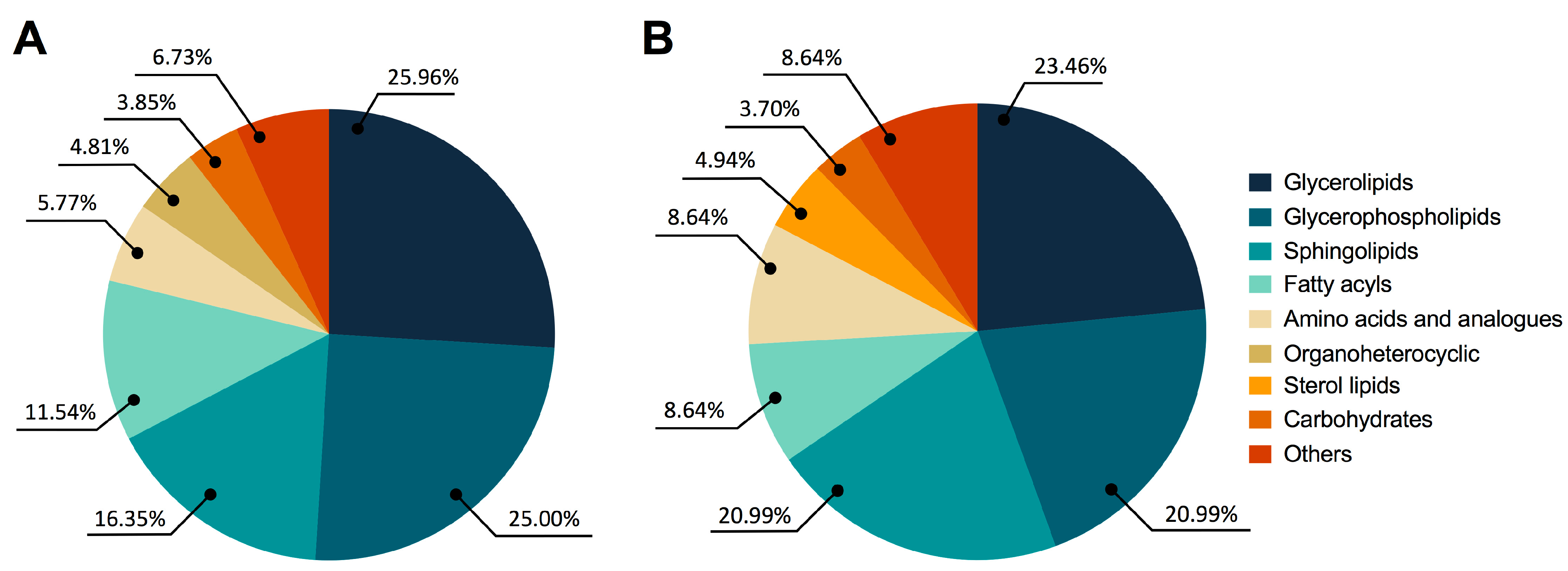
3.2. Untargeted Metabolomic and Lipidomic Analyses
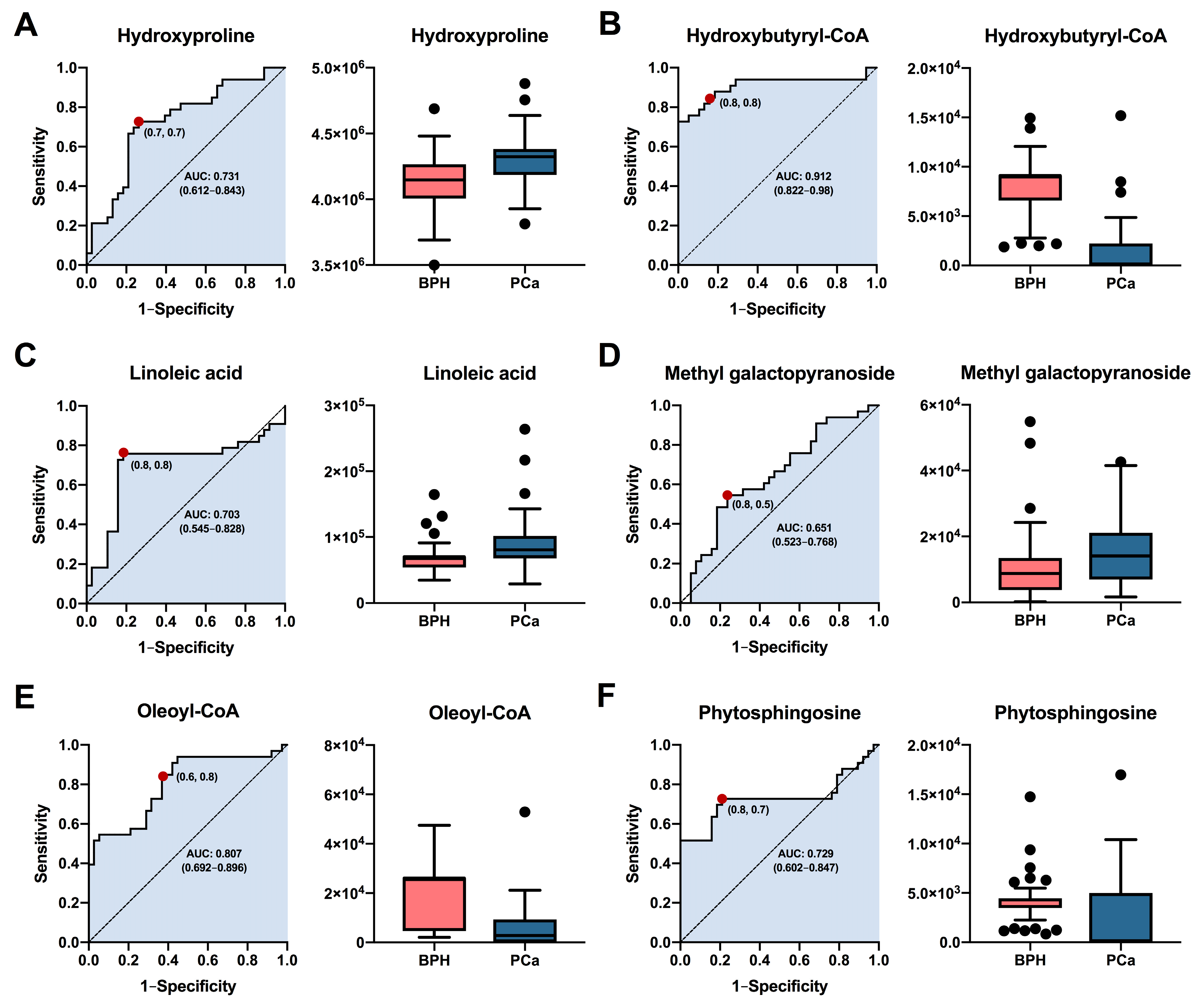
3.3. Potential Diagnostic Biomarkers among BPH and PCa Patients
3.4. Subgrouping of the Samples Belonging to the PCa Group
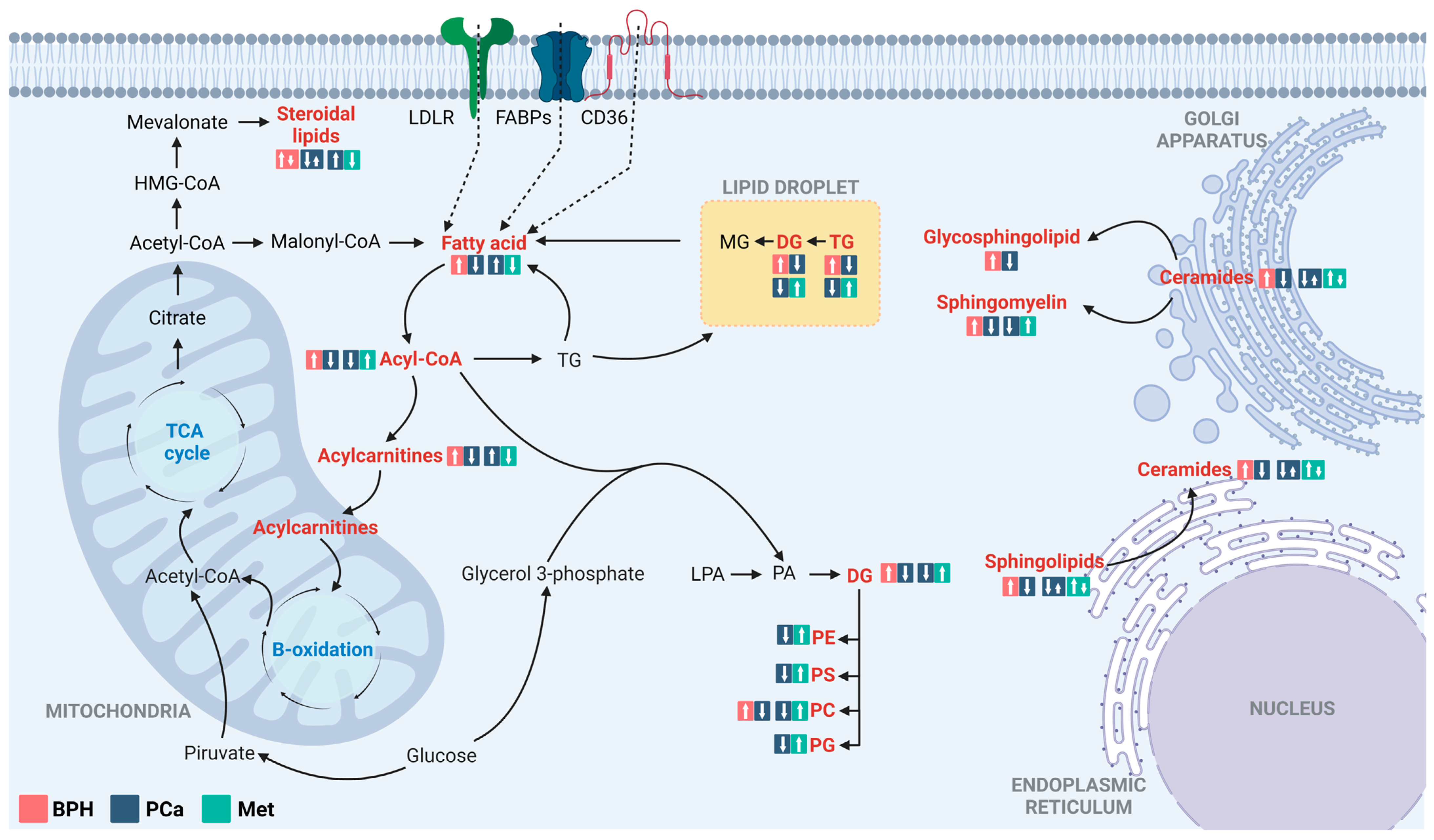
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Cancer Observatory. Available online: https://gco.iarc.fr/today (accessed on 17 May 2023).
- Sharma, S.; Zapatero-Rodríguez, J.; O’Kennedy, R. Prostate Cancer Diagnostics: Clinical Challenges and the Ongoing Need for Disruptive and Effective Diagnostic Tools. Biotechnol. Adv. 2017, 35, 135–149. [Google Scholar] [CrossRef] [PubMed]
- De Koning, H.J.; Auvinen, A.; Sanchez, A.B.; Da Silva, F.C.; Ciatto, S.; Denis, L.; Gohagan, J.K.; Hakama, M.; Hugosson, J.; Kranse, R.; et al. Large-Scale Randomized Prostate Cancer Screening Trials: Program Performances in the European Randomized Screening for Prostate Cancer Trial and the Prostate, Lung, Colorectal and Ovary Cancer Trial. Int. J. Cancer 2002, 97, 237–244. [Google Scholar] [CrossRef]
- Bangma, C.H.; Roemeling, S.; Schröder, F.H. Overdiagnosis and Overtreatment of Early Detected Prostate Cancer. World J. Urol. 2007, 25, 3–9. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Lin, C.C.; Mariotto, A.B.; Siegel, R.L.; Stein, K.D.; Kramer, J.L.; Alteri, R.; Robbins, A.S.; Jemal, A. Cancer Treatment and Survivorship Statistics, 2014. CA Cancer J. Clin. 2014, 64, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Faulds, M.H.; Dahlman-Wright, K. Metabolic Diseases and Cancer Risk. Curr. Opin. Oncol. 2012, 24, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Pakstis, A.J.; Gandotra, N.; Cowan, T.M.; Zhao, H.; Kidd, K.K.; Scharfe, C. Metabolic Diversity in Human Populations and Correlation with Genetic and Ancestral Geographic Distances. Mol. Genet. Metab. 2022, 137, 292–300. [Google Scholar] [CrossRef]
- Mullins, J.K.; Loeb, S. Environmental Exposures and Prostate Cancer. Urol. Oncol. Semin. Orig. Investig. 2012, 30, 216–219. [Google Scholar] [CrossRef]
- Hinata, N.; Fujisawa, M. Racial Differences in Prostate Cancer Characteristics and Cancer-Specific Mortality: An Overview. World J. Men’s Health 2022, 40, 217–227. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Gaude, E.; Hilvo, M.; Frezza, C. The Metabolic Alterations of Cancer Cells. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 542, pp. 1–23. ISBN 9780124166189. [Google Scholar]
- Oermann, E.K.; Wu, J.; Guan, K.L.; Xiong, Y. Alterations of Metabolic Genes and Metabolites in Cancer. Semin Cell Dev. Biol. 2012, 23, 370–380. [Google Scholar] [CrossRef]
- Suri, G.S.; Kaur, G.; Carbone, G.M.; Shinde, D. Metabolomics in Oncology. Cancer Rep. 2023, 6, e1795. [Google Scholar] [CrossRef]
- Subramani, R.; Poudel, S.; Smith, K.D.; Estrada, A.; Lakshmanaswamy, R. Metabolomics of Breast Cancer: A Review. Metabolites 2022, 12, 643. [Google Scholar] [CrossRef] [PubMed]
- Kdadra, M.; Höckner, S.; Leung, H.; Kremer, W.; Schiffer, E. Metabolomics Biomarkers of Prostate Cancer: A Systematic Review. Diagnostics 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- EAU Guidelines. 2023. Available online: http://uroweb.org/guidelines/compilations-of-all-guidelines/ (accessed on 16 June 2023).
- Kirwan, J.A.; Gika, H.; Beger, R.D.; Bearden, D.; Dunn, W.B.; Goodacre, R.; Theodoridis, G.; Witting, M.; Yu, L.R.; Wilson, I.D. Quality Assurance and Quality Control Reporting in Untargeted Metabolic Phenotyping: MQACC Recommendations for Analytical Quality Management. Metabolomics 2022, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Kind, T.; Cajka, T.; Hazen, S.L.; Tang, W.H.W.; Kaddurah-Daouk, R.; Irvin, M.R.; Arnett, D.K.; Barupal, D.K.; Fiehn, O. Systematic Error Removal Using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 2019, 91, 3590–3596. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass Spectral and Retention Index Libraries for Metabolomics Based on Quadrupole and Time-of-Flight Gas Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef]
- Stattin, P.; Holmberg, E.; Johansson, J.E.; Holmberg, L.; Adolfsson, J.; Hugosson, J. Outcomes in Localized Prostate Cancer: National Prostate Cancer Register of Sweden Follow-up Study. J. Natl. Cancer Inst. 2010, 102, 950–958. [Google Scholar] [CrossRef]
- Tolkach, Y.; Kristiansen, G. The Heterogeneity of Prostate Cancer: A Practical Approach. Pathobiology 2018, 85, 108–116. [Google Scholar] [CrossRef]
- Scattoni, V.; Maccagnano, C.; Capitanio, U.; Gallina, A.; Briganti, A.; Montorsi, F. Random Biopsy: When, How Many and Where to Take the Cores? World J. Urol. 2014, 32, 859–869. [Google Scholar] [CrossRef]
- Gerlinger, M.; Catto, J.W.; Orntoft, T.F.; Real, F.X.; Zwarthoff, E.C.; Swanton, C. Intratumour Heterogeneity in Urologic Cancers: From Molecular Evidence to Clinical Implications. Eur. Urol. 2015, 67, 729–737. [Google Scholar] [CrossRef]
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial Interindividual and Limited Intraindividual Genomic Diversity among Tumors from Men with Metastatic Prostate Cancer. Nat. Med. 2016, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, P.; Baade, P.D.; Aitken, J.F.; Ralph, N.; Chambers, S.K.; Dunn, J. Geographical Variations in Prostate Cancer Outcomes: A Systematic Review of International Evidence. Front. Oncol. 2019, 9, 238. [Google Scholar] [CrossRef]
- Center, M.M.; Jemal, A.; Lortet-Tieulent, J.; Ward, E.; Ferlay, J.; Brawley, O.; Bray, F. International Variation in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2012, 61, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Wang, Z.; Cheng, L. Tumor Microenvironment Heterogeneity an Important Mediator of Prostate Cancer Progression and Therapeutic Resistance. NPJ Precis Oncol. 2022, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Deberardinis, R.J.; Thompson, C.B. Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us? Cell 2012, 148, 1132–1144. [Google Scholar] [CrossRef]
- Fu, Y.; Zou, T.; Shen, X.; Nelson, P.J.; Li, J.; Wu, C.; Yang, J.; Zheng, Y.; Bruns, C.; Zhao, Y.; et al. Lipid Metabolism in Cancer Progression and Therapeutic Strategies. MedComm 2021, 2, 27–59. [Google Scholar] [CrossRef]
- Bian, X.; Liu, R.; Meng, Y.; Xing, D.; Xu, D.; Lu, Z. Lipid Metabolism and Cancer. J. Exp. Med. 2021, 218, e20201606. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhao, X.; Cheng, C.; Li, N.; Liu, Y.; Cao, Y. The Implications of Signaling Lipids in Cancer Metastasis. Exp. Mol. Med. 2018, 50, 1–10. [Google Scholar] [CrossRef]
- Vasseur, S.; Guillaumond, F. Lipids in Cancer: A Global View of the Contribution of Lipid Pathways to Metastatic Formation and Treatment Resistance. Oncogenesis 2022, 11, 46. [Google Scholar] [CrossRef]
- Crowe, F.L.; Allen, N.E.; Appleby, P.N.; Overvad, K.; Aardestrup, I.V.; Johnsen, N.F.; Tjønneland, A.; Linseisen, J.; Kaaks, R.; Boeing, H.; et al. Fatty Acid Composition of Plasma Phospholipids and Risk of Prostate Cancer in a Case-Control Analysis Nested within the European Prospective Investigation into Cancer and Nutrition. Am. J. Clin. Nutr. 2008, 88, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Mao, J.; Ai, J.; Deng, Y.; Roth, M.R.; Pound, C.; Henegar, J.; Welti, R.; Bigler, S.A. Identification of Plasma Lipid Biomarkers for Prostate Cancer by Lipidomics and Bioinformatics. PLoS ONE 2012, 7, e48889. [Google Scholar] [CrossRef]
- Freeman, V.L.; Flanigan, R.C.; Meydani, M. Prostatic Fatty Acids and Cancer Recurrence after Radical Prostatectomy for Early-Stage Prostate Cancer. Cancer Causes Control 2007, 18, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Siltari, A.; Syvälä, H.; Lou, Y.R.; Gao, Y.; Murtola, T.J. Role of Lipids and Lipid Metabolism in Prostate Cancer Progression and the Tumor’s Immune Environment. Cancers 2022, 14, 4293. [Google Scholar] [CrossRef] [PubMed]
- Vignozzi, L.; Gacci, M.; Cellai, I.; Santi, R.; Corona, G.; Morelli, A.; Rastrelli, G.; Comeglio, P.; Sebastanelli, A.; Maneschi, E.; et al. Fat Boosts, While Androgen Receptor Activation Counteracts, BPH-Associated Prostate Inflammation. Prostate 2013, 73, 789–800. [Google Scholar] [CrossRef]
- Zhu, C.; Wu, J.; Wu, Y.; Guo, W.; Lu, J.; Zhu, W.; Li, X.; Xu, N.; Zhang, Q. Triglyceride to High-Density Lipoprotein Cholesterol Ratio and Total Cholesterol to High-Density Lipoprotein Cholesterol Ratio and Risk of Benign Prostatic Hyperplasia in Chinese Male Subjects. Front. Nutr. 2022, 9, 999995. [Google Scholar] [CrossRef]
- Chokkalingam, A.P.; Nyrén, O.; Johansson, J.E.; Gridley, G.; McLaughlin, J.K.; Adami, H.O.; Hsing, A.W. Prostate Carcinoma Risk Subsequent to Diagnosis of Benign Prostatic Hyperplasia: A Population-Based Cohort Study in Sweden. Cancer 2003, 98, 1727–1734. [Google Scholar] [CrossRef]
- Kim, S.H.; Kwon, W.A.; Joung, J.Y. Impact of Benign Prostatic Hyperplasia and/or Prostatitis on the Risk of Prostate Cancer in Korean Patients. World J. Men’s Health 2020, 38, 358–365. [Google Scholar] [CrossRef]
- Schenk, J.M.; Kristal, A.R.; Arnold, K.B.; Tangen, C.M.; Neuhouser, M.L.; Lin, D.W.; White, E.; Thompson, I.M. Association of Symptomatic Benign Prostatic Hyperplasia and Prostate Cancer: Results from the Prostate Cancer Prevention Trial. Am. J. Epidemiol. 2011, 173, 1419–1428. [Google Scholar] [CrossRef]
- Ørsted, D.D.; Bojesen, S.E. The Link between Benign Prostatic Hyperplasia and Prostate Cancer. Nat. Rev. Urol. 2013, 10, 49–54. [Google Scholar] [CrossRef]
- Ørsted, D.D.; Bojesen, S.E.; Nielsen, S.F.; Nordestgaard, B.G. Association of Clinical Benign Prostate Hyperplasia with Prostate Cancer Incidence and Mortality Revisited: A Nationwide Cohort Study of 3,009,258 Men. Eur. Urol. 2011, 60, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.S.; de Marzo, A.M. Prostate Cancer and Inflammation: The Evidence. Histopathology 2012, 60, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Martin-Perez, M.; Urdiroz-Urricelqui, U.; Bigas, C.; Benitah, S.A. Lipid Metabolism in Metastasis and Therapy. Curr. Opin. Syst. Biol. 2021, 28, 100401. [Google Scholar] [CrossRef]
- Bergers, G.; Fendt, S.M. The Metabolism of Cancer Cells during Metastasis. Nat. Rev. Cancer 2021, 21, 162–180. [Google Scholar] [CrossRef]
- Corbet, C.; Bastien, E.; Santiago de Jesus, J.P.; Dierge, E.; Martherus, R.; Vander Linden, C.; Doix, B.; Degavre, C.; Guilbaud, C.; Petit, L.; et al. TGFβ2-Induced Formation of Lipid Droplets Supports Acidosis-Driven EMT and the Metastatic Spreading of Cancer Cells. Nat. Commun. 2020, 11, 454. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, Y.D.; Yu, G.Y.; Cui, J.R.; Lou, Z.; Zhang, H.; Huang, Y.; Bai, C.G.; Deng, L.L.; Liu, P.; et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell 2020, 38, 734–747.e9. [Google Scholar] [CrossRef]
- Jia, S.; Zhou, L.; Shen, T.; Zhou, S.; Ding, G.; Cao, L. Down-Expression of CD36 in Pancreatic Adenocarcinoma and Its Correlation with Clinicopathological Features and Prognosis. J. Cancer 2018, 9, 578–583. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A Promising Cancer Biomarker. Clin. Transl. Med. 2018, 7, 578–583. [Google Scholar] [CrossRef]
- Wenk, M.R. The Emerging Field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Xu, B.; Chen, Y.; Chen, X.; Gan, L.; Zhang, Y.; Feng, J.; Yu, L. Metabolomics Profiling Discriminates Prostate Cancer from Benign Prostatic Hyperplasia within the Prostate-Specific Antigen Gray Zone. Front. Oncol. 2021, 11, 730638. [Google Scholar] [CrossRef]
- Zhang, X.; Xia, B.; Zheng, H.; Ning, J.; Zhu, Y.; Shao, X.; Liu, B.; Dong, B.; Gao, H. Identification of Characteristic Metabolic Panels for Different Stages of Prostate Cancer by 1H NMR-Based Metabolomics Analysis. J. Transl. Med. 2022, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Sebastiano, M.R.; Konstantinidou, G. Targeting Long Chain Acyl-Coa Synthetases for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 3624. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Y.; Wang, J.; Wen, X.; Marcus, M.T.; Daniels, G.; Zhang, D.Y.; Ye, F.; Wang, L.H.; Du, X.; et al. Long Chain Fatty Acyl-CoA Synthetase 4 Is a Biomarker for and Mediator of Hormone Resistance in Human Breast Cancer. PLoS ONE 2013, 8, e77060. [Google Scholar] [CrossRef]
- Li, C.J.; Chiu, Y.H.; Chang, C.; Chang, Y.C.I.; Sheu, J.J.C.; Chiang, A.J. Acetyl Coenzyme a Synthase 2 Acts as a Prognostic Biomarker Associated with Immune Infiltration in Cervical Squamous Cell Carcinoma. Cancers 2021, 13, 3125. [Google Scholar] [CrossRef] [PubMed]


| Variables | HV | BPH | PCa | Met | Overall |
|---|---|---|---|---|---|
| (N = 20) | (N = 37) | (N = 34) | (N = 17) | (N = 108) | |
| Age (years) | |||||
| Mean (SD) | 29.7 (3.20) | 66.4 (6.50) | 67.2 (7.81) | 73.0 (8.54) | 60.9 (16.60) |
| Median [Min, Max] | 29.0 [25.0, 34.0] | 67.0 [55.0, 80.0] | 68.0 [52.0, 82.0] | 74.0 [61.0, 88.0] | 64.0 [25.0, 88.0] |
| Anthropometric measurement | |||||
| Weight (kg) | |||||
| Mean (SD) | 79.6 (7.58) | 71.9 (9.63) | 73.6 (10.3) | 63.2 (8.52) | 72.5 (10.4) |
| Median [min, max] | 79.0 [65.0, 98.0] | 70.0 [49.0, 100] | 72.5 [48.0, 94.0] | 63.0 [42.0, 78.0] | 71.9 [42.0, 100] |
| Height (m) | |||||
| Mean (SD) | 1.75 (0.05) | 1.68 (0.06) | 1.69 (0.05) | 1.67 (0.06) | 1.70 (0.06) |
| Median [min, max] | 1.76 [1.65, 1.85] | 1.68 [1.56, 1.80] | 1.69 [1.53, 1.77] | 1.67 [1.57, 1.80] | 1.70 [1.53, 1.85] |
| Body Mass Index | |||||
| Mean (SD) | 26.0 (2.79) | 25.4 (2.82) | 25.9 (3.46) | 22.6 (2.58) | 25.2 (3.18) |
| Median [min, max] | 25.5 [22.5, 33.9] | 25.4 [16.8, 33.0] | 25.6 [19.2, 34.7] | 22.5 [16.0, 28.7] | 25.1 [16.0, 34.7] |
| Prostate-Specific Antigen (ng/mL) | |||||
| Mean (SD) | 0.558 (0.28) | 9.08 (4.30) | 17.3 (15.0) | 247 (418) | 43.8 (174) |
| Median [min, max] | 0.470 [0.250, 1.2] | 8.00 [3.10, 22.5] | 11.7 [1.09, 64.0] | 64.0 [0.15, 1590] | 8.05 [0.15, 1590] |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) | 2 (11.8%) | 2 (1.9%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardo-Rodriguez, D.; Santamaría-Torres, M.; Salinas, A.; Jiménez-Charris, E.; Mosquera, M.; Cala, M.P.; García-Perdomo, H.A. Unveiling Disrupted Lipid Metabolism in Benign Prostate Hyperplasia, Prostate Cancer, and Metastatic Patients: Insights from a Colombian Nested Case–Control Study. Cancers 2023, 15, 5465. https://doi.org/10.3390/cancers15225465
Pardo-Rodriguez D, Santamaría-Torres M, Salinas A, Jiménez-Charris E, Mosquera M, Cala MP, García-Perdomo HA. Unveiling Disrupted Lipid Metabolism in Benign Prostate Hyperplasia, Prostate Cancer, and Metastatic Patients: Insights from a Colombian Nested Case–Control Study. Cancers. 2023; 15(22):5465. https://doi.org/10.3390/cancers15225465
Chicago/Turabian StylePardo-Rodriguez, Daniel, Mary Santamaría-Torres, Angela Salinas, Eliécer Jiménez-Charris, Mildrey Mosquera, Mónica P. Cala, and Herney Andrés García-Perdomo. 2023. "Unveiling Disrupted Lipid Metabolism in Benign Prostate Hyperplasia, Prostate Cancer, and Metastatic Patients: Insights from a Colombian Nested Case–Control Study" Cancers 15, no. 22: 5465. https://doi.org/10.3390/cancers15225465
APA StylePardo-Rodriguez, D., Santamaría-Torres, M., Salinas, A., Jiménez-Charris, E., Mosquera, M., Cala, M. P., & García-Perdomo, H. A. (2023). Unveiling Disrupted Lipid Metabolism in Benign Prostate Hyperplasia, Prostate Cancer, and Metastatic Patients: Insights from a Colombian Nested Case–Control Study. Cancers, 15(22), 5465. https://doi.org/10.3390/cancers15225465