Expression of Soluble Form of Aurora A as a Predictive Factor for Neoadjuvant Therapy in Breast Cancer Patients: A Single-Center Pilot Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Patients
- Age above 18 years.
- Patients with a confirmed diagnosis of breast cancer on histopathological examination from a core needle biopsy.
- Qualified for NAT treatment by MDT (multidisciplinary team) within BCU according to ESMO and PTO (Polish oncological society) recommendations for neoadjuvant treatment.
- With complete medical records and results to assess the response to NAT in the RCB classification.
- Given informed consent to participate in the study.
- Lack of MDT qualification for neoadjuvant treatment.
- Lack of documentation, which prevents the assessment of response to treatment.
2.3. Methods
Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Univariate Analysis
3.3. Multivariate Analysis
3.4. Exploratory Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: Meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018, 19, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, E.; Bossuyt, V.; Viale, G.; Cameron, D.; Badve, S.; Denkert, C.; MacGrogan, G.; Penault-Llorca, F.; Boughey, J.; Curigliano, G.; et al. Standardization of pathologic evaluation and reporting of postneoadjuvant specimens in clinical trials of breast cancer: Recommendations from an international working group. Mod. Pathol. 2015, 28, 1185–1201. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.D.; Posch, F.; Suppan, C.; Bargfrieder, U.; Gumpoldsberger, M.; Hammer, R.; Hauser, H.; Dandachi, N.; Prein, K.; Stoeger, H.; et al. Validation of Residual Cancer Burden as Prognostic Factor for Breast Cancer Patients After Neoadjuvant Therapy. Ann. Surg. Oncol. 2019, 26, 4274–4283. [Google Scholar] [CrossRef] [PubMed]
- Symmans, W.F.; Wei, C.; Gould, R.; Yu, X.; Zhang, Y.; Liu, M.; Walls, A.; Bousamra, A.; Ramineni, M.; Sinn, B.; et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated with Residual Cancer Burden and Breast Cancer Subtype. J. Clin. Oncol. 2017, 35, 1049–1060. [Google Scholar] [CrossRef]
- Campbell, J.I.; Yau, C.; Krass, P.; Moore, D.; Carey, L.A.; Au, A.; Chhieng, D.; Giri, D.; Livasy, C.; Mies, C.; et al. Comparison of residual cancer burden, American Joint Committee on Cancer staging and pathologic complete response in breast cancer after neoadjuvant chemotherapy: Results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res. Treat. 2017, 165, 181–191. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Donovan, N.; McDermott, E.; Crown, J. Validated biomarkers: The key to precision treatment in patients with breast cancer. Breast 2016, 29, 192–201. [Google Scholar] [CrossRef]
- Naito, Y.; Urasaki, T. Precision medicine in breast cancer. Chin. Clin. Oncol. 2018, 7, 29. [Google Scholar] [CrossRef]
- Loke, S.Y.; Lee, A.S.G. The future of blood-based biomarkers for the early detection of breast cancer. Eur. J. Cancer 2018, 92, 54–68. [Google Scholar] [CrossRef]
- Seale, K.N.; Tkaczuk, K.H.R. Circulating Biomarkers in Breast Cancer. Clin. Breast Cancer 2022, 22, e319–e331. [Google Scholar] [CrossRef]
- Jassem, J.; Krzakowski, M. Breast cancer. Oncol. Clin. Pract. 2018, 14, 171–215. [Google Scholar]
- Lin, D.C.; Genzen, J.R. Concordance analysis of paired cancer antigen (CA) 15-3 and 27.29 testing. Breast Cancer Res. Treat. 2018, 167, 269–276. [Google Scholar] [CrossRef]
- Van Poznak, C.; Somerfield, M.R.; Bast, R.C.; Cristofanilli, M.; Goetz, M.P.; Gonzalez-Angulo, A.M.; Hicks, D.G.; Hill, E.G.; Liu, M.C.; Lucas, W.; et al. Use of Biomarkers to Guide Decisions on Systemic Therapy for Women with Metastatic Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2015, 33, 2695–2704. [Google Scholar] [CrossRef] [PubMed]
- Borah, N.A.; Reddy, M.M. Aurora Kinase B Inhibition: A Potential Therapeutic Strategy for Cancer. Molecules 2021, 26, 1981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, C.; Li, H.; Lv, F.; Li, X.; Qian, X.; Fu, L.; Xu, B.; Guo, X. Elevated Aurora B expression contributes to chemoresistance and poor prognosis in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 751–757, eCollection 2015. [Google Scholar] [PubMed]
- Guarino Almeida, E.; Renaudin, X.; Venkitaraman, A.R. A kinase-independent function for AURORA-A in replisome assembly during DNA replication initiation. Nucleic Acids Res. 2020, 48, 7844–7855. [Google Scholar] [CrossRef]
- Zheng, F.; Yue, C.; Li, G.; He, B.; Cheng, W.; Wang, X.; Yan, M.; Long, Z.; Qiu, W.; Yuan, Z.; et al. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat. Commun. 2016, 7, 10180. [Google Scholar] [CrossRef]
- Lykkesfeldt, A.E.; Iversen, B.R.; Jensen, M.B.; Ejlertsen, B.; Giobbie-Hurder, A.; Reiter, B.E.; Kirkegaard, T.; Rasmussen, B.B. Aurora kinase A as a possible marker for endocrine resistance in early estrogen receptor positive breast cancer. Acta Oncol. 2018, 57, 67–73. [Google Scholar] [CrossRef]
- Wander, S.A.; Cohen, O.; Gong, X.; Johnson, G.N.; Buendia-Buendia, J.E.; Lloyd, M.R.; Kim, D.; Luo, F.; Mao, P.; Helvie, K.; et al. The Genomic Landscape of Intrinsic and Acquired Resistance to Cyclin-Dependent Kinase 4/6 Inhibitors in Patients with Hormone Receptor–Positive Metastatic Breast Cancer. Cancer Discov. 2020, 10, 1174–1193. [Google Scholar] [CrossRef]
- Jiang, J.; Guo, Z.; Xu, J.; Sun, T.; Zheng, X. Identification of Aurora Kinase A as a Biomarker for Prognosis in Obesity Patients with Early Breast Cancer. Onco Targets Ther. 2020, 13, 4971–4985. [Google Scholar] [CrossRef]
- Lin, X.; Xiang, X.; Hao, L.; Wang, T.; Lai, Y.; Abudoureyimu, M.; Zhou, H.; Feng, B.; Chu, X.; Wang, R. The role of Aurora-A in human cancers and future therapeutics. Am. J. Cancer Res. 2020, 10, 2705–2729. [Google Scholar] [PubMed]
- Ali, H.R.; Dawson, S.J.; Blows, F.M.; Provenzano, E.; Pharoah, P.D.; Caldas, C. Aurora kinase A outperforms Ki67 as a prognostic marker in ER-positive breast cancer. Br. J. Cancer 2012, 106, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Alanazi, S.; Yuan, L.; Solomon, T.; Thaker, T.M.; Jura, N.; Garrett, J.T. Activating HER3 mutations in breast cancer. Oncotarget 2018, 9, 27773–27788. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.A.; Gilliver, S.C.; Borre, M.; Sundquist, J.; Sundquist, K.; Nexo, E.; Sorensen, B.S. Soluble HER3 predicts survival in bladder cancer patients. Oncol. Lett. 2018, 15, 1783–1788. [Google Scholar] [CrossRef]
- Lee, H.; Akita, R.W.; Sliwkowski, M.X.; Maihle, N.J. A naturally occurring secreted human ErbB3 receptor isoform inhibits heregulin-stimulated activation of ErbB2, ErbB3, and ErbB4. Cancer Res. 2001, 61, 4467–4473. [Google Scholar]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.-I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef]
- Black, L.E.; Longo, J.F.; Carroll, S.L. Mechanisms of Receptor Tyrosine-Protein Kinase ErbB-3 (ERBB3) Action in Human Neoplasia. Am. J. Pathol. 2019, 189, 1898–1912. [Google Scholar] [CrossRef]
- Menendez, J.A.; Mehmi, I.; Papadimitropoulou, A.; Vander Steen, T.; Cuyàs, E.; Verdura, S.; Espinoza, I.; Vellon, L.; Atlas, E.; Lupu, R. Fatty Acid Synthase Is a Key Enabler for Endocrine Resistance in Heregulin-Overexpressing Luminal B-Like Breast Cancer. Int. J. Mol. Sci. 2020, 21, 7661. [Google Scholar] [CrossRef]
- Mizuno, T.; Kojima, Y.; Yonemori, K.; Yoshida, H.; Sugiura, Y.; Ohtake, Y.; Okuma, H.S.; Nishikawa, T.; Tanioka, M.; Sudo, K.; et al. Neoadjuvant chemotherapy promotes the expression of HER3 in patients with ovarian cancer. Oncol. Lett. 2020, 20, 336. [Google Scholar] [CrossRef]
- Karachaliou, N.; Lazzari, C.; Verlicchi, A.; Sosa, A.E.; Rosell, R. HER3 as a Therapeutic Target in Cancer. BioDrugs 2017, 31, 63–73. [Google Scholar] [CrossRef] [PubMed]
- D’Abronzo, L.S.; Pan, C.X.; Ghosh, P.M. Evaluation of protein levels of the receptor tyrosine kinase ERBB3 in serum. Methods Mol. Biol. 2018, 1655, 319–334. [Google Scholar] [PubMed]
- Jagarlamudi, K.K.; Shaw, M. Thymidine kinase 1 as a tumor biomarker: Technical advances offer new potential to an old biomarker. Biomark. Med. 2018, 12, 1035–1048. [Google Scholar] [CrossRef]
- Weagel, E.G.; Burrup, W.; Kovtun, R.; Velazquez, E.J.; Felsted, A.M.; Townsend, M.H.; Ence, Z.E.; Suh, E.; Piccolo, S.R.; Weber, K.S.; et al. Membrane expression of thymidine kinase 1 and potential clinical relevance in lung, breast, and colorectal malignancies. Cancer Cell Int. 2018, 18, 135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, W.; Huo, B.; Dong, L.; Zhang, J. Relationship between thymidine kinase 1 before radiotherapy and prognosis in breast cancer patients with diabetes. Biosci. Rep. 2020, 40, BSR20192813. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Fornander, T.; Johansson, H.; Johansson, U.; Hu, G.Z.; Rutqvist, L.E.; Skog, S. Thymidine kinase 1 in serum predicts increased risk of distant or loco-regional recurrence following surgery in patients with early breast cancer. Anticancer Res. 2006, 26, 4753–4759. [Google Scholar]
- Bagegni, N.; Thomas, S.; Liu, N.; Luo, J.; Hoog, J.; Northfelt, D.W.; Goetz, M.P.; Forero, A.; Bergqvist, M.; Karen, J.; et al. Serum thymidine kinase 1 activity as a pharmacodynamic marker of cyclin-dependent kinase 4/6 inhibition in patients with early-stage breast cancer receiving neoadjuvant palbociclib. Breast Cancer Res. 2017, 19, 123. [Google Scholar] [CrossRef]
- Tribukait, B. Early prediction of pathologic response to neoadjuvant treatment of breast cancer: Use of a cell-loss metric based on serum thymidine kinase 1 and tumour volume. BMC Cancer 2020, 20, 440. [Google Scholar] [CrossRef]
- Chung, L.; Moore, K.; Phillips, L.; Boyle, F.M.; Marsh, D.J.; Baxter, R.C. Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Res. 2014, 16, R63. [Google Scholar] [CrossRef]
- Kazarian, A.; Blyuss, O.; Metodieva, G.; Gentry-Maharaj, A.; Ryan, A.; Kiseleva, E.M.; Prytomanova, O.M.; Jacobs, I.J.; Widschwendter, M.; Menon, U.; et al. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br. J. Cancer 2017, 116, 501–508. [Google Scholar] [CrossRef]
- Zeillinger, R.; Kury, F.; Czerwenka, K.; Kubista, E.; Sliutz, G.; Knogler, W.; Huber, J.; Zielinski, C.; Reiner, G.; Jakesz, R.; et al. HER-2 amplification, steroid receptors and epidermal growth factor receptor in primary breast cancer. Oncogene 1989, 4, 109–114. [Google Scholar] [PubMed]
- Doroshow, D.B.; Doroshow, J.H. Genomics and the History of Precision Oncology. Surg. Oncol. Clin. N. Am. 2020, 29, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Božović, A.; Mandušić, V.; Todorović, L.; Krajnović, M. Estrogen Receptor Beta: The Promising Biomarker and Potential Target in Metastases. Int. J. Mol. Sci. 2021, 22, 1656. [Google Scholar] [CrossRef] [PubMed]
- Telli, M.L.; Nagata, H.; Wapnir, I.L.; Acharya, C.; Zablotsky, K.E.; Fox, B.A.; Bifulco, C.B.; Jensen, S.M.; Ballesteros-Merino, C.; Le, M.H.; et al. Intratumoral plasmid IL-12 expands CD8+ T cells and induces a CXCR3 gene signature in triple-negative breast tumors that sensitizes patients to anti-PD-1 therapy. Clin. Cancer Res. 2021, 27, 2481–2493. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.; Hilton, J.; Clemons, M.; Mazzarello, S.; Hutton, B.; Haggar, F.; Addison, C.L.; Kuchuk, I.; Zhu, X.; Gelmon, K.; et al. Estrogen, progesterone, and HER2/neu receptor discordance between primary and metastatic breast tumors—A review. Cancer Metastasis Rev. 2016, 35, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Amir, E.; Miller, N.; Geddie, W.; Freedman, O.; Kassam, F.; Simmons, C.; Oldfield, M.; Dranitsaris, G.; Tomlinson, G.; Laupacis, A.; et al. Prospective Study Evaluating the Impact of Tissue Confirmation of Metastatic Disease in Patients with Breast Cancer. J. Clin. Oncol. 2012, 30, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Steenbruggen, T.G.; van Seijen, M.; Janssen, L.M.; van Ramshorst, M.S.; van Werkhoven, E.; Vrancken Peeters, M.-J.T.D.F.; Wesseling, J.; Lips, E.H.; Sonke, G.S. Prognostic Value of Residual Disease after Neoadjuvant Therapy in HER2-Positive Breast Cancer Evaluated by Residual Cancer Burden, Neoadjuvant Response Index, and Neo-Bioscore. Clin. Cancer Res. 2019, 25, 4985–4992. [Google Scholar] [CrossRef]
- Nowecki, Z.; Jagiello-Gruszfeld, A.; Pogoda, K.; Niwińska, A.; Olszewski, W.P.; Winter, P.; Matkowski, R.; Wysocki, W.M. Neoadjuvant therapy for breast cancer patients and its impact on surgical treatment and radiotherapy (part 1.). Nowotw. J. Oncol. 2021, 71, 17–25. [Google Scholar] [CrossRef]
- Livingston-Rosanoff, D.; Schumacher, J.; Vande Walle, K.; Stankowski-Drengler, T.; Greenberg, C.C.; Neuman, H.; Wilke, L.G. Does Tumor Size Predict Response to Neoadjuvant Chemotherapy in the Modern Era of Biologically Driven Treatment? A Nationwide Study of US Breast Cancer Patients. Clin. Breast Cancer 2019, 19, e741–e747. [Google Scholar] [CrossRef]
- Katayama, A.; Miligy, I.M.; Shiino, S.; Toss, M.S.; Eldib, K.; Kurozumi, S.; Quinn, C.M.; Badr, N.; Murray, C.; Provenzano, E.; et al. Predictors of pathological complete response to neoadjuvant treatment and changes to post-neoadjuvant HER2 status in HER2-positive invasive breast cancer. Mod. Pathol. 2021, 34, 1271–1281. [Google Scholar] [CrossRef]
- Jagarlamudi, K.K.; Hansson, L.O.; Eriksson, S. Breast and prostate cancer patients differ significantly in their serum Thymidine kinase 1 (TK1) specific activities compared with those hematological malignancies and blood donors: Implications of using serum TK1 as a biomarker. BMC Cancer 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhang, P.; Zou, L.; Li, H.; Wang, X.; Zhou, S.; Fornander, T.; Skog, S. Concentration of thymidine kinase 1 in serum (S-TK1) is a more sensitive proliferation marker in human solid tumors than its activity. Oncol. Rep. 2005, 14, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Bitter, E.E.; Townsend, M.H.; Erickson, R.; Allen, C.; O’Neill, K.L. Thymidine kinase 1 through the ages: A comprehensive review. Cell Biosci. 2020, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- McCartney, A.; Biagioni, C.; Schiavon, G.; Bergqvist, M.; Mattsson, K.; Migliaccio, I.; Benelli, M.; Romagnoli, D.; Bonechi, M.; Boccalini, G.; et al. Prognostic role of serum thymidine kinase 1 activity in patients with hormone receptor–positive metastatic breast cancer: Analysis of the randomised phase III Evaluation of Faslodex versus Exemestane Clinical Trial (EFECT). Eur. J. Cancer 2019, 114, 55–66. [Google Scholar] [CrossRef]
- Bonechi, M.; Galardi, F.; Biagioni, C.; De Luca, F.; Bergqvist, M.; Neumüller, M.; Guarducci, C.; Boccalini, G.; Gabellini, S.; Migliaccio, I.; et al. Plasma thymidine kinase-1 activity predicts outcome in patients with hormone receptor positive and HER2 negative metastatic breast cancer treated with endocrine therapy. Oncotarget 2018, 9, 16389–16399. [Google Scholar] [CrossRef]
- Broughton, M.N.; Westgaard, A.; Paus, E.; Øijordsbakken, M.; Henanger, K.J.; Naume, B.; Bjøro, T. Specific antibodies and sensitive immunoassays for the human epidermal growth factor receptors (HER2, HER3, and HER4). Tumor Biol. 2017, 39, 6. [Google Scholar] [CrossRef]
- Li, C.; Wang, S.; Xing, Z.; Lin, A.; Liang, K.; Song, J.; Hu, Q.; Yao, J.; Chen, Z.; Park, P.K.; et al. A ROR1–HER3–lncRNA signalling axis modulates the Hippo–YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017, 19, 106–119. [Google Scholar] [CrossRef]
- Gutsch, D.; Jenke, R.; Büch, T.; Aigner, A. Inhibition of HER Receptors Reveals Distinct Mechanisms of Compensatory Upregulation of Other HER Family Members: Basis for Acquired Resistance and for Combination Therapy. Cells 2021, 10, 272. [Google Scholar] [CrossRef]
- Du, R.; Huang, C.; Liu, K.; Li, X.; Dong, Z. Targeting AURKA in Cancer: Molecular mechanisms and opportunities for Cancer therapy. Mol. Cancer 2021, 20, 15. [Google Scholar] [CrossRef]
- Cirak, Y.; Furuncuoglu, Y.; Yapicier, O.; Aksu, A.; Cubukcu, E. Aurora a overexpression in breast cancer patients induces taxane resistance and results in worse prognosis. J. BUON 2015, 20, 1414–1419. [Google Scholar]

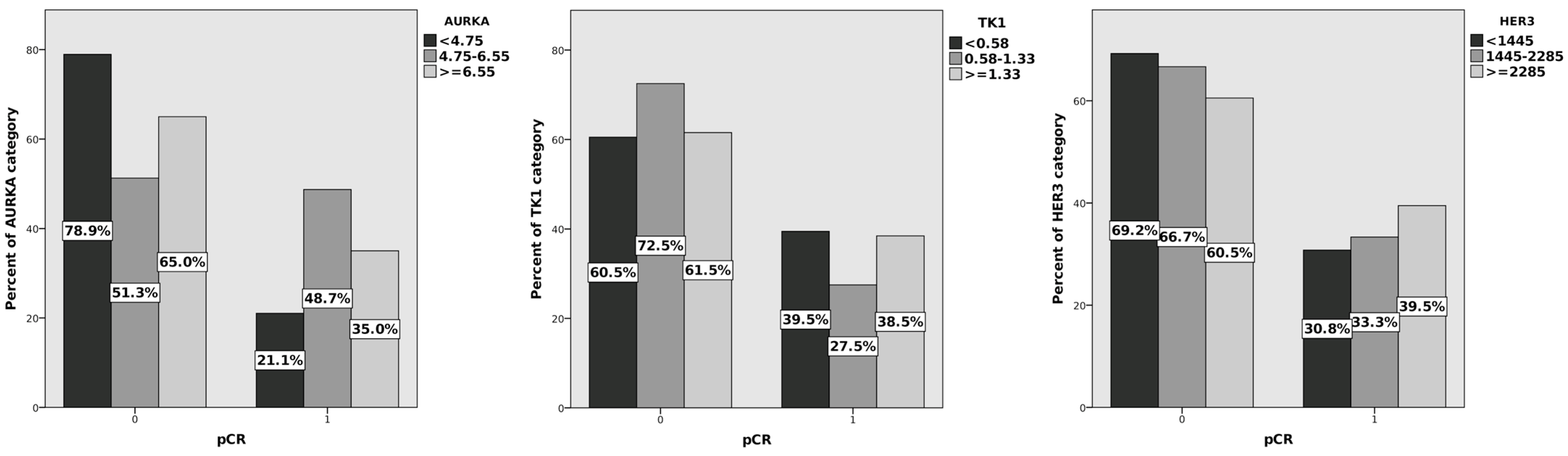
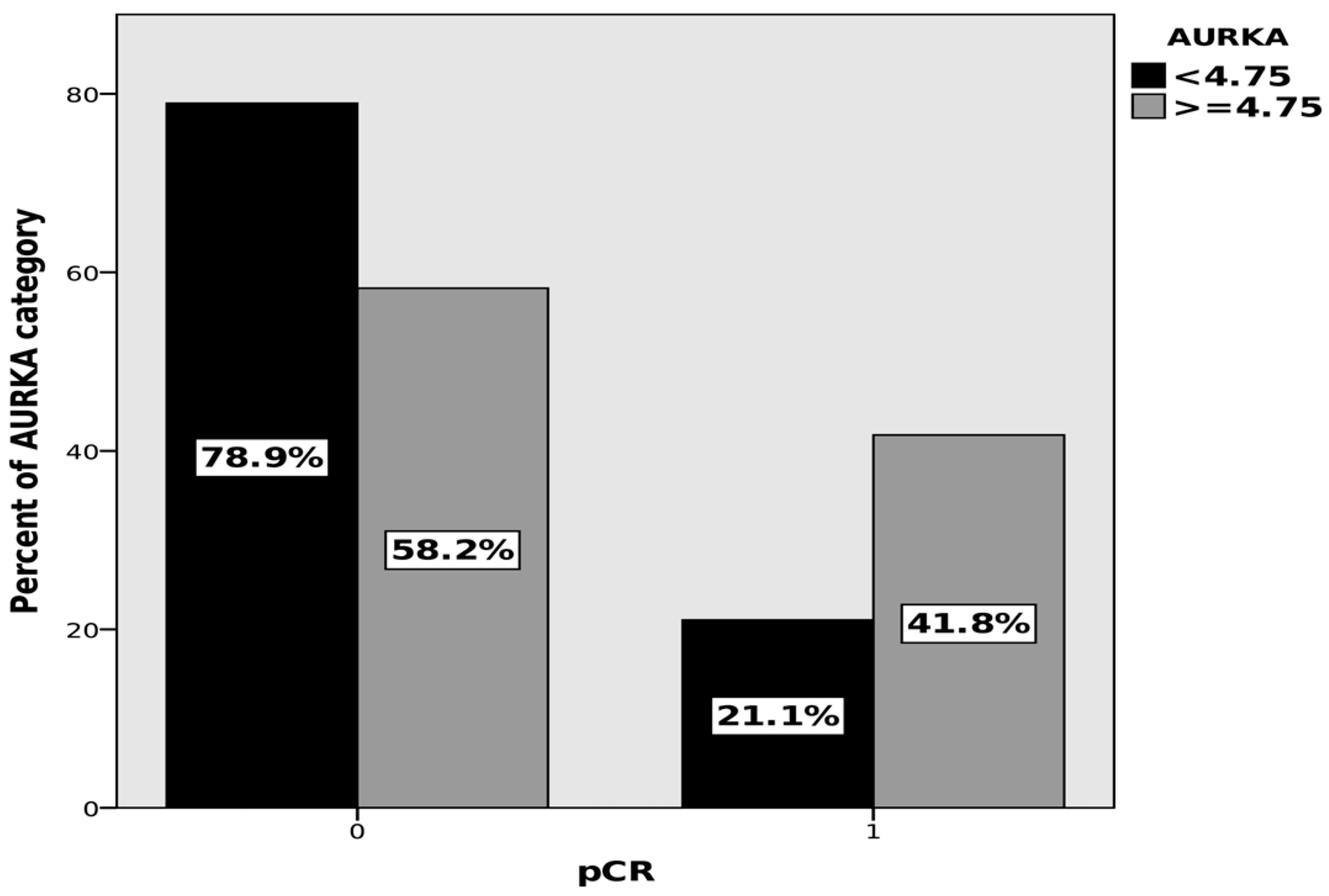
| Characteristics | N = 119 (%) | pCR | |
|---|---|---|---|
| % of Patients | p | ||
| Age (median, range) | 53 (31–77) | ||
| ER positive | 74 (62.18%) | ||
| PR positive | 61 (51.26%) | ||
| HER2 positive | 45 (37.82%) | ||
| Ki67 ≤ 20 | 23 (19.33%) | ||
| Ki67 > 20 | 96 (80.67%) | ||
| Treatment | |||
| TCH/TCH + P * | 44 (36.97%) | ||
| 4xAC + 12xPXL ** | 61 (51.26%) | ||
| Hormonal | 14 (11.76%) | ||
| Tumor size (T) | |||
| T1 | 8 (6.72%) | 5 (62.50%) | 0.3584 |
| T2 | 73 (61.34%) | 25 (34.25%) | |
| T3 | 25 (21.01%) | 7 (28.00%) | |
| T4 | 11 (9.24%) | 4 (36.25%) | |
| Tx | 2 (1.68%) | 0 (0%) | |
| Lymph node status (N) | |||
| N0 | 52 (43.70%) | 21 (40.38%) | 0.2435 |
| N1 | 44 (36.97%) | 15 (34.09%) | |
| N2 | 19 (15.97%) | 3 (15.78%) | |
| N3 | 4 (3.36%) | 2 (50.00%) | |
| Distance metastasis status (M) | |||
| M0 | 111 (93.28%) | 40 (34.48%) | 0.1670 |
| M1 | 8 (6.62%) | 1 (12.50%) | |
| Tumor grade (G) | |||
| G1 | 8 (6.72%) | 1 (12.50%) | 0.0078 |
| G2 | 61 (51.26%) | 15 (24.59%) | |
| G3 | 50 (42.02%) | 25 (50.00%) | |
| Biological subtype | |||
| Luminal A | 13 (10.92%) | 0 (0%) | 0.0003 |
| Luminal B | 37 (31.09%) | 6 (16.66%) | |
| Luminal B HER2-enriched | 26 (21.85%) | 10 (38.46%) | |
| Non-Luminal HER2-positive | 19 (15.97%) | 10 (52.63%) | |
| TNBC /triple negative | 24 (20.17%) | 15 (62.50%) | |
| Parameters | β Coefficient | Standard Error | p Value | Odds Ratio | 95%—Confidence Interval | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| N0 * | −0.687 | 0.332 | 0.039 | 0.503 | 0.263 | 0.965 |
| PR (-) ** | −2.259 | 0.511 | <0.001 | 0.104 | 0.038 | 0.284 |
| Ki67 > 20% | 1.694 | 0.755 | 0.025 | 5.441 | 1.239 | 23.893 |
| AURKA ≥ 4.75 ng/mL | 1.244 | 0.545 | 0.023 | 3.470 | 1.192 | 10.105 |
| Constant | −3.178 | 1.546 | 0.040 | 0.042 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winter, P.; Fuksiewicz, M.; Jagiello-Gruszfeld, A.; Nowecki, Z.; Kotowicz, B. Expression of Soluble Form of Aurora A as a Predictive Factor for Neoadjuvant Therapy in Breast Cancer Patients: A Single-Center Pilot Study. Cancers 2023, 15, 5446. https://doi.org/10.3390/cancers15225446
Winter P, Fuksiewicz M, Jagiello-Gruszfeld A, Nowecki Z, Kotowicz B. Expression of Soluble Form of Aurora A as a Predictive Factor for Neoadjuvant Therapy in Breast Cancer Patients: A Single-Center Pilot Study. Cancers. 2023; 15(22):5446. https://doi.org/10.3390/cancers15225446
Chicago/Turabian StyleWinter, Pawel, Malgorzata Fuksiewicz, Agnieszka Jagiello-Gruszfeld, Zbigniew Nowecki, and Beata Kotowicz. 2023. "Expression of Soluble Form of Aurora A as a Predictive Factor for Neoadjuvant Therapy in Breast Cancer Patients: A Single-Center Pilot Study" Cancers 15, no. 22: 5446. https://doi.org/10.3390/cancers15225446
APA StyleWinter, P., Fuksiewicz, M., Jagiello-Gruszfeld, A., Nowecki, Z., & Kotowicz, B. (2023). Expression of Soluble Form of Aurora A as a Predictive Factor for Neoadjuvant Therapy in Breast Cancer Patients: A Single-Center Pilot Study. Cancers, 15(22), 5446. https://doi.org/10.3390/cancers15225446







