Real-World Outcomes of Immunotherapy in Second- or Later-Line Non-Small Cell Lung Cancer with Actionable Genetic Alterations
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Design and Study Population
2.2. Outcomes
2.3. Identification of Actionable Genetic Alteration, PD-L1, and TMB
2.4. Statistical Analyses
3. Results
3.1. Clinical Characteristics
3.2. Molecular Biomarkers (PD-L1, TMB, and Co-Existing Mutations)
3.3. Treatment Outcomes of Immune Checkpoint Inhibitors
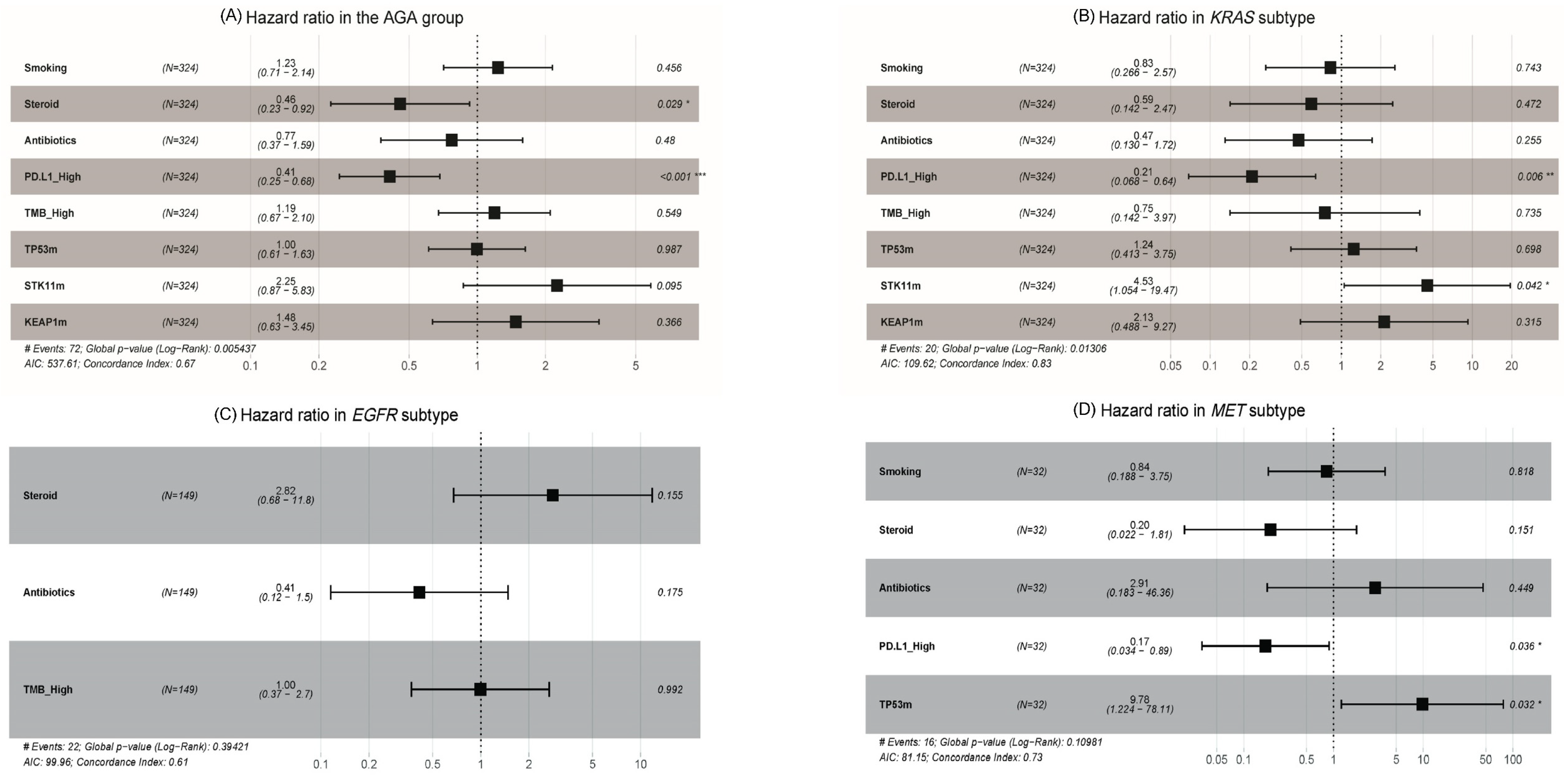
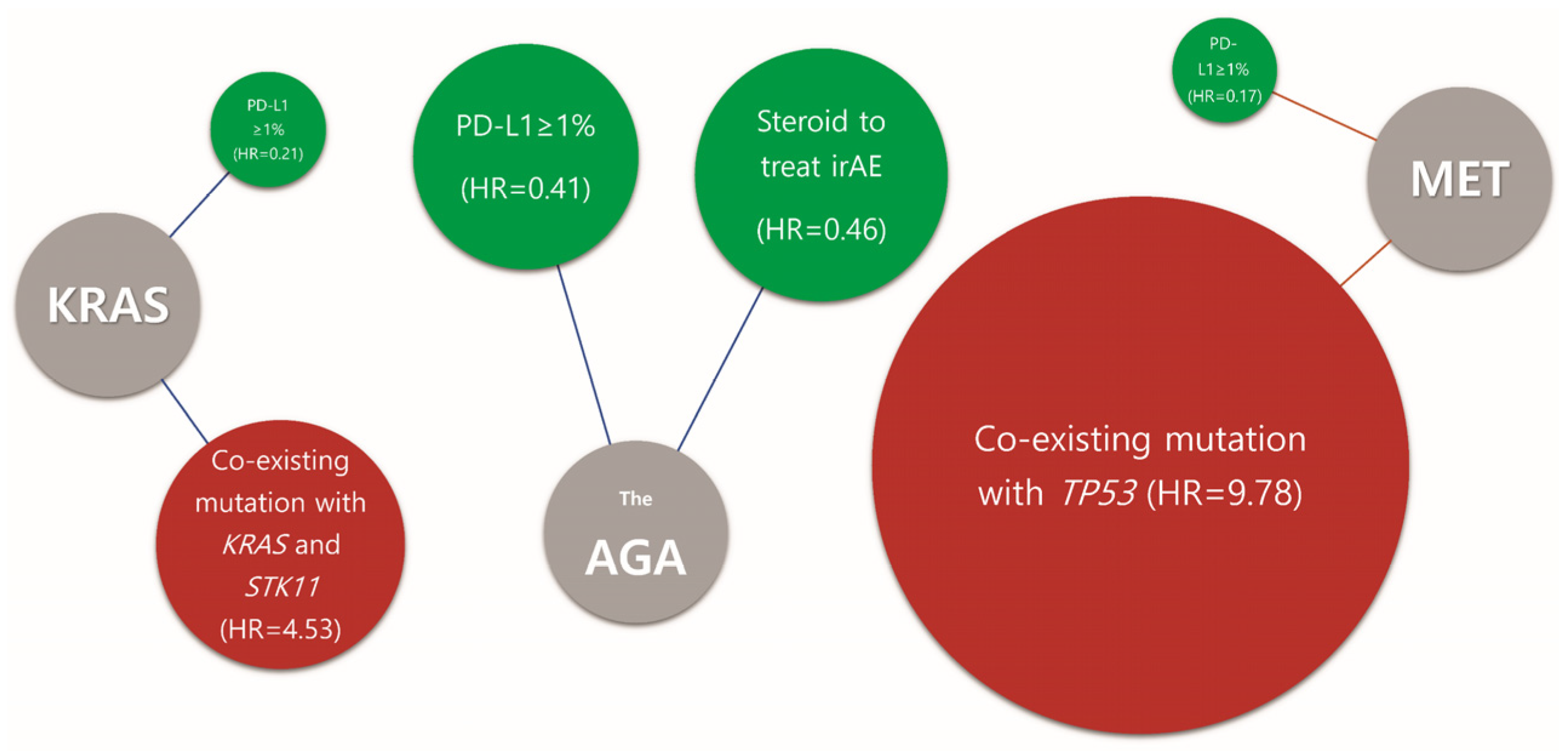
3.4. Clinical and Molecular Predictive Markers for Immune Checkpoint Inhibitor
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015, 21, 687–692. [Google Scholar] [CrossRef]
- Rodak, O.; Peris-Diaz, M.D.; Olbromski, M.; Podhorska-Okolow, M.; Dziegiel, P. Current Landscape of Non-Small Cell Lung Cancer: Epidemiology, Histological Classification, Targeted Therapies, and Immunotherapy. Cancers 2021, 13, 5789. [Google Scholar] [CrossRef]
- Negrao, M.V.; Skoulidis, F.; Montesion, M.; Schulze, K.; Bara, I.; Shen, V.; Xu, H.; Hu, S.; Sui, D.; Elamin, Y.Y.; et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J. Immunother. Cancer 2021, 9, e002891. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Aguilar, E.J.; Ricciuti, B.; Gainor, J.F.; Kehl, K.L.; Kravets, S.; Dahlberg, S.; Nishino, M.; Sholl, L.M.; Adeni, A.; Subegdjo, S.; et al. Outcomes to first-line pembrolizumab in patients with non-small-cell lung cancer and very high PD-L1 expression. Ann. Oncol. 2019, 30, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.K.; Lopes, G.; Cho, B.C.; Kowalski, D.M.; Kasahara, K.; Wu, Y.L.; de Castro, G., Jr.; Turna, H.Z.; Cristescu, R.; Aurora-Garg, D.; et al. Associations of tissue tumor mutational burden and mutational status with clinical outcomes in KEYNOTE-042: Pembrolizumab versus chemotherapy for advanced PD-L1-positive NSCLC. Ann. Oncol. 2023, 34, 377–388. [Google Scholar] [CrossRef]
- Cordeiro de Lima, V.C.; Corassa, M.; Saldanha, E.; Freitas, H.; Arrieta, O.; Raez, L.; Samtani, S.; Ramos, M.; Rojas, C.; Burotto, M.; et al. STK11 and KEAP1 mutations in non-small cell lung cancer patients: Descriptive analysis and prognostic value among Hispanics (STRIKE registry-CLICaP). Lung Cancer 2022, 170, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Pavan, A.; Bragadin, A.B.; Calvetti, L.; Ferro, A.; Zulato, E.; Attili, I.; Nardo, G.; Dal Maso, A.; Frega, S.; Menin, A.G.; et al. Role of next generation sequencing-based liquid biopsy in advanced non-small cell lung cancer patients treated with immune checkpoint inhibitors: Impact of STK11, KRAS and TP53 mutations and co-mutations on outcome. Transl. Lung Cancer Res. 2021, 10, 202–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Man, J.; Lord, S.; Cooper, W.; Links, M.; Gebski, V.; Herbst, R.S.; Gralla, R.J.; Mok, T.; Yang, J.C. Clinical and Molecular Characteristics Associated with Survival Among Patients Treated with Checkpoint Inhibitors for Advanced Non-Small Cell Lung Carcinoma: A Systematic Review and Meta-analysis. JAMA Oncol. 2018, 4, 210–216. [Google Scholar] [CrossRef]
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Non-Small Cell Lung Cancer; NCCN: Fort Washington, PA, USA, 2023.
- Seegobin, K.; Majeed, U.; Wiest, N.; Manochakian, R.; Lou, Y.; Zhao, Y. Immunotherapy in Non-Small Cell Lung Cancer with Actionable Mutations Other Than EGFR. Front Oncol. 2021, 11, 750657. [Google Scholar] [CrossRef]
- Cortinovis, D.; Gregorc, V.; Migliorino, M.R.; Abate, M.I.; Manzo, A.; Malapelle, U.; Morabito, A.; Italian Lung Cancer Working Group. New perspectives in the second-line treatment of non squamous NSCLC patients: Results from a large Italian Lung Cancer Working Group. Crit. Rev. Oncol. Hematol. 2017, 109, 35–41. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Ward, E.; Jemal, A.; Cokkinides, V.; Singh, G.K.; Cardinez, C.; Ghafoor, A.; Thun, M. Cancer disparities by race/ethnicity and socioeconomic status. CA Cancer J. Clin. 2004, 54, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Kim, H.C.; Choi, C.M. Recent Trends of Lung Cancer in Korea. Tuberc. Respir. Dis. 2021, 84, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.A.; Jeong, O.; Chang, D.K.; Park, S.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Ahn, M.J.; Park, K. Real-time autOmatically updated data warehOuse in healThcare (ROOT): An innovative and automated data collection system. Transl. Lung Cancer Res. 2021, 10, 3865–3874. [Google Scholar] [CrossRef] [PubMed]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Xu, Y.; Huang, Z.; Cheng, G.; Xie, M.; Zhou, Z.; Yu, Y.; Xi, W.; Fan, Y. Tumor immune microenvironment and immunotherapy efficacy in BRAF mutation non-small-cell lung cancer. Cell Death Dis. 2022, 13, 1064. [Google Scholar] [CrossRef]
- Liu, C.; Zheng, S.; Jin, R.; Wang, X.; Wang, F.; Zang, R.; Xu, H.; Lu, Z.; Huang, J.; Lei, Y.; et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. 2020, 470, 95–105. [Google Scholar] [CrossRef]
- Jeanson, A.; Tomasini, P.; Souquet-Bressand, M.; Brandone, N.; Boucekine, M.; Grangeon, M.; Chaleat, S.; Khobta, N.; Milia, J.; Mhanna, L.; et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J. Thorac. Oncol. 2019, 14, 1095–1101. [Google Scholar] [CrossRef]
- Wu, S.G.; Liao, W.Y.; Su, K.Y.; Yu, S.L.; Huang, Y.L.; Yu, C.J.; Chih-Hsin Yang, J.; Shih, J.Y. Prognostic Characteristics and Immunotherapy Response of Patients with Nonsquamous NSCLC with KRAS Mutation in East Asian Populations: A Single-Center Cohort Study in Taiwan. JTO Clin. Res. Rep. 2021, 2, 100140. [Google Scholar] [CrossRef]
- Wiest, N.; Majeed, U.; Seegobin, K.; Zhao, Y.; Lou, Y.; Manochakian, R. Role of Immune Checkpoint Inhibitor Therapy in Advanced EGFR-Mutant Non-Small Cell Lung Cancer. Front. Oncol. 2021, 11, 751209. [Google Scholar] [CrossRef]
- Lee, C.K.; Man, J.; Lord, S.; Links, M.; Gebski, V.; Mok, T.; Yang, J.C. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J. Thorac. Oncol. 2017, 12, 403–407. [Google Scholar] [CrossRef]
- Guo, X.; Du, H.; Li, J.; Yang, M.; Xiong, A.; Zhang, H.; Wu, F. Efficacy of ICIs on patients with oncogene-driven non-small cell lung cancer: A retrospective study. Cancer Drug Resist. 2022, 5, 15–24. [Google Scholar] [CrossRef]
- Albitar, M.; Sudarsanam, S.; Ma, W.; Jiang, S.; Chen, W.; Funari, V.; Blocker, F.; Agersborg, S. Correlation of MET gene amplification and TP53 mutation with PD-L1 expression in non-small cell lung cancer. Oncotarget 2018, 9, 13682–13693. [Google Scholar] [CrossRef] [PubMed]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]
- Di Federico, A.; De Giglio, A.; Parisi, C.; Gelsomino, F. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? Eur. J. Cancer 2021, 157, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Brueckl, W.M.; Ficker, J.H.; Zeitler, G. Clinically relevant prognostic and predictive markers for immune-checkpoint-inhibitor (ICI) therapy in non-small cell lung cancer (NSCLC). BMC Cancer 2020, 20, 1185. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Jia, X.; Jiang, P.; Mao, Z.; Liang, T.; Du, Y.; Zhang, J.; Zhang, G.; Niu, G.; et al. Effect of Immune-Related Adverse Events and Pneumonitis on Prognosis in Advanced Non-Small Cell Lung Cancer: A Comprehensive Systematic Review and Meta-analysis. Clin. Lung Cancer 2021, 22, e889–e900. [Google Scholar] [CrossRef]
- Paderi, A.; Gambale, E.; Botteri, C.; Giorgione, R.; Lavacchi, D.; Brugia, M.; Mazzoni, F.; Giommoni, E.; Bormioli, S.; Amedei, A.; et al. Association of Systemic Steroid Treatment and Outcome in Patients Treated with Immune Checkpoint Inhibitors: A Real-World Analysis. Molecules 2021, 26, 5789. [Google Scholar] [CrossRef] [PubMed]
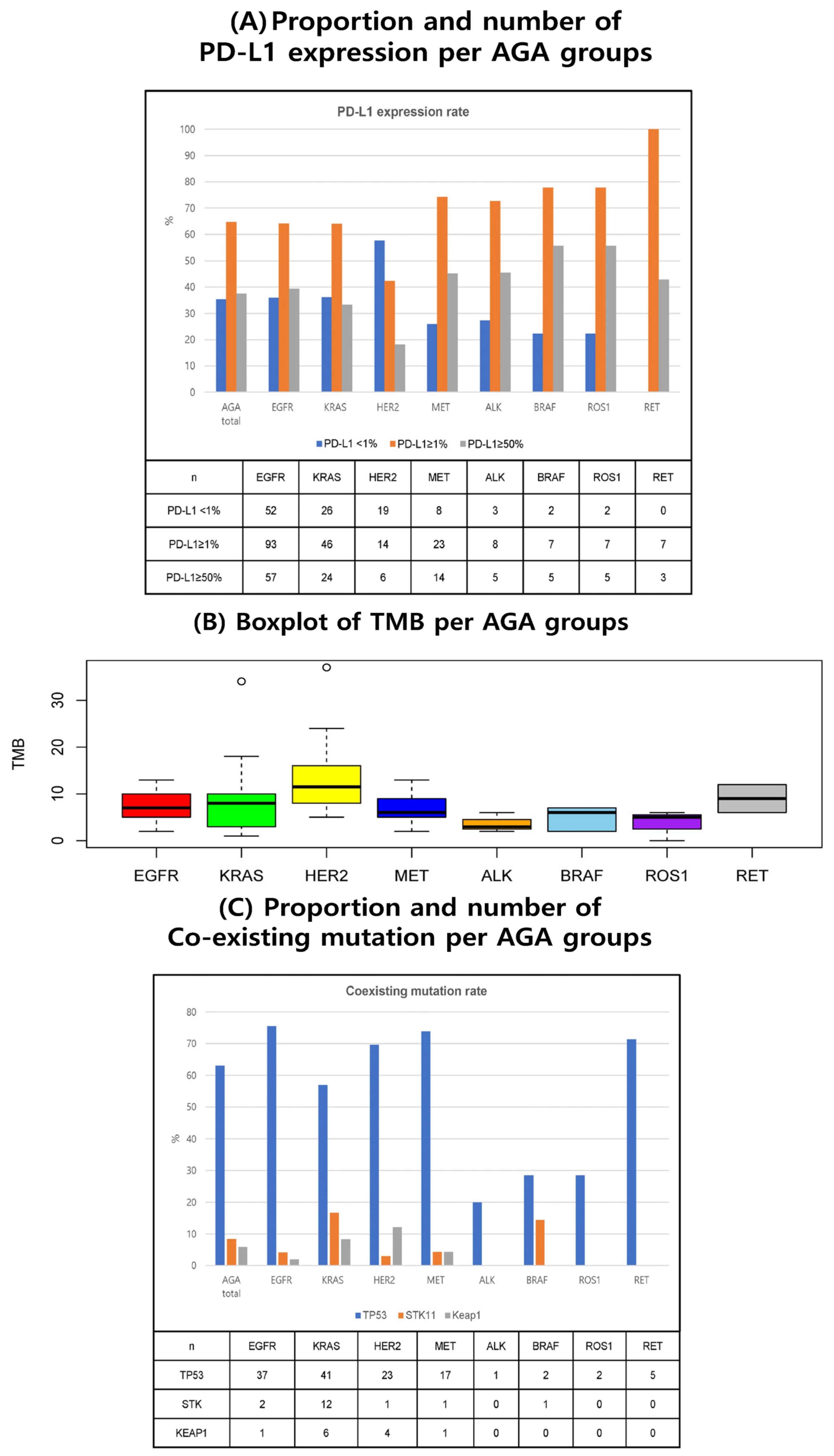

| AGA Type n (%) | EGFR | KRAS | HER2 | MET | ALK | BRAF | ROS1 | RET | p | |
|---|---|---|---|---|---|---|---|---|---|---|
| n = 149 (46.0) | n = 72 (22.2) | n = 34 (10.5) | n = 32 (9.9) | n = 12 (3.7) | n = 9 (2.8) | n = 9 (2.8) | n = 7 (2.2) | |||
| Age 1 (years) | Median age (95% range) | 62.2 (39.1–81.7) | 67.2 (44.4–81.7) | 66.1 (47.3–76.5) | 64.3 (44.1–78.4) | 60.1 (53.8–82.0) | 60.1 (54.0–73.60) | 62.4 (44.4–80.9) | 59.7 (30.8–65.9) | |
| <65 | 90 (60.4) | 30 (41.7) | 17 (50.0) | 18 (56.2) | 9 (75.0) | 7 (77.8) | 7 (77.8) | 5 (71.4) | 0.06 | |
| ≥65 | 59 (39.6) | 42 (58.3) | 17 (50.0) | 14 (43.8) | 3 (25.0) | 2 (22.2) | 2 (22.2) | 2 (28.6) | ||
| Sex | Male | 70 (47.0) | 41 (56.9) | 22 (64.7) | 24 (75.0) | 7 (58.3) | 5 (55.6) | 2 (22.2) | 1 (14.3) | 0.01 |
| Female | 79 (53.0) | 31 (43.1) | 12 (35.3) | 8 (25.0) | 5 (41.7) | 4 (44.4) | 7 (77.8) | 6 (85.7) | ||
| Smoking | Never | 90 (60.4) | 34 (47.2) | 13 (38.2) | 11 (34.4) | 6 (50.0) | 5 (55.6) | 8 (88.9) | 7 (100) | 0.002 |
| Ex or Current | 59 (39.6) | 38 (52.8) | 21 (61.8) | 21 (65.6) | 6 (50.0) | 4 (44.4) | 1 (11.1) | 0 (0.0) | ||
| ECOG | 0–1 | 147 (98.7) | 67 (93.1) | 33 (97.1) | 31 (96.9) | 12 (100) | 9 (100) | 9 (100) | 7 (100) | 0.45 |
| ≥2 | 2 (1.3) | 5 (6.9) | 1 (2.9) | 1 (3.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Histology | Non-squamous | 139 (93.3) | 65 (90.3) | 27 (79.4) | 26 (81.2) | 10 (83.3) | 8 (88.9) | 9 (100) | 6 (85.7) | 0.19 |
| Others | 10 (6.7) | 7 (9.7) | 7 (20.6) | 6 (18.8) | 2 (16.7) | 1 (11.1) | 0 (0.0) | 1 (14.3) | ||
| Line of Therapy | 2nd line | 19 (12.8) | 61 (84.7) | 3 (25.0) | 29 (90.6) | 3 (25.0) | 4 (44.4) | 5 (55.6) | 6 (85.7) | <0.001 |
| Later line | 130 (87.2) | 11 (15.3) | 9 (75.0) | 3 (9.4) | 9 (75.0) | 5 (55.6) | 4 (44.4) | 1 (14.3) | ||
| Concomitant disease and therapy | ||||||||||
| HTN | No | 94 (63.1) | 42 (58.3) | 21 (61.8) | 17 (53.1) | 8 (66.7) | 3 (33.3) | 4 (44.4) | 4 (57.1) | 0.82 |
| Yes | 54 (36.2) | 30 (41.7) | 13 (38.2) | 14 (43.8) | 4 (33.3) | 6 (66.7) | 5 (55.6) | 3 (42.9) | ||
| NI | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (3.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| DM | No | 118 (79.2) | 55 (76.4) | 21 (61.8) | 23 (71.9) | 9 (75.0) | 4 (44.4) | 8 (88.9) | 6 (85.7) | 0.14 |
| Yes | 31 (20.8) | 17 (23.6) | 13 (38.2) | 8 (25.0) | 3 (25.0) | 5 (55.6) | 1 (11.1) | 1 (14.3) | ||
| NI | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (3.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| RT * | No | 130 (87.2) | 64 (88.9) | 25 (73.5) | 29 (90.6) | 10 (83.3) | 9 (100) | 7 (77.8) | 6 (85.7) | 0.35 |
| Yes | 19 (12.8) | 8 (11.1) | 9 (26.5) | 3 (9.4) | 2 (16.7) | 0 (0.0) | 2 (22.2) | 1 (14.3) | ||
| Steroid † | No | 113 (75.8) | 55 (76.4) | 24 (70.6) | 26 (81.2) | 8 (66.7) | 9 (100) | 7 (77.8) | 6 (85.7) | 0.68 |
| Yes | 36 (24.2) | 17 (23.6) | 10 (29.4) | 6 (18.8) | 4 (33.3) | 0 (0.0) | 2 (22.2) | 1 (14.3) | ||
| Antibiotics $ | No | 126 (84.6) | 56 (77.8) | 31 (91.2) | 29 (90.6) | 7 (58.3) | 9 (100) | 9 (100) | 6 (85.7) | 0.05 |
| Yes | 23 (15.4) | 16 (22.2) | 3 (8.8) | 3 (9.4) | 5 (41.7) | 0 (0.0) | 0 (0.0) | 1 (14.3) | ||
| Biomarkers N (%) | AGA n = 324 | Wild Type n = 602 | p | EGFR n = 149 | KRAS n = 72 | HER2 n = 34 | MET n = 32 | ALK n = 12 | BRAF n = 9 | ROS1 n = 9 | RET n = 7 | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PD-L1 expression | 317 (97.8) | 596 (99.0) | 145 (97.3) | 72 (100) | 33 (97.1) | 31 (96.9) | 11 (91.7) | 9 (100) | 9 (100) | 7 (100) | |||
| Median (95% CI) | 10 (0–100) | 9 (0–100) | 10 (0–90) | 7.5 (0–100) | 0 (0–82) | 10 (0–100) | 25 (0–90) | 50 (0–98) | 50 (0–94) | 10 (1.6–90) | |||
| Cutoff | H (≥1%) | 205 (64.7) | 355 (59.6) | 0.15 | 93 (64.1) | 46 (63.9) | 14 (42.4) | 23 (74.2) | 8 (72.7) | 7 (77.8) | 7 (77.8) | 7 (100) | 0.05 |
| L (<1%) | 112 (35.3) | 241 (40.4) | 52 (35.9) | 26 (36.1) | 19 (57.6) | 8 (25.8) | 3 (27.3) | 2 (22.2) | 2 (22.2) | 0 (0.0) | |||
| TMB | 92 (28.4) | 92 (15.3) | 22 (14.8) | 29 (40.3) | 16 (47.1) | 12 (37.5) | 3 (25.0) | 5 (55.6) | 3 (33.3) | 2 (28.6) | |||
| Median (95% CI) | 7.0 (1.3–22.6) | 10.5 (2.3–40.1) | 7.0 (2.5–12.5) | 8.0 (1.0–22.8) | 11.5 (5.0–32.13) | 6.0 (2.3–11.9) | 3.0 (2.1–5.9) | 6.0 (2.0–7.0) | 5.0 (0.3–6.0) | 9.0 (6.2–11.9) | |||
| Cutoff | H (≥10) | 26 (28.3) | 49 (53.3) | 0.001 | 6 (27.3) | 7 (24.1) | 11 (68.8) | 1 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0.01 |
| Muts/Mb | L (<10) | 66 (71.7) | 43 (46.7) | 16 (72.7) | 22 (75.9) | 5 (31.2) | 11 (91.7) | 3 (100) | 5 (100) | 3 (100) | 1 (50.0) | ||
| TP53 | 203 (62.7) | 208 (34.6) | 49 (32.9) | 72 (100) | 33 (97.1) | 23 (71.9) | 5 (41.7) | 7 (77.8) | 7 (77.8) | 7 (100) | |||
| Mutation (+) | 128 (63.1) | 165 (79.3) | <0.001 | 37 (75.5) | 41 (56.9) | 23 (69.7) | 17 (73.9) | 1 (20.0) | 2 (28.6) | 2 (28.6) | 5 (71.4) | 0.01 | |
| Mutation (−) | 75 (36.9) | 43 (20.7) | 12 (24.5) | 31 (43.1) | 10 (30.3) | 6 (26.1) | 4 (80.0) | 5 (71.4) | 5 (71.4) | 2 (28.6) | |||
| STK11 | 202 (62.3) | 208 (34.6) | 49 (32.9) | 72 (100) | 33 (97.1) | 23 (71.9) | 4 (41.7) | 7 (77.8) | 7 (77.8) | 7 (100) | |||
| Mutation (+) | 17 (8.4) | 25 (12.0) | 0.30 | 2 (4.1) | 12 (16.7) | 1 (3.0) | 1 (4.3) | 0 (0.0) | 1 (14.3) | 0 (0.0) | 0 (0.0) | 0.13 | |
| Mutation (−) | 185 (91.6) | 183 (88.0) | 47 (95.9) | 60 (83.3) | 32 (97.0) | 22 (95.7) | 4 (100) | 6 (85.7) | 7 (100) | 7 (100) | |||
| KEAP 1 | 202 (62.3) | 208 (34.6) | 49 (32.9) | 72 (100) | 33 (97.1) | 23 (71.9) | 4 (41.7) | 7 (77.8) | 7 (77.8) | 7 (100) | |||
| Mutation (+) | 12 (5.9) | 26 (12.5) | 0.03 | 1 (2.0) | 6 (8.3) | 4 (12.1) | 1 (4.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.54 | |
| Mutation (−) | 190 (94.1) | 182 (87.5) | 48 (98.0) | 66 (91.7) | 29 (87.9) | 22 (95.7) | 4 (100) | 7 (100) | 7 (100) | 7 (100) | |||
| AGA | Wild Type | EGFR | KRAS | HER2 | MET | ALK | BRAF | ROS1 | RET | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 324 | n = 602 | p | n = 149 | n = 72 | n = 34 | n = 32 | n = 12 | n = 9 | n = 9 | n = 7 | ||
| Best response | ||||||||||||
| CR | 1 (0.3) | 6 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (11.1) | 0 (0.0) | ||
| PR | 44 (13.6) | 128 (21.3) | 14 (9.4) | 16 (22.2) | 1 (2.9) | 8 (25.0) | 1 (8.3) | 1 (11.1) | 1 (11.1) | 2 (28.6) | ||
| SD | 75 (23.1) | 147 (24.4) | 23 (15.4) | 23 (31.9) | 10 (29.4) | 7 (21.9) | 4 (33.3) | 3 (33.3) | 2 (22.2) | 3 (42.9) | ||
| PD | 192 (59.3) | 303 (50.3) | 105 (70.5) | 32 (44.4) | 21 (61.8) | 16 (50.0) | 6 (50.0) | 5 (55.6) | 5 (55.6) | 2 (28.6) | ||
| NE | 12 (3.7) | 18 (3.0) | 7 (4.7) | 1 (1.4) | 2 (5.9) | 1 (3.1) | 1 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| ORR | CR + PR | 45 (13.9) | 134 (22.3) | 0.82 | 14 (9.4) | 16 (22.2) | 1 (2.9) | 8 (25.0) | 1 (8.3) | 1 (11.1) | 2 (22.2) | 2 (28.6) |
| PFS | Median (95% CI) (months) | 2.0 (2.0–2.0) | 2.1 (2.0–3.0) | <0.001 | 2.0 (2.00–2.03) | 2.1 (2.0–3.1) | 2.0 (2.0–3.0) | 3.1 (2.0–10.1) | 2.0 (2.0-NR) | 2.0 (2.0-NR) | 3.0 (2.0-NR) | 2.0 (1.0-NR) |
| OS | Median (95% CI) (months) | 12.2 (10.1–15.3) | 10.1 (8.2–11.2) | 0.06 | 9.2 (7.1–13.2) | 12.2 (9.1–22.3) | 10.1 (8.1-NR) | 22.3 (11.2-NR) | 5.0 (3.0-NR) | 11.1 (6.0-NR) | NR (34.6-NR) | 27.3 (18.3-NR) |
| The 12-month PFS rate | 11.3% (8.1–15.7) | 18.2% (15.2–21.8) | 6.4% (3.35–12.38) | 17.4% (10.19–29.57) | 5.4% (0.89–32.66) | 23.5% (11.91–46.20) | 0% NA | 22.2% (6.55–75.44) | 22.2% (6.55–75.44) | 0% NA | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jun, S.; Park, S.; Sun, J.-M.; Lee, S.-H.; Ahn, J.S.; Ahn, M.-J.; Cho, J.; Jung, H.A. Real-World Outcomes of Immunotherapy in Second- or Later-Line Non-Small Cell Lung Cancer with Actionable Genetic Alterations. Cancers 2023, 15, 5450. https://doi.org/10.3390/cancers15225450
Jun S, Park S, Sun J-M, Lee S-H, Ahn JS, Ahn M-J, Cho J, Jung HA. Real-World Outcomes of Immunotherapy in Second- or Later-Line Non-Small Cell Lung Cancer with Actionable Genetic Alterations. Cancers. 2023; 15(22):5450. https://doi.org/10.3390/cancers15225450
Chicago/Turabian StyleJun, Soojin, Sehhoon Park, Jong-Mu Sun, Se-Hoon Lee, Jin Seok Ahn, Myung-Ju Ahn, Juhee Cho, and Hyun Ae Jung. 2023. "Real-World Outcomes of Immunotherapy in Second- or Later-Line Non-Small Cell Lung Cancer with Actionable Genetic Alterations" Cancers 15, no. 22: 5450. https://doi.org/10.3390/cancers15225450
APA StyleJun, S., Park, S., Sun, J.-M., Lee, S.-H., Ahn, J. S., Ahn, M.-J., Cho, J., & Jung, H. A. (2023). Real-World Outcomes of Immunotherapy in Second- or Later-Line Non-Small Cell Lung Cancer with Actionable Genetic Alterations. Cancers, 15(22), 5450. https://doi.org/10.3390/cancers15225450






