A Radiomics-Based Machine Learning Perspective on the Parotid Gland as a Potential Surrogate Marker for HPV in Oropharyngeal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
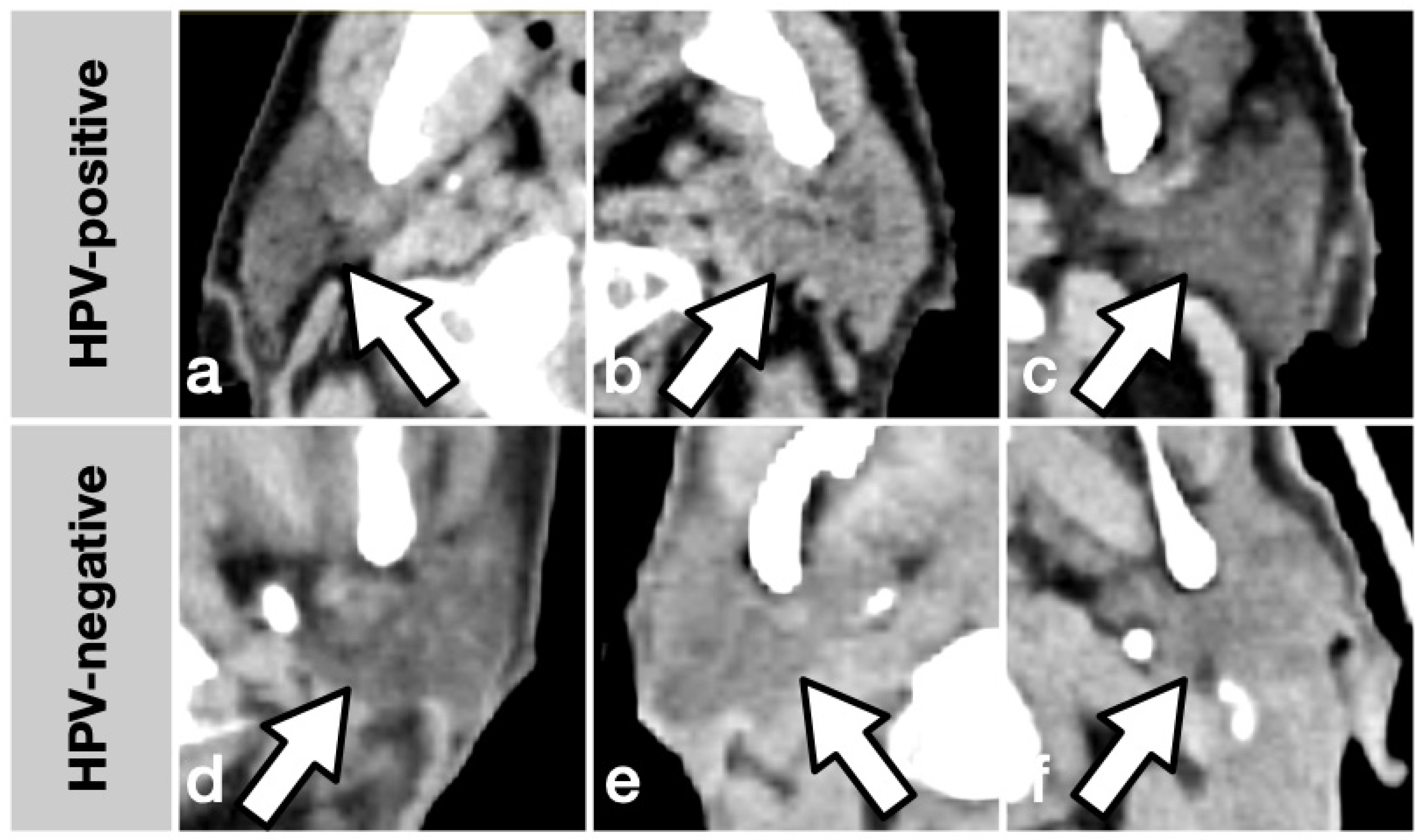
2.2. Imaging and Feature Extraction
2.3. Machine Learning Methods
2.4. Dataset Creation and Model Evaluation
3. Results
3.1. Patients and Datasets
3.2. ML Model Performances
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human Papillomavirus and Survival of Patients with Oropharyngeal Cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Semrau, R.; Duerbaum, H.; Temming, S.; Huebbers, C.; Stenner, M.; Drebber, U.; Klussmann, J.P.; Müller, R.-P.; Preuss, S.F. Prognostic Impact of Human Papillomavirus Status, Survivin, and Epidermal Growth Factor Receptor Expression on Survival in Patients Treated with Radiochemotherapy for Very Advanced Nonresectable Oropharyngeal Cancer. Head Neck 2013, 35, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human Papillomavirus and Rising Oropharyngeal Cancer Incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; D’Souza, G.; Westra, W.; Sugar, E.; Xiao, W.; Begum, S.; Viscidi, R. Distinct Risk Factor Profiles for Human Papillomavirus Type 16-Positive and Human Papillomavirus Type 16-Negative Head and Neck Cancers. J. Natl. Cancer Inst. 2008, 100, 407–420. [Google Scholar] [CrossRef]
- O’Rorke, M.A.; Ellison, M.V.; Murray, L.J.; Moran, M.; James, J.; Anderson, L.A. Human Papillomavirus Related Head and Neck Cancer Survival: A Systematic Review and Meta-Analysis. Oral Oncol. 2012, 48, 1191–1201. [Google Scholar] [CrossRef]
- Curado, M.P.; Boyle, P. Epidemiology of Head and Neck Squamous Cell Carcinoma Not Related to Tobacco or Alcohol. Curr. Opin. Oncol. 2013, 25, 229–234. [Google Scholar] [CrossRef]
- Young, D.; Xiao, C.C.; Murphy, B.; Moore, M.; Fakhry, C.; Day, T.A. Increase in Head and Neck Cancer in Younger Patients Due to Human Papillomavirus (HPV). Oral Oncol. 2015, 51, 727–730. [Google Scholar] [CrossRef]
- Touska, P.; Connor, S. Imaging of Human Papilloma Virus Associated Oropharyngeal Squamous Cell Carcinoma and Its Impact on Diagnosis, Prognostication, and Response Assessment. Br. J. Radiol. 2022, 95, 20220149. [Google Scholar] [CrossRef]
- Lambin, P.; Rios Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Non-invasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Caprini, E.; D’Agnese, G.; Brennan, P.A.; Rahimi, S. Human Papilomaviru-Related Oropharyngeal Squamous Cell Carcinoma and Radiomics: A New Era? J. Oral Pathol. Med. 2023, 52, 300–304. [Google Scholar] [CrossRef]
- Vageli, D.; Sourvinos, G.; Ioannou, M.; Koukoulis, G.K.; Spandidos, D.A. High-Risk Human Papillomavirus (HPV) in Parotid Lesions. Int. J. Biol. Markers 2007, 22, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.G.; Tong, T.; Maghami, E. Diagnosis and Management of Malignant Salivary Gland Tumors of the Parotid Gland. Otolaryngol. Clin. N. Am. 2016, 49, 343–380. [Google Scholar] [CrossRef]
- Lewis, J.S. P16 Immunohistochemistry as a Standalone Test for Risk Stratification in Oropharyngeal Squamous Cell Carcinoma. Head Neck Pathol. 2012, 6, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; van Griethuysen, J.J.M.; Parmar, C.; Aerts, H.J.W.L.; Fedorov, A.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Aucoin, N.; Narayan, V.; Pieper, S. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- GitHub—Dmlc/Xgboost. Available online: https://github.com/dmlc/xgboost/ (accessed on 7 February 2023).
- Rosenblatt, F. The Perceptron: A Probabilistic Model for Information Storage and Organization in the Brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; ISBN 9780262337373. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Smola, A.J.; Schölkopf, B. A Tutorial on Support Vector Regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Štrumbelj, E.; Kononenko, I. Explaining Prediction Models and Individual Predictions with Feature Contributions. Knowl. Inf. Syst. 2014, 41, 647–665. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems 30; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: New York, NY, USA, 2017; pp. 4765–4774. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Halligan, S.; Menu, Y.; Mallett, S. Why Did European Radiology Reject My Radiomic Biomarker Paper? How to Correctly Evaluate Imaging Biomarkers in a Clinical Setting. Eur. Radiol. 2021, 31, 9361–9368. [Google Scholar] [CrossRef]
- Ghorbani, A.; Zou, J. Data Shapley: Equitable Valuation of Data for Machine Learning. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Chaudhuri, K., Salakhutdinov, R., Eds.; PMLR: London, UK, 2019; Volume 97, pp. 2242–2251. [Google Scholar]
- Kwon, Y.; Zou, J. Beta Shapley: A Unified and Noise-Reduced Data Valuation Framework for Machine Learning. arXiv 2021, arXiv:2110.14049. [Google Scholar]
- Hamerla, G.; Meyer, H.-J.; Hambsch, P.; Wolf, U.; Kuhnt, T.; Hoffmann, K.-T.; Surov, A. Radiomics Model Based on Non-Contrast CT Shows No Predictive Power for Complete Pathological Response in Locally Advanced Rectal Cancer. Cancers 2019, 11, 1680. [Google Scholar] [CrossRef]
- Ojala, M.; Garriga, G.C. Permutation Tests for Studying Classifier Performance. J. Mach. Learn. Res. 2010, 11, 1833–1863. [Google Scholar]
- Isayeva, T.; Said-Al-Naief, N.; Ren, Z.; Li, R.; Gnepp, D.; Brandwein-Gensler, M. Salivary Mucoepidermoid Carcinoma: Demonstration of Transcriptionally Active Human Papillomavirus 16/18. Head Neck Pathol. 2013, 7, 135–148. [Google Scholar] [CrossRef]
- Teng, W.-Q.; Chen, X.-P.; Xue, X.-C.; Zhang, Y.; Tan, X.-J.; Sun, G.; Wang, Y.; Wang, L. Distribution of 37 Human Papillomavirus Types in Parotid Gland Tumor Tissues. Oncol. Lett. 2014, 7, 834–838. [Google Scholar] [CrossRef]
- Descamps, G.; Duray, A.; Rodriguez, A.; Chantrain, G.; Depuydt, C.E.; Delvenne, P.; Saussez, S. Detection and Quantification of Human Papillomavirus in Benign and Malignant Parotid Lesions. Anticancer Res. 2012, 32, 3929–3932. [Google Scholar]
- Bishop, J.A.; Yonescu, R.; Batista, D.; Yemelyanova, A.; Ha, P.K.; Westra, W.H. Mucoepidermoid Carcinoma Does Not Harbor Transcriptionally Active High Risk Human Papillomavirus Even in the Absence of the MAML2 Translocation. Head Neck Pathol. 2014, 8, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Miccai, M.D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group Matched Computed Tomography Segmentation and Demographic Data for Oropharyngeal Cancer Radiomics Challenges. Sci. Data 2017, 4, 170077. [Google Scholar] [CrossRef]
- Yu, K.; Zhang, Y.; Yu, Y.; Huang, C.; Liu, R.; Li, T.; Yang, L.; Morris, J.S.; Baladandayuthapani, V.; Zhu, H. Radiomic Analysis in Prediction of Human Papilloma Virus Status. Clin. Transl. Radiat. Oncol. 2017, 7, 49–54. [Google Scholar] [CrossRef]
- Kang, J.J.; Yu, Y.; Chen, L.; Zakeri, K.; Gelblum, D.Y.; McBride, S.M.; Riaz, N.; Tsai, C.J.; Kriplani, A.; Hung, T.K.W.; et al. Consensuses, Controversies, and Future Directions in Treatment Deintensification for Human Papillomavirus-Associated Oropharyngeal Cancer. CA Cancer J. Clin. 2022, 73, 164–197. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S., Jr. Human Papillomavirus Testing in Head and Neck Squamous Cell Carcinoma in 2020: Where Are We Now and Where Are We Going? Head Neck Pathol. 2020, 14, 321–329. [Google Scholar] [CrossRef]
- Damgacioglu, H.; Sonawane, K.; Zhu, Y.; Li, R.; Balasubramanian, B.A.; Lairson, D.R.; Giuliano, A.R.; Deshmukh, A.A. Oropharyngeal Cancer Incidence and Mortality Trends in All 50 States in the US, 2001–2017. JAMA Otolaryngol. Head Neck Surg. 2022, 148, 155–165. [Google Scholar] [CrossRef]
- Yin, P.; Mao, N.; Zhao, C.; Wu, J.; Sun, C.; Chen, L.; Hong, N. Comparison of Radiomics Machine-Learning Classifiers and Feature Selection for Differentiation of Sacral Chordoma and Sacral Giant Cell Tumour Based on 3D Computed Tomography Features. Eur. Radiol. 2019, 29, 1841–1847. [Google Scholar] [CrossRef]
- Pfister, D.G.; Ang, K.-K.; Brizel, D.M.; Burtness, B.A.; Cmelak, A.J.; Colevas, A.D.; Dunphy, F.; Eisele, D.W.; Gilbert, J.; Gillison, M.L.; et al. Head and Neck Cancers. J. Natl. Compr. Cancer Netw. 2011, 9, 596–650. [Google Scholar] [CrossRef]
- Pfister, D.G.; Spencer, S.; Adelstein, D.; Adkins, D.; Anzai, Y.; Brizel, D.M.; Bruce, J.Y.; Busse, P.M.; Caudell, J.J.; Cmelak, A.J.; et al. Head and Neck Cancers, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 873–898. [Google Scholar] [CrossRef]



| All | Train | Test | |
|---|---|---|---|
| Patients [n] | 53 | 39 | 14 |
| Mean age [years] (range) | 60.8 (41–91) | 60.1 (45–80) | 62.6 (41–91) |
| Male [%] | 90.6 | 94.9 | 78.6 |
| HPV positive [%] | 37.7 | 33.3 | 50.0 |
| T stage | |||
| T4 | 35 | 27 | 8 |
| T3 | 9 | 5 | 4 |
| T2 | 5 | 4 | 1 |
| T1 | 4 | 3 | 1 |
| N stage | |||
| N2 | 38 | 28 | 10 |
| N3 | 5 | 4 | 1 |
| N1 | 5 | 3 | 2 |
| N0 | 5 | 4 | 1 |
| M stage | |||
| M0 | 49 | 36 | 13 |
| M1 | 3 | 2 | 1 |
| Grading | |||
| G2 | 31 | 21 | 10 |
| G3 | 21 | 17 | 4 |
| G1 | 1 | 1 | 0 |
| Scanners | |||
| No. 1 | 20 | 14 | 6 |
| No. 2 | 19 | 11 | 8 |
| No. 3 | 14 | 14 | 0 |
| ML Model | LR | MLP | RF | SVC | XGB | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | AUC | p-Value | AUC | p-Value | AUC | p-Value | AUC | p-Value | AUC | p-Value |
| All original_ | 0.59 [0.50–0.75] | 0.066 | 0.61 [0.50–0.75] | 0.102 | 0.57 [0.50–0.75] | 0.185 | 0.51 [0.50–0.62] | 0.092 | 0.57 [0.50–0.69] | 0.101 |
| All original_firstorder_ | 0.98 [0.75–1.00] | 0.168 | 0.92 [0.56–1.00] | 0.129 | 0.77 [0.50–1.00] | 0.137 | 0.71 [0.50–1.00] | 0.703 | 0.86 [0.50–1.00] | 0.039 * |
| All original_shape_ | 0.72 [0.50–0.88] | 0.131 | 0.71 [0.50–0.88] | 0.048 * | 0.64 [0.50–0.88] | 0.248 | 0.51 [0.50–0.62] | 0.109 | 0.68 [0.50–0.97] | 0.066 |
| GTV LNM original_ | 0.60 [0.50–0.70] | 0.019 * | 0.63 [0.50–0.76] | 0.054 | 0.60 [0.50–0.74] | 0.025 * | 0.51 [0.50–0.56] | 0.012 * | 0.57 [0.50–0.69] | 0.007 * |
| GTV LNM original_firstorder_ | 0.76 [0.51–0.89] | 0.032 * | 0.81 [0.65–0.96] | 0.023 * | 0.67 [0.51–0.83] | 0.018 * | 0.51 [0.50–0.56] | 0.006 * | 0.60 [0.50–0.76] | 0.005 * |
| GTV LNM original_shape_ | 0.60 [0.51–0.67] | 0.005 * | 0.82 [0.56–0.98] | 0.005 * | 0.72 [0.60–0.81] | 0.009 * | 0.51 [0.50–0.56] | 0.004 * | 0.74 [0.62–0.87] | 0.003 * |
| GTV TM original_ | 0.65 [0.50–0.84] | 0.065 | 0.62 [0.50–0.79] | 0.030 * | 0.61 [0.51–0.77] | 0.007 * | 0.59 [0.50–0.74] | 0.033 * | 0.71 [0.51–0.85] | 0.008 * |
| GTV TM original_firstorder_ | 0.61 [0.51–0.73] | 0.182 | 0.61 [0.50–0.78] | 0.389 | 0.59 [0.50–0.75] | 0.149 | 0.55 [0.50–0.57] | 0.071 | 0.60 [0.50–0.77] | 0.047 * |
| GTV TM original_shape_ | 0.61 [0.50–0.73] | 0.582 | 0.60 [0.50–0.80] | 0.036 * | 0.59 [0.50–0.75] | 0.210 | 0.56 [0.50–0.70] | 0.795 | 0.61 [0.50–0.80] | 0.240 |
| Parotid original_ | 0.68 [0.56–0.76] | 0.020 * | 0.75 [0.57–0.85] | 0.004 * | 0.67 [0.59–0.77] | 0.001 * | 0.54 [0.50–0.60] | 0.002 * | 0.66 [0.51–0.80] | 0.001 * |
| Parotid original_firstorder_ | 0.63 [0.52–0.72] | 0.006 * | 0.68 [0.55–0.81] | 0.001 * | 0.72 [0.62–0.82] | 0.002 * | 0.54 [0.50–0.62] | 0.019 * | 0.66 [0.55–0.79] | 0.002 * |
| Parotid original_shape_ | 0.57 [0.51–0.74] | 0.002 * | 0.67 [0.50–0.91] | 0.008 * | 0.76 [0.55–0.87] | 0.001 * | 0.52 [0.50–0.58] | 0.025 * | 0.69 [0.51–0.85] | 0.002 * |
| Dataset | n Training (% Positive) | n Training Data Shapley (% Positive) | n Test (% Positive) | n Selected Features (n All Features) |
|---|---|---|---|---|
| All original_ | 29 (66%) | 29 (66%) | 6 (33%) | 2 (321) |
| All original_firstorder_ | 29 (66%) | 29 (66%) | 6 (33%) | 2 (60) |
| All original_shape_ | 29 (66%) | 29 (66%) | 6 (33%) | 2 (48) |
| GTV LNM original_ | 81 (72%) | 65 (89%) | 15 (40%) | 8 (107) |
| GTV LNM original_firstorder_ | 81 (72%) | 67 (87%) | 15 (40%) | 8 (20) |
| GTV LNM original_shape_ | 81 (72%) | 62 (94%) | 15 (40%) | 8 (16) |
| GTV TM original_ | 39 (64%) | 39 (64%) | 13 (62%) | 3 (107) |
| GTV TM original_firstorder_ | 39 (64%) | 39 (64%) | 13 (62%) | 3 (20) |
| GTV TM original_shape_ | 39 (64%) | 39 (64%) | 13 (62%) | 3 (16) |
| Parotid original_ | 78 (67%) | 71 (73%) | 24 (50%) | 7 (107) |
| Parotid original_firstorder_ | 78 (67%) | 73 (71%) | 24 (50%) | 7 (20) |
| Parotid original_shape_ | 78 (67%) | 66 (79%) | 24 (50%) | 7 (16) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasse, G.; Glaas, A.; Meyer, H.-J.; Zebralla, V.; Dietz, A.; Hering, K.; Kuhnt, T.; Denecke, T. A Radiomics-Based Machine Learning Perspective on the Parotid Gland as a Potential Surrogate Marker for HPV in Oropharyngeal Cancer. Cancers 2023, 15, 5425. https://doi.org/10.3390/cancers15225425
Prasse G, Glaas A, Meyer H-J, Zebralla V, Dietz A, Hering K, Kuhnt T, Denecke T. A Radiomics-Based Machine Learning Perspective on the Parotid Gland as a Potential Surrogate Marker for HPV in Oropharyngeal Cancer. Cancers. 2023; 15(22):5425. https://doi.org/10.3390/cancers15225425
Chicago/Turabian StylePrasse, Gordian, Agnes Glaas, Hans-Jonas Meyer, Veit Zebralla, Andreas Dietz, Kathrin Hering, Thomas Kuhnt, and Timm Denecke. 2023. "A Radiomics-Based Machine Learning Perspective on the Parotid Gland as a Potential Surrogate Marker for HPV in Oropharyngeal Cancer" Cancers 15, no. 22: 5425. https://doi.org/10.3390/cancers15225425
APA StylePrasse, G., Glaas, A., Meyer, H.-J., Zebralla, V., Dietz, A., Hering, K., Kuhnt, T., & Denecke, T. (2023). A Radiomics-Based Machine Learning Perspective on the Parotid Gland as a Potential Surrogate Marker for HPV in Oropharyngeal Cancer. Cancers, 15(22), 5425. https://doi.org/10.3390/cancers15225425








