Unveiling a Biomarker Signature of Meningioma: The Need for a Panel of Genomic, Epigenetic, Proteomic, and RNA Biomarkers to Advance Diagnosis and Prognosis
Abstract
Simple Summary
Abstract
1. Introduction
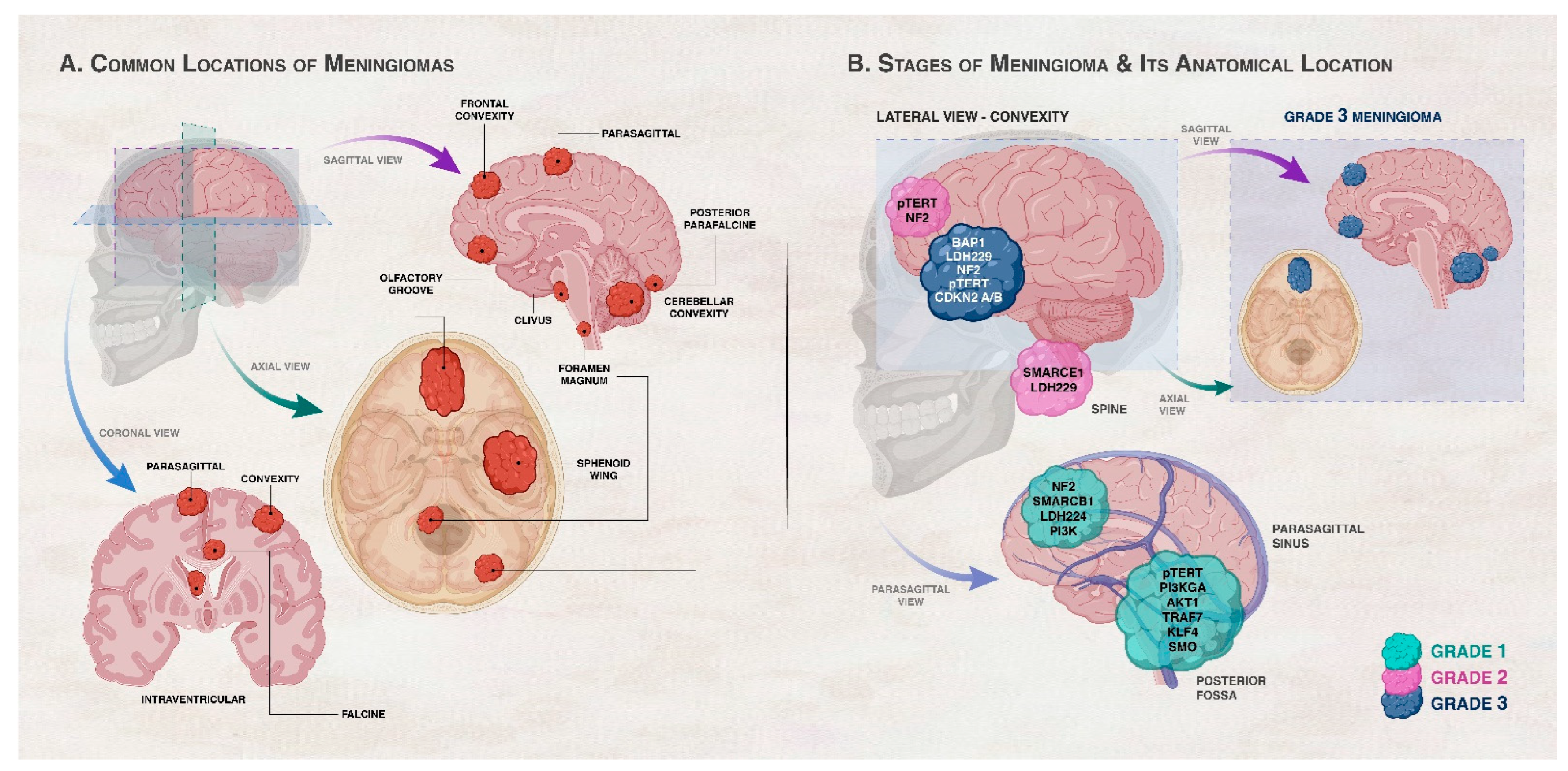
2. Grading of Meningiomas
3. Genomic Alterations and Epigenetic Modifications in Meningiomas
4. NF2/Merlin Signaling Pathways in Meningiomas
5. Biomarkers of Meningiomas
5.1. Current Diagnosis and Prognosis
5.2. The Need for a Profile of Biomarkers of Different Types
5.3. Exploring Protein Biomarkers as Meningioma Biomarkers
- Serum Protein Biomarkers
- b.
- Cerebrospinal Fluid Protein Biomarkers
5.4. LncRNA and miRNA in Diagnosis and Prognosis of Meningiomas
6. Animal Models for Discovery of Meningioma Biomarkers
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gittleman, H.R.; Ostrom, Q.T.; Rouse, C.D.; Dowling, J.A.; de Blank, P.M.; Kruchko, C.A.; Elder, J.B.; Rosenfeld, S.S.; Selman, W.R.; Sloan, A.E.; et al. Trends in central nervous system tumor incidence relative to other common cancers in adults, adolescents, and children in the United States, 2000 to 2010. Cancer 2015, 121, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Vecchione-Koval, T.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncology 2017, 19, v1–v88. [Google Scholar] [CrossRef] [PubMed]
- Flint-Richter, P.; Mandelzweig, L.; Oberman, B.; Sadetzki, S. Possible interaction between ionizing radiation, smoking, and gender in the causation of meningioma. Neuro-Oncology 2011, 13, 345–352. [Google Scholar] [CrossRef]
- Schneider, B.; Pülhorn, H.; Röhrig, B.; Rainov, N.G. Predisposing conditions and risk factors for development of symptomatic meningioma in adults. Cancer Detect. Prev. 2005, 29, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Achey, R.L.; Gittleman, H.; Schroer, J.; Khanna, V.; Kruchko, C.; Barnholtz-Sloan, J.S. Nonmalignant and malignant meningioma incidence and survival in the elderly, 2005–2015, using the Central Brain Tumor Registry of the United States. Neuro-Oncology 2019, 21, 380–391. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Yang, S.Y.; Park, C.K.; Park, S.H.; Kim, D.G.; Chung, Y.S.; Jung, H.W. Atypical and anaplastic meningiomas: Prognostic implications of clinicopathological features. J. Neurol. Neurosurg. Psychiatry 2008, 79, 574–580. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Loewenstern, J.; Rutland, J.; Gill, C.; Arib, H.; Pain, M.; Umphlett, M.; Kinoshita, Y.; McBride, R.; Donovan, M.; Sebra, R.; et al. Comparative genomic analysis of driver mutations in matched primary and recurrent meningiomas. Oncotarget 2019, 10, 3506–3517. [Google Scholar] [CrossRef]
- Dumanski, J.P.; Carlbom, E.; Collins, V.P.; Nordenskjöld, M. Deletion mapping of a locus on human chromosome 22 involved in the oncogenesis of meningioma. Proc. Natl. Acad. Sci. USA 1987, 84, 9275–9279. [Google Scholar] [CrossRef]
- Schneider, G.; Lutz, S.; Henn, W.; Zang, K.D.; Blin, N. Search for putative suppressor genes in meningioma: Significance of chromosome 22. Hum. Genet. 1992, 88, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Wellenreuther, R.; Kraus, J.A.; Lenartz, D.; Menon, A.G.; Schramm, J.; Louis, D.N.; Ramesh, V.; Gusella, J.F.; Wiestler, O.D.; von Deimling, A. Analysis of the neurofibromatosis 2 gene reveals molecular variants of meningioma. Am. J. Pathol. 1995, 146, 827–832. [Google Scholar] [PubMed]
- Gutmann, D.H.; Giordano, M.J.; Fishback, A.S.; Guha, A. Loss of merlin expression in sporadic meningiomas, ependymomas and schwannomas. Neurology 1997, 49, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Ruttledge, M.H.; Sarrazin, J.; Rangaratnam, S.; Phelan, C.M.; Twist, E.; Merel, P.; Delattre, O.; Thomas, G.; Nordenskjöld, M.; Collins, V.P.; et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat. Genet. 1994, 6, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, M.D.; Espinosa, A.B.; Maíllo, A.; Sayagués, J.M.; Alguero, M.d.C.; Lumbreras, E.; Díaz, P.; Gonçalves, J.M.; Onzain, I.; Merino, M.; et al. Characterization of chromosome 14 abnormalities by interphase in situ hybridization and comparative genomic hybridization in 124 meningiomas: Correlation with clinical, histopathologic, and prognostic features. Am. J. Clin. Pathol. 2005, 123, 744–751. [Google Scholar] [CrossRef]
- Sayagués, J.M.; Tabernero, M.D.; Maíllo, A.; Espinosa, A.; Rasillo, A.; Díaz, P.; Ciudad, J.; López, A.; Merino, M.; Gonçalves, J.M.; et al. Intratumoral Patterns of Clonal Evolution in Meningiomas as Defined by Multicolor Interphase Fluorescence in Situ Hybridization (FISH). J. Mol. Diagn. 2004, 6, 316–325. [Google Scholar] [CrossRef]
- Strickland, M.R.; Gill, C.M.; Nayyar, N.; D’Andrea, M.R.; Thiede, C.; Juratli, T.A.; Schackert, G.; Borger, D.R.; Santagata, S.; Frosch, M.P.; et al. Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J. Neurosurg. 2017, 127, 438–444. [Google Scholar] [CrossRef]
- Clark, V.E.; Harmancı, A.S.; Bai, H.; Youngblood, M.W.; Lee, T.I.; Baranoski, J.F.; Ercan-Sencicek, A.G.; Abraham, B.J.; Weintraub, A.S.; Hnisz, D.; et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat. Genet. 2016, 48, 1253–1259. [Google Scholar] [CrossRef]
- Brastianos, P.K.; Horowitz, P.M.; Santagata, S.; Jones, R.T.; McKenna, A.; Getz, G.; Ligon, K.L.; Palescandolo, E.; Van Hummelen, P.; Ducar, M.D.; et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 2013, 45, 285–289. [Google Scholar] [CrossRef]
- Sahm, F.; Bissel, J.; Koelsche, C.; Schweizer, L.; Capper, D.; Reuss, D.; Böhmer, K.; Lass, U.; Göck, T.; Kalis, K.; et al. AKT1E17K mutations cluster with meningothelial and transitional meningiomas and can be detected by SFRP1 immunohistochemistry. Acta Neuropathol. 2013, 126, 757–762. [Google Scholar] [CrossRef]
- Reuss, D.E.; Piro, R.M.; Jones, D.T.W.; Simon, M.; Ketter, R.; Kool, M.; Becker, A.; Sahm, F.; Pusch, S.; Meyer, J.; et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013, 125, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.E.; Erson-Omay, E.Z.; Serin, A.; Yin, J.; Cotney, J.; Ozduman, K.; Avsar, T.; Li, J.; Murray, P.B.; Henegariu, O.; et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 2013, 339, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Aldape, K.; Nejad, R.; Louis, D.N.; Zadeh, G. Integrating molecular markers into the World Health Organization classification of CNS tumors: A survey of the neuro-oncology community. Neuro Oncol. 2017, 19, 336–344. [Google Scholar] [CrossRef]
- Papaioannou, M.-D.; Djuric, U.; Kao, J.; Karimi, S.; Zadeh, G.; Aldape, K.; Diamandis, P. Proteomic analysis of meningiomas reveals clinically distinct molecular patterns. Neuro-Oncology 2019, 21, 1028–1038. [Google Scholar] [CrossRef]
- Nassiri, F.; Wang, J.Z.; Singh, O.; Karimi, S.; Dalcourt, T.; Ijad, N.; Pirouzmand, N.; Ng, H.K.; Saladino, A.; Pollo, B.; et al. Loss of H3K27me3 in meningiomas. Neuro Oncol. 2021, 23, 1282–1291. [Google Scholar] [CrossRef]
- Katz, L.M.; Hielscher, T.; Liechty, B.; Silverman, J.; Zagzag, D.; Sen, R.; Wu, P.; Golfinos, J.G.; Reuss, D.; Neidert, M.C.; et al. Loss of histone H3K27me3 identifies a subset of meningiomas with increased risk of recurrence. Acta Neuropathol. 2018, 135, 955–963. [Google Scholar] [CrossRef]
- Bello, M.J.; Amiñoso, C.; Lopez-Marin, I.; Arjona, D.; Gonzalez-Gomez, P.; Alonso, M.E.; Lomas, J.; de Campos, J.M.; Kusak, M.E.; Vaquero, J.; et al. DNA methylation of multiple promoter-associated CpG islands in meningiomas: Relationship with the allelic status at 1p and 22q. Acta Neuropathol. 2004, 108, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Barski, D.; Wolter, M.; Reifenberger, G.; Riemenschneider, M.J. Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain. Pathol. 2010, 20, 623–631. [Google Scholar] [CrossRef]
- Nazem, A.A.; Ruzevick, J.; Ferreira, M.J., Jr. Advances in meningioma genomics, proteomics, and epigenetics: Insights into biomarker identification and targeted therapies. Oncotarget 2020, 11, 4544–4553. [Google Scholar] [CrossRef]
- Gritsch, S.; Batchelor, T.T.; Gonzalez Castro, L.N. Diagnostic, therapeutic, and prognostic implications of the 2021 World Health Organization classification of tumors of the central nervous system. Cancer 2022, 128, 47–58. [Google Scholar] [CrossRef]
- Kshettry, V.R.; Ostrom, Q.T.; Kruchko, C.; Al-Mefty, O.; Barnett, G.H.; Barnholtz-Sloan, J.S. Descriptive epidemiology of World Health Organization grades II and III intracranial meningiomas in the United States. Neuro-Oncology 2015, 17, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Goldbrunner, R.; Minniti, G.; Preusser, M.; Jenkinson, M.D.; Sallabanda, K.; Houdart, E.; von Deimling, A.; Stavrinou, P.; Lefranc, F.; Lund-Johansen, M.; et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016, 17, e383–e391. [Google Scholar] [CrossRef] [PubMed]
- Hashiba, T.; Hashimoto, N.; Izumoto, S.; Suzuki, T.; Kagawa, N.; Maruno, M.; Kato, A.; Yoshimine, T. Serial volumetric assessment of the natural history and growth pattern of incidentally discovered meningiomas. J. Neurosurg. 2009, 110, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Buerki, R.A.; Horbinski, C.M.; Kruser, T.; Horowitz, P.M.; James, C.D.; Lukas, R.V. An overview of meningiomas. Future Oncology 2018, 14, 2161–2177. [Google Scholar] [CrossRef] [PubMed]
- Chotai, S.; Schwartz, T.H. The Simpson Grading: Is It Still Valid? Cancers 2022, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Surov, A.; Gottschling, S.; Bolz, J.; Kornhuber, M.; Alfieri, A.; Holzhausen, H.-J.; Abbas, J.; Kösling, S. Distant metastases in meningioma: An underestimated problem. J. Neuro-Oncol. 2013, 112, 323–327. [Google Scholar] [CrossRef]
- Enomoto, T.; Aoki, M.; Kouzaki, Y.; Abe, H.; Imamura, N.; Iwasaki, A.; Inoue, T.; Nabeshima, K. WHO Grade I Meningioma Metastasis to the Lung 26 Years after Initial Surgery: A Case Report and Literature Review. NMC Case Rep. J. 2019, 6, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Paix, A.; Waissi, W.; Antoni, D.; Adeduntan, R.; Noël, G. Visceral and bone metastases of a WHO grade 2 meningioma: A case report and review of the literature. Cancer/Radiothérapie 2017, 21, 55–59. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Duan, Y.; Ye, Y.; Xiao, L.; Mao, R. Subcutaneous Metastasis of Atypical Meningioma: Case Report and Literature Review. World Neurosurg. 2020, 138, 182–186. [Google Scholar] [CrossRef]
- Tsitsikov, E.N.; Hameed, S.; Tavakol, S.A.; Stephens, T.M.; Tsytsykova, A.V.; Garman, L.; Bi, W.L.; Dunn, I.F. Specific gene expression signatures of low grade meningiomas. Front. Oncol. 2023, 13, 1126550. [Google Scholar] [CrossRef]
- Harmancı, A.S.; Youngblood, M.W.; Clark, V.E.; Coşkun, S.; Henegariu, O.; Duran, D.; Erson-Omay, E.Z.; Kaulen, L.D.; Lee, T.I.; Abraham, B.J.; et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat. Commun. 2017, 8, 14433. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.J.; Wan, Y.W.; Al-Ouran, R.; Revelli, J.P.; Cardenas, M.F.; Oneissi, M.; Xi, L.; Jalali, A.; Magnotti, J.F.; Muzny, D.M.; et al. Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc. Natl. Acad. Sci. USA 2019, 116, 21715–21726. [Google Scholar] [CrossRef]
- Nassiri, F.; Liu, J.; Patil, V.; Mamatjan, Y.; Wang, J.Z.; Hugh-White, R.; Macklin, A.M.; Khan, S.; Singh, O.; Karimi, S.; et al. A clinically applicable integrative molecular classification of meningiomas. Nature 2021, 597, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Bayley, J.C.t.; Hadley, C.C.; Harmanci, A.O.; Harmanci, A.S.; Klisch, T.J.; Patel, A.J. Multiple approaches converge on three biological subtypes of meningioma and extract new insights from published studies. Sci. Adv. 2022, 8, eabm6247. [Google Scholar] [CrossRef] [PubMed]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classification of Central Nervous System tumours: A practical update on what neurosurgeons need to know-a minireview. Acta Neurochir. 2022, 164, 2453–2464. [Google Scholar] [CrossRef]
- Marastoni, E.; Barresi, V. Meningioma Grading beyond Histopathology: Relevance of Epigenetic and Genetic Features to Predict Clinical Outcome. Cancers 2023, 15, 2945. [Google Scholar] [CrossRef] [PubMed]
- Pawloski, J.A.; Fadel, H.A.; Huang, Y.W.; Lee, I.Y. Genomic Biomarkers of Meningioma: A Focused Review. Int. J. Mol. Sci. 2021, 22, 10222. [Google Scholar] [CrossRef]
- Ruttledge, M.H.; Xie, Y.G.; Han, F.Y.; Giovannini, M.; Janson, M.; Fransson, I.; Werelius, B.; Delattre, O.; Thomas, G.; Evans, G.; et al. Physical mapping of the NF2/meningioma region on human chromosome 22q12. Genomics 1994, 19, 52–59. [Google Scholar] [CrossRef]
- Ichimura, K.; Yuasa, Y. Molecular biological analysis of neurofibromatosis type 2 gene. Nihon. Rinsho 1993, 51, 2462–2466. [Google Scholar]
- Cui, Y.; Ma, L.; Schacke, S.; Yin, J.C.; Hsueh, Y.P.; Jin, H.; Morrison, H. Merlin cooperates with neurofibromin and Spred1 to suppress the Ras-Erk pathway. Hum. Mol. Genet. 2021, 29, 3793–3806. [Google Scholar] [CrossRef]
- Maggio, I.; Franceschi, E.; Tosoni, A.; Nunno, V.D.; Gatto, L.; Lodi, R.; Brandes, A.A. Meningioma: Not always a benign tumor. A review of advances in the treatment of meningiomas. CNS Oncol. 2021, 10, Cns72. [Google Scholar] [CrossRef] [PubMed]
- Moussalem, C.; Massaad, E.; Minassian, G.B.; Ftouni, L.; Bsat, S.; Houshiemy, M.N.E.; Alomari, S.; Sarieddine, R.; Kobeissy, F.; Omeis, I. Meningioma genomics: A therapeutic challenge for clinicians. J. Integr. Neurosci. 2021, 20, 463–469. [Google Scholar] [CrossRef]
- Deng, J.; Hua, L.; Han, T.; Tian, M.; Wang, D.; Tang, H.; Sun, S.; Chen, H.; Cheng, H.; Zhang, T.; et al. The CREB-binding protein inhibitor ICG-001: A promising therapeutic strategy in sporadic meningioma with NF2 mutations. Neurooncol. Adv. 2020, 2, vdz055. [Google Scholar] [CrossRef]
- Youngblood, M.W.; Miyagishima, D.F.; Jin, L.; Gupte, T.; Li, C.; Duran, D.; Montejo, J.D.; Zhao, A.; Sheth, A.; Tyrtova, E.; et al. Associations of meningioma molecular subgroup and tumor recurrence. Neuro Oncol. 2021, 23, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Abedalthagafi, M.; Bi, W.L.; Aizer, A.A.; Merrill, P.H.; Brewster, R.; Agarwalla, P.K.; Listewnik, M.L.; Dias-Santagata, D.; Thorner, A.R.; Van Hummelen, P.; et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016, 18, 649–655. [Google Scholar] [CrossRef]
- Bi, W.L.; Zhang, M.; Wu, W.W.; Mei, Y.; Dunn, I.F. Meningioma Genomics: Diagnostic, Prognostic, and Therapeutic Applications. Front. Surg. 2016, 3, 40. [Google Scholar] [CrossRef]
- Zotti, T.; Scudiero, I.; Vito, P.; Stilo, R. The Emerging Role of TRAF7 in Tumor Development. J. Cell Physiol. 2017, 232, 1233–1238. [Google Scholar] [CrossRef]
- Zhou, M.; Yuan, J.; Deng, Y.; Fan, X.; Shen, J. Emerging role of SWI/SNF complex deficiency as a target of immune checkpoint blockade in human cancers. Oncogenesis 2021, 10, 3. [Google Scholar] [CrossRef]
- Ross, J.P.; Rand, K.N.; Molloy, P.L. Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2010, 2, 245–269. [Google Scholar] [CrossRef] [PubMed]
- Robert, S.M.; Vetsa, S.; Nadar, A.; Vasandani, S.; Youngblood, M.W.; Gorelick, E.; Jin, L.; Marianayagam, N.; Erson-Omay, E.Z.; Günel, M.; et al. The integrated multiomic diagnosis of sporadic meningiomas: A review of its clinical implications. J. Neurooncol. 2022, 156, 205–214. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, K. Pan-cancer analysis of frequent DNA co-methylation patterns reveals consistent epigenetic landscape changes in multiple cancers. BMC Genom. 2017, 18, 1045. [Google Scholar] [CrossRef]
- Nassiri, F.; Mamatjan, Y.; Suppiah, S.; Badhiwala, J.H.; Mansouri, S.; Karimi, S.; Saarela, O.; Poisson, L.; Gepfner-Tuma, I.; Schittenhelm, J.; et al. DNA methylation profiling to predict recurrence risk in meningioma: Development and validation of a nomogram to optimize clinical management. Neuro Oncol. 2019, 21, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Sharma, R.; Shukla, N.; Narwal, P.; Katiyar, A.; Mahajan, S.; Sahu, S.; Garg, A.; Sharma, M.C.; Suri, A.; et al. DNA methylation profiling of meningiomas highlights clinically distinct molecular subgroups. J. Neurooncol. 2023, 161, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Magill, S.T.; Eaton, C.D.; Prager, B.C.; Chen, W.C.; Cady, M.A.; Seo, K.; Lucas, C.G.; Casey-Clyde, T.J.; Vasudevan, H.N.; et al. Meningioma DNA methylation groups identify biological drivers and therapeutic vulnerabilities. Nat. Genet. 2022, 54, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Abbritti, R.V.; Polito, F.; Cucinotta, M.; Lo Giudice, C.; Caffo, M.; Tomasello, C.; Germanò, A.; Aguennouz, M. Meningiomas and Proteomics: Focus on New Potential Biomarkers and Molecular Pathways. Cancer Genom. Proteom. 2016, 13, 369–379. [Google Scholar]
- Sievers, P.; Hielscher, T.; Schrimpf, D.; Stichel, D.; Reuss, D.E.; Berghoff, A.S.; Neidert, M.C.; Wirsching, H.G.; Mawrin, C.; Ketter, R.; et al. CDKN2A/B homozygous deletion is associated with early recurrence in meningiomas. Acta Neuropathol. 2020, 140, 409–413. [Google Scholar] [CrossRef]
- Zhi, F.; Zhou, G.; Wang, S.; Shi, Y.; Peng, Y.; Shao, N.; Guan, W.; Qu, H.; Zhang, Y.; Wang, Q.; et al. A microRNA expression signature predicts meningioma recurrence. Int. J. Cancer 2013, 132, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Shao, N.; Li, B.; Xue, L.; Deng, D.; Xu, Y.; Lan, Q.; Peng, Y.; Yang, Y. A serum 6-miRNA panel as a novel non-invasive biomarker for meningioma. Sci. Rep. 2016, 6, 32067. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, G.; Huang, H.; Li, H.; Lin, S.; Wang, Y. Differentially Expressed MicroRNAs in Radioresistant and Radiosensitive Atypical Meningioma: A Clinical Study in Chinese Patients. Front. Oncol. 2020, 10, 501. [Google Scholar] [CrossRef]
- Carneiro, V.; Cirino, M.; Panepucci, R.; Peria, F.; Tirapelli, D.; Colli, B.; Carlotti, C.G., Jr. The Role of MicroRNA 181d as a Possible Biomarker Associated With Tumor Progression in Meningiomas. Cureus 2021, 13, e19158. [Google Scholar] [CrossRef]
- Ding, C.; Yi, X.; Xu, J.; Huang, Z.; Bu, X.; Wang, D.; Ge, H.; Zhang, G.; Gu, J.; Kang, D.; et al. Long Non-Coding RNA MEG3 Modifies Cell-Cycle, Migration, Invasion, and Proliferation Through AKAP12 by Sponging miR-29c in Meningioma Cells. Front. Oncol. 2020, 10, 537763. [Google Scholar] [CrossRef] [PubMed]
- Slavik, H.; Balik, V.; Kokas, F.Z.; Slavkovsky, R.; Vrbkova, J.; Rehulkova, A.; Lausova, T.; Ehrmann, J.; Gurska, S.; Uberall, I.; et al. Transcriptomic Profiling Revealed Lnc-GOLGA6A-1 as a Novel Prognostic Biomarker of Meningioma Recurrence. Neurosurgery 2022, 91, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, S.; Li, Q.; Ma, Y.; Sun, P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed. Pharmacother. 2018, 105, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Eraky, A.M. Non-coding RNAs as Genetic Biomarkers for the Diagnosis, Prognosis, Radiosensitivity, and Histopathologic Grade of Meningioma. Cureus 2023, 15, e34593. [Google Scholar] [CrossRef] [PubMed]
- Murnyák, B.; Bognár, L.; Klekner, Á.; Hortobágyi, T. Epigenetics of Meningiomas. Biomed. Res. Int. 2015, 2015, 532451. [Google Scholar] [CrossRef]
- Shen, L.; Lin, D.; Cheng, L.; Tu, S.; Wu, H.; Xu, W.; Pan, Y.; Wang, X.; Zhang, J.; Shao, A. Is DNA Methylation a Ray of Sunshine in Predicting Meningioma Prognosis? Front. Oncol. 2020, 10, 1323. [Google Scholar] [CrossRef]
- Li, Z.; Gao, Y.; Zhang, J.; Han, L.; Zhao, H. DNA methylation meningioma biomarkers: Attributes and limitations. Front. Mol. Neurosci. 2023, 16, 1182759. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Hielscher, T.; Ricken, G.; Furtner, J.; Schrimpf, D.; Widhalm, G.; Rajky, U.; Marosi, C.; Hainfellner, J.A.; von Deimling, A.; et al. Prognostic impact of genetic alterations and methylation classes in meningioma. Brain. Pathol. 2022, 32, e12970. [Google Scholar] [CrossRef]
- Sahm, F.; Schrimpf, D.; Olar, A.; Koelsche, C.; Reuss, D.; Bissel, J.; Kratz, A.; Capper, D.; Schefzyk, S.; Hielscher, T.; et al. TERT Promoter Mutations and Risk of Recurrence in Meningioma. J. Natl. Cancer Inst. 2016, 108, djv377. [Google Scholar] [CrossRef]
- Mirian, C.; Duun-Henriksen, A.K.; Juratli, T.; Sahm, F.; Spiegl-Kreinecker, S.; Peyre, M.; Biczok, A.; Tonn, J.C.; Goutagny, S.; Bertero, L.; et al. Poor prognosis associated with TERT gene alterations in meningioma is independent of the WHO classification: An individual patient data meta-analysis. J. Neurol. Neurosurg Psychiatry 2020, 91, 378–387. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.K.; Yoo, Y.C.; Park, N.H.; Park, D.B.; Yoo, J.S.; An, H.J.; Park, Y.M.; Cho, K.G. Proteome analysis of human cerebrospinal fluid as a diagnostic biomarker in patients with meningioma. Med. Sci. Monit. 2012, 18, Br450–Br460. [Google Scholar] [CrossRef]
- Erkan, E.P.; Ströbel, T.; Dorfer, C.; Sonntagbauer, M.; Weinhäusel, A.; Saydam, N.; Saydam, O. Circulating Tumor Biomarkers in Meningiomas Reveal a Signature of Equilibrium Between Tumor Growth and Immune Modulation. Front. Oncol. 2019, 9, 1031. [Google Scholar] [CrossRef]
- Jia, L.; Liu, C.; Xin, Y.; Zhang, A.; Zhou, Y.; Dong, D.; Ren, L. Evaluating EFEMP1 in Cerebrospinal Fluid and Serum as a Potential Diagnosis Biomarker for Meningiomas. Clin. Lab. 2017, 63, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Anis, S.E.; Lotfalla, M.; Zain, M.; Kamel, N.N.; Soliman, A.A. Value of SSTR2A and Claudin—1 in Differentiating Meningioma from Schwannoma and Hemangiopericytoma. Open Access Maced J. Med. Sci. 2018, 6, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Baia, G.S.; Smith, J.S.; McDermott, M.W.; Bollen, A.W.; VandenBerg, S.R.; Lamborn, K.R.; Lal, A. Expression of the Hypoxia Marker Carbonic Anhydrase 9 Is Associated with Anaplastic Phenotypes in Meningiomas. Clin. Cancer Res. 2007, 13, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Ijare, O.B.; Hambarde, S.; Brasil da Costa, F.H.; Lopez, S.; Sharpe, M.A.; Helekar, S.A.; Hangel, G.; Bogner, W.; Widhalm, G.; Bachoo, R.M.; et al. Glutamine anaplerosis is required for amino acid biosynthesis in human meningiomas. Neuro Oncol. 2022, 24, 556–568. [Google Scholar] [CrossRef]
- Masalha, W.; Daka, K.; Woerner, J.; Pompe, N.; Weber, S.; Delev, D.; Krüger, M.T.; Schnell, O.; Beck, J.; Heiland, D.H.; et al. Metabolic alterations in meningioma reflect the clinical course. BMC Cancer 2021, 21, 211. [Google Scholar] [CrossRef]
- Safari Yazd, H.; Bazargani, S.F.; Fitzpatrick, G.; Yost, R.A.; Kresak, J.; Garrett, T.J. Metabolomic and Lipidomic Characterization of Meningioma Grades Using LC-HRMS and Machine Learning. J. Am. Soc. Mass. Spectrom. 2023, 34, 10. [Google Scholar] [CrossRef]
- Bender, L.; Somme, F.; Ruhland, E.; Cicek, A.E.; Bund, C.; Namer, I.J. Metabolomic Profile of Aggressive Meningiomas by Using High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance. J. Proteome Res. 2020, 19, 292–299. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Stichel, D.; Hielscher, T.; Sievers, P.; Berghoff, A.S.; Schrimpf, D.; Sill, M.; Euskirchen, P.; Blume, C.; Patel, A.; et al. Integrated Molecular-Morphologic Meningioma Classification: A Multicenter Retrospective Analysis, Retrospectively and Prospectively Validated. J. Clin. Oncol. 2021, 39, 3839–3852. [Google Scholar] [CrossRef]
- Driver, J.; Hoffman, S.E.; Tavakol, S.; Woodward, E.; Maury, E.A.; Bhave, V.; Greenwald, N.F.; Nassiri, F.; Aldape, K.; Zadeh, G.; et al. A molecularly integrated grade for meningioma. Neuro Oncol. 2022, 24, 796–808. [Google Scholar] [CrossRef]
- Jiang, C.; Song, T.; Li, J.; Ao, F.; Gong, X.; Lu, Y.; Zhang, C.; Chen, L.; Liu, Y.; He, H.; et al. RAS Promotes Proliferation and Resistances to Apoptosis in Meningioma. Mol. Neurobiol. 2017, 54, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Pecina-Slaus, N. Merlin, the NF2 gene product. Pathol. Oncol. Res. 2013, 19, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Mawrin, C.; Sasse, T.; Kirches, E.; Kropf, S.; Schneider, T.; Grimm, C.; Pambor, C.; Vorwerk, C.K.; Firsching, R.; Lendeckel, U.; et al. Different activation of mitogen-activated protein kinase and Akt signaling is associated with aggressive phenotype of human meningiomas. Clin. Cancer Res. 2005, 11, 4074–4082. [Google Scholar] [CrossRef]
- Mougel, G.M.; Appay, R.; Querdray, A.; Roche, C.; Jijon, A.; Konstantinova, J.; Soude, A.; Graillon, T.; Barlier, A. A Hippo signaling pathway is strongly involved in meningioma tumorigenesis. Neuro-Oncology 2022, 24, ii95. [Google Scholar] [CrossRef]
- Hong, A.W.; Meng, Z.; Plouffe, S.W.; Lin, Z.; Zhang, M.; Guan, K.L. Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes Dev. 2020, 34, 511–525. [Google Scholar] [CrossRef]
- Baia, G.S.; Caballero, O.L.; Riggins, G.J. The Hippo signaling pathway and translational opportunities for brain cancers. CNS Oncol. 2012, 1, 113–115. [Google Scholar] [CrossRef]
- Sievers, P.; Chiang, J.; Schrimpf, D.; Stichel, D.; Paramasivam, N.; Sill, M.; Gayden, T.; Casalini, B.; Reuss, D.E.; Dalton, J.; et al. YAP1-fusions in pediatric NF2-wildtype meningioma. Acta Neuropathol. 2020, 139, 215–218. [Google Scholar] [CrossRef]
- Laraba, L.; Hillson, L.; de Guibert, J.G.; Hewitt, A.; Jaques, M.R.; Tang, T.T.; Post, L.; Ercolano, E.; Rai, G.; Yang, S.M.; et al. Inhibition of YAP/TAZ-driven TEAD activity prevents growth of NF2-null schwannoma and meningioma. Brain 2022, 146, 1697–1713. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Rong, R.; Tang, X.; Gutmann, D.H.; Ye, K. Neurofibromatosis 2 (NF2) tumor suppressor merlin inhibits phosphatidylinositol 3-kinase through binding to PIKE-L. Proc. Natl. Acad. Sci. USA 2004, 101, 18200–18205. [Google Scholar] [CrossRef]
- Lee, S.; Karas, P.J.; Hadley, C.C.; Bayley, V.J.; Khan, A.B.; Jalali, A.; Sweeney, A.D.; Klisch, T.J.; Patel, A.J. The Role of Merlin/NF2 Loss in Meningioma Biology. Cancers 2019, 11, 1633. [Google Scholar] [CrossRef]
- Barresi, V.; Lionti, S.; La Rocca, L.; Caliri, S.; Caffo, M. High p-mTOR expression is associated with recurrence and shorter disease-free survival in atypical meningiomas. Neuropathology 2019, 39, 22–29. [Google Scholar] [CrossRef] [PubMed]
- James, M.F.; Han, S.; Polizzano, C.; Plotkin, S.R.; Manning, B.D.; Stemmer-Rachamimov, A.O.; Gusella, J.F.; Ramesh, V. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol. Cell Biol. 2009, 29, 4250–4261. [Google Scholar] [CrossRef] [PubMed]
- Shih, K.C.; Chowdhary, S.; Rosenblatt, P.; Weir, A.B., 3rd; Shepard, G.C.; Williams, J.T.; Shastry, M.; Burris, H.A., 3rd; Hainsworth, J.D. A phase II trial of bevacizumab and everolimus as treatment for patients with refractory, progressive intracranial meningioma. J. Neurooncol. 2016, 129, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.R.; Kumthekar, P.; Wen, P.Y.; Barker, F.G.; Stemmer-Rachamimov, A.; Beauchamp, R.L.; Jordan, J.T.; Muzikansky, A.; Ramesh, V. Multi-center, single arm phase II study of the dual mTORC1/mTORC2 inhibitor vistusertib for patients with recurrent or progressive grade II-III meningiomas. J. Clin. Oncol. 2021, 39, 2024. [Google Scholar] [CrossRef]
- Pecina-Slaus, N.; Kafka, A.; Lechpammer, M. Molecular Genetics of Intracranial Meningiomas with Emphasis on Canonical Wnt Signalling. Cancers 2016, 8, 67. [Google Scholar] [CrossRef]
- Lau, Y.K.; Murray, L.B.; Houshmandi, S.S.; Xu, Y.; Gutmann, D.H.; Yu, Q. Merlin is a potent inhibitor of glioma growth. Cancer Res. 2008, 68, 5733–5742. [Google Scholar] [CrossRef]
- Spille, D.C.; Sporns, P.B.; Heß, K.; Stummer, W.; Brokinkel, B. Prediction of High-Grade Histology and Recurrence in Meningiomas Using Routine Preoperative Magnetic Resonance Imaging: A Systematic Review. World Neurosurg. 2019, 128, 174–181. [Google Scholar] [CrossRef]
- Nowosielski, M.; Galldiks, N.; Iglseder, S.; Kickingereder, P.; von Deimling, A.; Bendszus, M.; Wick, W.; Sahm, F. Diagnostic challenges in meningioma. Neuro-Oncology 2017, 19, 1588–1598. [Google Scholar] [CrossRef]
- Tagle, P.; Villanueva, P.; Torrealba, G.; Huete, I. Intracranial metastasis or meningioma? Surg. Neurol. 2002, 58, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Bendszus, M.; Warmuth-Metz, M.; Burger, R.; Klein, R.; Tonn, J.C.; Solymosi, L. Diagnosing dural metastases: The value of 1 H magnetic resonance spectroscopy. Neuroradiology 2001, 43, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Bourekas, E.C.; Wildenhain, P.; Lewin, J.S.; Tarr, R.W.; Dastur, K.J.; Raji, M.R.; Lanzieri, C.F. The dural tail sign revisited. AJNR Am. J. Neuroradiol. 1995, 16, 1514–1516. [Google Scholar] [PubMed]
- Wilms, G.; Lammens, M.; Marchal, G.; Demaerel, P.; Verplancke, J.; Van Calenbergh, F.; Goffin, J.; Plets, C.; Baert, A.L. Prominent dural enhancement adjacent to nonmeningiomatous malignant lesions on contrast-enhanced MR images. AJNR Am. J. Neuroradiol. 1991, 12, 761–764. [Google Scholar]
- Huttner, H.B.; Bergmann, O.; Salehpour, M.; El Cheikh, R.; Nakamura, M.; Tortora, A.; Heinke, P.; Coras, R.; Englund, E.; Eyüpoglu, I.Y.; et al. Meningioma growth dynamics assessed by radiocarbon retrospective birth dating. EBioMedicine 2018, 27, 176–181. [Google Scholar] [CrossRef]
- Narai, A.; Hermann, P.; Auer, T.; Kemenczky, P.; Szalma, J.; Homolya, I.; Somogyi, E.; Vakli, P.; Weiss, B.; Vidnyanszky, Z. Movement-related artefacts (MR-ART) dataset of matched motion-corrupted and clean structural MRI brain scans. Sci. Data 2022, 9, 630. [Google Scholar] [CrossRef]
- Usman, M.; Latif, S.; Asim, M.; Lee, B.-D.; Qadir, J. Retrospective Motion Correction in Multishot MRI using Generative Adversarial Network. Sci. Rep. 2020, 10, 4786. [Google Scholar] [CrossRef]
- Harter, P.N.; Braun, Y.; Plate, K.H. Classification of meningiomas—Advances and controversies. Chin. Clin. Oncol. 2017, 6, S2. [Google Scholar] [CrossRef]
- Rogers, C.L.; Perry, A.; Pugh, S.; Vogelbaum, M.A.; Brachman, D.; McMillan, W.; Jenrette, J.; Barani, I.; Shrieve, D.; Sloan, A.; et al. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOG Trial 0539. Neuro-Oncology 2016, 18, 565–574. [Google Scholar] [CrossRef]
- Zhang, Q.; Jia, G.-J.; Zhang, G.-B.; Wang, L.; Wu, Z.; Jia, W.; Hao, S.-Y.; Ni, M.; Li, D.; Wang, K.; et al. A Logistic Regression Model for Detecting the Presence of Malignant Progression in Atypical Meningiomas. World Neurosurg. 2019, 126, e392–e401. [Google Scholar] [CrossRef]
- Parada, C.A.; Osbun, J.W.; Busald, T.; Karasozen, Y.; Kaur, S.; Shi, M.; Barber, J.; Adidharma, W.; Cimino, P.J.; Pan, C.; et al. Phosphoproteomic and Kinomic Signature of Clinically Aggressive Grade I (1.5) Meningiomas Reveals RB1 Signaling as a Novel Mediator and Biomarker. Clin. Cancer Res. 2020, 26, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Hua, L.; Bian, L.; Chen, H.; Chen, L.; Cheng, H.; Dou, C.; Geng, D.; Hong, T.; Ji, H.; et al. Molecular diagnosis and treatment of meningiomas: An expert consensus (2022). Chin. Med. J. 2022, 135, 1894–1912. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashed, M.; Foshay, K.; Abedalthagafi, M. Recent Advances in Meningioma Immunogenetics. Front. Oncol. 2019, 9, 1472. [Google Scholar] [CrossRef] [PubMed]
- Sofela, A.A.; McGavin, L.; Whitfield, P.C.; Hanemann, C.O. Biomarkers for differentiating grade II meningiomas from grade I: A systematic review. Br. J. Neurosurg. 2021, 35, 696–702. [Google Scholar] [CrossRef]
- Raad, M.; El Tal, T.; Gul, R.; Mondello, S.; Zhang, Z.; Boustany, R.M.; Guingab, J.; Wang, K.K.; Kobeissy, F. Neuroproteomics approach and neurosystems biology analysis: ROCK inhibitors as promising therapeutic targets in neurodegeneration and neurotrauma. Electrophoresis 2012, 33, 3659–3668. [Google Scholar] [CrossRef] [PubMed]
- Ottens, A.K.; Kobeissy, F.H.; Fuller, B.F.; Liu, M.C.; Oli, M.W.; Hayes, R.L.; Wang, K.K. Novel neuroproteomic approaches to studying traumatic brain injury. Prog. Brain. Res. 2007, 161, 401–418. [Google Scholar] [CrossRef]
- Kishida, Y.; Natsume, A.; Kondo, Y.; Takeuchi, I.; An, B.; Okamoto, Y.; Shinjo, K.; Saito, K.; Ando, H.; Ohka, F.; et al. Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis 2012, 33, 436–441. [Google Scholar] [CrossRef]
- Olar, A.; Wani, K.M.; Wilson, C.D.; Zadeh, G.; DeMonte, F.; Jones, D.T.; Pfister, S.M.; Sulman, E.P.; Aldape, K.D. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017, 133, 431–444. [Google Scholar] [CrossRef]
- Sahm, F.; Schrimpf, D.; Stichel, D.; Jones, D.T.W.; Hielscher, T.; Schefzyk, S.; Okonechnikov, K.; Koelsche, C.; Reuss, D.E.; Capper, D.; et al. DNA methylation-based classification and grading system for meningioma: A multicentre, retrospective analysis. Lancet. Oncol. 2017, 18, 682–694. [Google Scholar] [CrossRef]
- Liu, J.; Xia, C.; Wang, G. Multi-Omics Analysis in Initiation and Progression of Meningiomas: From Pathogenesis to Diagnosis. Front. Oncol. 2020, 10, 1491. [Google Scholar] [CrossRef]
- Agnihotri, S.; Suppiah, S.; Tonge, P.D.; Jalali, S.; Danesh, A.; Bruce, J.P.; Mamatjan, Y.; Klironomos, G.; Gonen, L.; Au, K.; et al. Therapeutic radiation for childhood cancer drives structural aberrations of NF2 in meningiomas. Nat. Commun. 2017, 8, 186. [Google Scholar] [CrossRef]
- Liu, F.; Qian, J.; Ma, C. MPscore: A Novel Predictive and Prognostic Scoring for Progressive Meningioma. Cancers 2021, 13, 1113. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Bowman-Kirigin, J.A.; Desai, R.; Kang, L.-I.; Patel, P.R.; Patel, B.; Khan, S.M.; Bender, D.; Marlin, M.C.; Liu, J.; et al. Single-cell profiling of human dura and meningioma reveals cellular meningeal landscape and insights into meningioma immune response. Genome Med. 2022, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Shi, J. Machine learning and bioinformatics approaches for classification and clinical detection of bevacizumab responsive glioblastoma subtypes based on miRNA expression. Sci. Rep. 2022, 12, 8685. [Google Scholar] [CrossRef] [PubMed]
- Kitano, Y.; Aoki, K.; Ohka, F.; Yamazaki, S.; Motomura, K.; Tanahashi, K.; Hirano, M.; Naganawa, T.; Iida, M.; Shiraki, Y.; et al. Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds. ACS Appl. Mater. Interfaces 2021, 13, 17316–17329. [Google Scholar] [CrossRef] [PubMed]
- Bhawal, R.; Oberg, A.L.; Zhang, S.; Kohli, M. Challenges and Opportunities in Clinical Applications of Blood-Based Proteomics in Cancer. Cancers 2020, 12, 2428. [Google Scholar] [CrossRef] [PubMed]
- Gomes Marin, J.F.; Nunes, R.F.; Coutinho, A.M.; Zaniboni, E.C.; Costa, L.B.; Barbosa, F.G.; Queiroz, M.A.; Cerri, G.G.; Buchpiguel, C.A. Theranostics in Nuclear Medicine: Emerging and Re-emerging Integrated Imaging and Therapies in the Era of Precision Oncology. Radiographics 2020, 40, 1715–1740. [Google Scholar] [CrossRef]
- Salgues, B.; Graillon, T.; Horowitz, T.; Chinot, O.; Padovani, L.; Taïeb, D.; Guedj, E. Somatostatin Receptor Theranostics for Refractory Meningiomas. Curr. Oncol. 2022, 29, 5550–5565. [Google Scholar] [CrossRef]
- Tubre, T.; Hacking, S.; Alexander, A.; Brickman, A.; Delalle, I.; Elinzano, H.; Donahue, J.E. Prostate-Specific Membrane Antigen Expression in Meningioma: A Promising Theranostic Target. J. Neuropathol. Exp. Neurol. 2022. [Google Scholar] [CrossRef]
- Zhang, D.; Ye, S.; Pan, T. The role of serum and urinary biomarkers in the diagnosis of early diabetic nephropathy in patients with type 2 diabetes. PeerJ 2019, 7, e7079. [Google Scholar] [CrossRef]
- Konstantinidou, A.E.; Givalos, N.; Gakiopoulou, H.; Korkolopoulou, P.; Kotsiakis, X.; Boviatsis, E.; Agrogiannis, G.; Mahera, H.; Patsouris, E. Caspase-3 immunohistochemical expression is a marker of apoptosis, increased grade and early recurrence in intracranial meningiomas. Apoptosis 2007, 12, 695–705. [Google Scholar] [CrossRef]
- Ghantasala, S.; Pai, M.G.J.; Biswas, D.; Gahoi, N.; Mukherjee, S.; Kp, M.; Nissa, M.U.; Srivastava, A.; Epari, S.; Shetty, P.; et al. Multiple Reaction Monitoring-Based Targeted Assays for the Validation of Protein Biomarkers in Brain Tumors. Front. Oncol. 2021, 11, 548243. [Google Scholar] [CrossRef]
- Abou Khouzam, R.; Brodaczewska, K.; Filipiak, A.; Zeinelabdin, N.A.; Buart, S.; Szczylik, C.; Kieda, C.; Chouaib, S. Tumor Hypoxia Regulates Immune Escape/Invasion: Influence on Angiogenesis and Potential Impact of Hypoxic Biomarkers on Cancer Therapies. Front. Immunol. 2020, 11, 613114. [Google Scholar] [CrossRef]
- El-Benhawy, S.A.; Sakr, O.A.; Fahmy, E.I.; Ali, R.A.; Hussein, M.S.; Nassar, E.M.; Salem, S.M.; Abu-Samra, N.; Elzawawy, S. Assessment of Serum Hypoxia Biomarkers Pre- and Post-radiotherapy in Patients with Brain Tumors. J. Mol. Neurosci. 2022, 81, 1008–1017. [Google Scholar] [CrossRef]
- Valvona, C.J.; Fillmore, H.L.; Nunn, P.B.; Pilkington, G.J. The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor. Brain Pathol. 2016, 26, 3–17. [Google Scholar] [CrossRef]
- Fattahi, M.J.; Sedaghat, F.; Malekzadeh, M.; Nejat, A.A.; Poostkar, M.; Saberi, Y.; Taghipour, M.; Ghaderi, A. Endocan serum levels in patients with low- and high-grade meningiomas: Does this biomarker have an indicative role? Egypt. J. Neurol. Psychiatry Neurosurg. 2021, 57, 92. [Google Scholar] [CrossRef]
- Atukeren, P.; Kunbaz, A.; Turk, O.; Kemerdere, R.; Ulu, M.O.; Turkmen Inanir, N.; Tanriverdi, T. Expressions of Endocan in Patients with Meningiomas and Gliomas. Dis. Markers 2016, 2016, 7157039. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, T.; Achini, F.; Grotzer, M.A. Targeting cerebrospinal fluid for discovery of brain cancer biomarkers. J. Cancer Metastasis Treat. 2016, 2, 176–187. [Google Scholar] [CrossRef]
- Choudhary, R.; Elabbas, A.; Vyas, A.; Osborne, D.; Chigurupati, H.D.; Abbas, L.F.; Kampa, P.; Farzana, M.H.; Sarwar, H.; Alfonso, M. Utilization of Cerebrospinal Fluid Proteome Analysis in the Diagnosis of Meningioma: A Systematic Review. Cureus 2021, 13, e20707. [Google Scholar] [CrossRef] [PubMed]
- Georgila, K.; Vyrla, D.; Drakos, E. Apolipoprotein A-I (ApoA-I), Immunity, Inflammation and Cancer. Cancers 2019, 11, 1097. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, M.U.; Hathout, Y.; MacDonald, T.J.; Kieran, M.W.; Gururangan, S.; Blaney, S.M.; Phillips, P.; Packer, R.; Gordish-Dressman, H.; Rood, B.R. Proteomic profiling of cerebrospinal fluid identifies prostaglandin D2 synthase as a putative biomarker for pediatric medulloblastoma: A pediatric brain tumor consortium study. Proteomics 2011, 11, 935–943. [Google Scholar] [CrossRef]
- Xiao, F.; Lv, S.; Zong, Z.; Wu, L.; Tang, X.; Kuang, W.; Zhang, P.; Li, X.; Fu, J.; Xiao, M.; et al. Cerebrospinal fluid biomarkers for brain tumor detection: Clinical roles and current progress. Am. J. Transl. Res. 2020, 12, 1379–1396. [Google Scholar] [PubMed]
- Wang, P.; Piao, Y.; Zhang, X.; Li, W.; Hao, X. The concentration of CYFRA 21-1, NSE and CEA in cerebro-spinal fluid can be useful indicators for diagnosis of meningeal carcinomatosis of lung cancer. Cancer Biomark 2013, 13, 123–130. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Kopkova, A.; Sana, J.; Fadrus, P.; Slaby, O. Cerebrospinal fluid microRNAs as diagnostic biomarkers in brain tumors. Clin. Chem. Lab. Med. 2018, 56, 869–879. [Google Scholar] [CrossRef]
- Slavik, H.; Balik, V.; Vrbkova, J.; Rehulkova, A.; Vaverka, M.; Hrabalek, L.; Ehrmann, J.; Vidlarova, M.; Gurska, S.; Hajduch, M.; et al. Identification of Meningioma Patients at High Risk of Tumor Recurrence Using MicroRNA Profiling. Neurosurgery 2020, 87, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Gareev, I.; Beylerli, O.; Liang, Y.; Xiang, H.; Liu, C.; Xu, X.; Yuan, C.; Ahmad, A.; Yang, G. The Role of MicroRNAs in Therapeutic Resistance of Malignant Primary Brain Tumors. Front. Cell Dev. Biol. 2021, 9, 740303. [Google Scholar] [CrossRef]
- Urbschat, S.; Landau, B.; Bewersdorf, N.C.; Schuster, C.; Wagenpfeil, G.; Schulz-Schaeffer, W.J.; Oertel, J.; Ketter, R. MicroRNA 200a as a histologically independent marker for meningioma recurrence: Results of a four microRNA panel analysis in meningiomas. Cancer Med. 2022, 12, 8433–8444. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Kim, J.H. Diverse Effects of Exosomes on COVID-19: A Perspective of Progress From Transmission to Therapeutic Developments. Front. Immunol. 2021, 12, 716407. [Google Scholar] [CrossRef]
- Saugstad, J.A.; Lusardi, T.A.; Van Keuren-Jensen, K.R.; Phillips, J.I.; Lind, B.; Harrington, C.A.; McFarland, T.J.; Courtright, A.L.; Reiman, R.A.; Yeri, A.S.; et al. Analysis of extracellular RNA in cerebrospinal fluid. J. Extracell Vesicles 2017, 6, 1317577. [Google Scholar] [CrossRef]
- Negroni, C.; Hilton, D.A.; Ercolano, E.; Adams, C.L.; Kurian, K.M.; Baiz, D.; Hanemann, C.O. GATA-4, a potential novel therapeutic target for high-grade meningioma, regulates miR-497, a potential novel circulating biomarker for high-grade meningioma. EBioMedicine 2020, 59, 102941. [Google Scholar] [CrossRef]
- Ricklefs, F.L.; Maire, C.L.; Wollmann, K.; Dührsen, L.; Fita, K.D.; Sahm, F.; Herold-Mende, C.; von Deimling, A.; Kolbe, K.; Holz, M.; et al. Diagnostic potential of extracellular vesicles in meningioma patients. Neuro Oncol. 2022, 24, 2078–2090. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Zhang, G.; Lan, Y.; Xie, A.; Shi, J.; Zhao, H.; Xu, L.; Zhu, S.; Luo, T.; Zhao, T.; Xiao, Y.; et al. Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements. J. Biol. Chem. 2019, 294, 15613–15622. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ge, Y.; Wang, D.; Liu, Q.; Sun, S.; Hua, L.; Deng, J.; Luan, S.; Cheng, H.; Xie, Q.; et al. LncRNA-IMAT1 Promotes Invasion of Meningiomas by Suppressing KLF4/hsa-miR22-3p/Snai1 Pathway. Mol. Cells 2022, 45, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ren, J.; Ma, J.; Wu, J.; Zhang, R.; Yuan, H.; Han, X. LINC00702/miR-4652-3p/ZEB1 axis promotes the progression of malignant meningioma through activating Wnt/beta-catenin pathway. Biomed. Pharmacother. 2019, 113, 108718. [Google Scholar] [CrossRef]
- Rana, M.W.; Pinkerton, H.; Thornton, H.; Nagy, D. Heterotransplantation of human glioblastoma multiforme and meningioma to nude mice. Proc. Soc. Exp. Biol. Med. 1977, 155, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.L.; Leppla, D.; Rokosz, N.; Wurster, R.D. Matrigel augments xenograft transplantation of meningioma cells into athymic mice. Neurosurgery 1998, 42, 130–135, discussion 135–136. [Google Scholar] [CrossRef]
- McCutcheon, I.E.; Friend, K.E.; Gerdes, T.M.; Zhang, B.M.; Wildrick, D.M.; Fuller, G.N. Intracranial injection of human meningioma cells in athymic mice: An orthotopic model for meningioma growth. J. Neurosurg. 2000, 92, 306–314. [Google Scholar] [CrossRef]
- Boetto, J.; Peyre, M.; Kalamarides, M. Mouse Models in Meningioma Research: A Systematic Review. Cancers 2021, 13, 3712. [Google Scholar] [CrossRef]
- Püttmann, S.; Senner, V.; Braune, S.; Hillmann, B.; Exeler, R.; Rickert, C.H.; Paulus, W. Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab. Investig. 2005, 85, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- van Furth, W.R.; Laughlin, S.; Taylor, M.D.; Salhia, B.; Mainprize, T.; Henkelman, M.; Cusimano, M.D.; Ackerley, C.; Rutka, J.T. Imaging of murine brain tumors using a 1.5 Tesla clinical MRI system. Can. J. Neurol. Sci. 2003, 30, 326–332. [Google Scholar] [CrossRef]
- Baia, G.S.; Dinca, E.B.; Ozawa, T.; Kimura, E.T.; McDermott, M.W.; James, C.D.; VandenBerg, S.R.; Lal, A. An orthotopic skull base model of malignant meningioma. Brain Pathol. 2008, 18, 172–179. [Google Scholar] [CrossRef]
- Castle, K.D.; Chen, M.; Wisdom, A.J.; Kirsch, D.G. Genetically engineered mouse models for studying radiation biology. Transl. Cancer Res. 2017, 6, S900–s913. [Google Scholar] [CrossRef] [PubMed]
- Peyre, M.; Salaud, C.; Clermont-Taranchon, E.; Niwa-Kawakita, M.; Goutagny, S.; Mawrin, C.; Giovannini, M.; Kalamarides, M. PDGF activation in PGDS-positive arachnoid cells induces meningioma formation in mice promoting tumor progression in combination with Nf2 and Cdkn2ab loss. Oncotarget 2015, 6, 32713–32722. [Google Scholar] [CrossRef]
- Kawashima, M.; Suzuki, S.O.; Yamashima, T.; Fukui, M.; Iwaki, T. Prostaglandin D synthase (beta-trace) in meningeal hemangiopericytoma. Mod. Pathol. 2001, 14, 197–201. [Google Scholar] [CrossRef]
- Urade, Y.; Kitahama, K.; Ohishi, H.; Kaneko, T.; Mizuno, N.; Hayaishi, O. Dominant expression of mRNA for prostaglandin D synthase in leptomeninges, choroid plexus, and oligodendrocytes of the adult rat brain. Proc. Natl. Acad. Sci. USA 1993, 90, 9070–9074. [Google Scholar] [CrossRef]
- Yamashima, T.; Sakuda, K.; Tohma, Y.; Yamashita, J.; Oda, H.; Irikura, D.; Eguchi, N.; Beuckmann, C.T.; Kanaoka, Y.; Urade, Y.; et al. Prostaglandin D synthase (beta-trace) in human arachnoid and meningioma cells: Roles as a cell marker or in cerebrospinal fluid absorption, tumorigenesis, and calcification process. J. Neurosci. 1997, 17, 2376–2382. [Google Scholar] [CrossRef] [PubMed]
- von Werder, A.; Seidler, B.; Schmid, R.M.; Schneider, G.; Saur, D. Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. Nat. Protoc. 2012, 7, 1167–1183. [Google Scholar] [CrossRef]
- Kalamarides, M.; Niwa-Kawakita, M.; Leblois, H.; Abramowski, V.; Perricaudet, M.; Janin, A.; Thomas, G.; Gutmann, D.H.; Giovannini, M. Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev. 2002, 16, 1060–1065. [Google Scholar] [CrossRef]
- Kalamarides, M.; Stemmer-Rachamimov, A.O.; Takahashi, M.; Han, Z.Y.; Chareyre, F.; Niwa-Kawakita, M.; Black, P.M.; Carroll, R.S.; Giovannini, M. Natural history of meningioma development in mice reveals: A synergy of Nf2 and p16(Ink4a) mutations. Brain Pathol. 2008, 18, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Peyre, M.; Stemmer-Rachamimov, A.; Clermont-Taranchon, E.; Quentin, S.; El-Taraya, N.; Walczak, C.; Volk, A.; Niwa-Kawakita, M.; Karboul, N.; Giovannini, M.; et al. Meningioma progression in mice triggered by Nf2 and Cdkn2ab inactivation. Oncogene 2013, 32, 4264–4272. [Google Scholar] [CrossRef] [PubMed]
- Mandara, M.T.; Ricci, G.; Rinaldi, L.; Sarli, G.; Vitellozzi, G. Immunohistochemical identification and image analysis quantification of oestrogen and progesterone receptors in canine and feline meningioma. J. Comp. Pathol. 2002, 127, 214–218. [Google Scholar] [CrossRef]
- Michelhaugh, S.K.; Guastella, A.R.; Varadarajan, K.; Klinger, N.V.; Parajuli, P.; Ahmad, A.; Sethi, S.; Aboukameel, A.; Kiousis, S.; Zitron, I.M.; et al. Development of patient-derived xenograft models from a spontaneously immortal low-grade meningioma cell line, KCI-MENG1. J. Transl. Med. 2015, 13, 227. [Google Scholar] [CrossRef][Green Version]
- Kim, H.; Park, K.J.; Ryu, B.K.; Park, D.H.; Kong, D.S.; Chong, K.; Chae, Y.S.; Chung, Y.G.; Park, S.I.; Kang, S.H. Forkhead box M1 (FOXM1) transcription factor is a key oncogenic driver of aggressive human meningioma progression. Neuropathol. Appl. Neurobiol. 2020, 46, 125–141. [Google Scholar] [CrossRef]
- Saydam, O.; Shen, Y.; Würdinger, T.; Senol, O.; Boke, E.; James, M.F.; Tannous, B.A.; Stemmer-Rachamimov, A.O.; Yi, M.; Stephens, R.M.; et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol. Cell Biol. 2009, 29, 5923–5940. [Google Scholar] [CrossRef] [PubMed]
- Tuchen, M.; Wilisch-Neumann, A.; Daniel, E.A.; Baldauf, L.; Pachow, D.; Scholz, J.; Angenstein, F.; Stork, O.; Kirches, E.; Mawrin, C. Receptor tyrosine kinase inhibition by regorafenib/sorafenib inhibits growth and invasion of meningioma cells. Eur. J. Cancer 2017, 73, 9–21. [Google Scholar] [CrossRef]
- Juratli, T.A.; Prilop, I.; Saalfeld, F.C.; Herold, S.; Meinhardt, M.; Wenzel, C.; Zeugner, S.; Aust, D.E.; Barker, F.G., 2nd; Cahill, D.P.; et al. Sporadic multiple meningiomas harbor distinct driver mutations. Acta Neuropathol. Commun. 2021, 9, 8. [Google Scholar] [CrossRef]

| Biomarker Type | Known Biomarkers | Study Design | Clinical Use | Correlation with Grade | Description of Marker Usage and Its Effects | Reference and Year |
|---|---|---|---|---|---|---|
| Genomics | NF2, TRAF7, AKT1, SMO, and PIK3CA | Review | Diagnosis/Therapy | - | - | [29], 2020 |
| SMARCB1 | Review | Diagnosis/Therapy | Grades 1 and 2 | Genetic risk factor for sporadic multiple meningiomas | [29], 2020 | |
| KLF4 | Review | Diagnosis/Therapy | Grade 1 | Downregulated in anaplastic meningiomas | [29], 2020; [65], 2017 | |
| CDKN2A/B homozygous deletion | Cohort of 528 meningioma patients | Diagnostic/Prognostic | Grade 3 > Grade 2; absent in Grade 1 | Faster progression to recurrence Higher mortality | [66], 2020 | |
| miRNA | miR-29c-3p and miR-219-5p | A study of 50 meningioma patients training set and 60 meningioma patients validation set compared to normal adjacent tissue | Diagnosis, Prognosis, and Therapy Response | Grades 1 > 2 > 3 | Downregulation associated with advanced clinical stages of meningiomas and significant correlation with higher recurrence rates | [67], 2013 |
| miR-190a | A study of 50 meningioma patients training set and 60 meningioma patients validation set compared to normal adjacent tissue | Prognosis | Grades 1 < 2 < 3 | Upregulation associated with advanced clinical stages of meningiomas, independent of other clinicopathological factors | [67], 2013 | |
| miR-17-5p, miR-199a, miR-190a, miR-186-5p, miR-155-5p, miR-22-3p, miR-24-3p, miR-26b-5p, miR-27a-3p, miR-27b-3p, miR-96-5p, and miR-146a-5p | A study of 50 meningioma patients training set and 60 meningioma patients validation set compared to normal adjacent tissue | Diagnosis, Prognosis, Histological grade, and Radio-sensitivity | - | Significantly upregulated in meningioma samples | [67], 2013 | |
| miR-219-5p, miR-106a-5p, miR-375, and miR-409-3p | 20 pre-operative meningiomas and 20 healthy controls as discovery set Candidate miRNAs were validated individually in another 210 meningioma and 210 healthy controls | Non-invasive Diagnostic/Prognostic | miR-219-5p: Grades 3 > 2 > 1 | Serum levels of the miRNA panel significantly increased in meningioma cases Serum levels of miR-219-5p positively correlated with higher meningioma grade | [68], 2016 | |
| miR-197 and miR-224 | 20 pre-operative meningiomas and 20 healthy controls as discovery set Candidate miRNAs were validated individually in another 210 meningiomas and 210 healthy controls | Non-invasive Diagnostic/Prognostic | - | Serum levels significantly decreased in meningioma cases High serum miR-409-3p and low miR-224 expression significantly correlated with higher recurrence rates | [68], 2016 | |
| Upregulation of miR-4286, miR-4695-5p, miR-6732-5p, miR-6855-5p, miR-7977, miR-6765-3p, and miR-6787-5p and downregulation of miR-1275, miR-30c-1-3p, miR-4449, miR-4539, miR-4684-3p, miR-6129, and miR-6891-5p | Study of 55 atypical meningioma patients (43 radio-sensitive and 12 radio-resistant meningiomas) and 6 arachnoid samples as control | Prognosis/response to radiotherapy | Grade 2 | 14 miRNAs significantly dysregulated in meningiomas Prediction of individual sensitivity to radiotherapy in patients resistant to radiotherapy Dysregulated miRNAs enriched in fatty acid biosynthesis and metabolism and TGFβ signaling pathways | [69], 2020 | |
| miR-181d | Study collected meningioma tissues and plasma of 40 meningioma patients (16 Grade 1, 16 Grade 2, and 8 Grade 3 patients) | Non-invasive Diagnosis/Prognosis | Grades 1 < 2 < 3 | Associated with tumor progression in plasma and tumor tissues | [70], 2021 | |
| LncRNA | LncRNA-LINC00460 | A study of tissues from 32 meningioma patients and 5 normal control cases, in addition to in vitro studies in meningioma cell lines | Diagnosis | Grades 2 < 3 | Upregulated in meningioma tissues and malignant cell lines | [71], 2020 |
| ISLR2, Lnc-GOLGA6A-1, AMH, and Grades 1 > 2 | A study of 64 meningioma patients (with and without recurrence and of different WHO grades) that were subjected to RNA-seq; 90 samples validated using RT-qPCR | Prognosis and Pathogenesis | Lnc-MAST4-5: Grades 1 > 2, 3 | ISLR2, Lnc-GOLGA6A-1, and AMH associated with recurrence risk | [72], 2022 | |
| Lnc-00460 | A study of 33 human meningioma tumor tissues and 10 normal meninges tissues, in addition to meningioma cell lines | Diagnosis | - | Upregulated in meningioma tissues and cell lines | [73], 2018 | |
| LncRNA-NUP210, LncRNA-SPIRE2, LncRNA-SLC7A1, and LncRNA-DMTN | Review | Diagnosis/Prognosis | - | Upregulated in meningiomas Target microRNA-195 | [74], 2023 | |
| Epigenetic | TIMP3, HOXA7, HOXA9, and HOXA10 | Review | Prognosis | - | Hypermethylation associated with tumor progression and malignant transformation | [75], 2015; [76], 2020; [77], 2023 |
| TRAF7, KLF4, NF2, TRAKL, ARID1A, and AKT1 | Retrospective analysis of formalin-fixed paraffin-embedded sections of 126 meningioma patients of different grades | Prognosis | - | Aberrant DNA methylation of these genes may be involved in the development and progression of meningiomas | [78], 2022; [77], 2023 | |
| TIMP3, CDKN2A, and NDRG2 | Review | Prognosis | - | Faster recurrence | [76], 2020; [77], 2023 | |
| TP73, RSSF1A, and MAL2 | Review | Prognosis | - | Hypermethylation increases risk of malignancy | [76], 2020; [77], 2023 | |
| H3K27me3 histone modification | Retrospective study of 232 meningioma patients | Diagnosis/Prognosis | Grades 1 < 2 < 3 | Loss of H3K27me3 methylation patterns correlated with high recurrence | [26], 2018 | |
| Mutations in hTERT gene promoter | Study of 252 meningioma patients | Diagnosis/Prognosis | Grade 3 (aggressive) | Presence of hTERT promoter mutations means shorter time to progression | [79], 2016 | |
| Mutations in hTERT gene promoter | Meta-analysis of 8 clinical trials | Diagnosis/Prognosis | Grades 1 < 2 < 3 | Presence of hTERT promoter mutations resulted in higher recurrence rates and mortality This was a better prediction than WHO grading system | [80], 2019 | |
| Proteomic | APO-E and APO-J | Proteomic analysis of CSF from 4 meningioma patients and 4 patients with a non-brain | Diagnosis | Grade 2 | Tumor progression marker | [81], 2012 |
| PTGDS | Clinical Study | Diagnosis | Grade 1 | Associated with higher grade and early recurrence in intracranial meningiomas | [82], 2019 | |
| Caspase-3, Amphiregulin, and VEFG-D | Screening cohort followed by a validation set of meningioma tissues and serum | Non-invasive diagnosis and prognosis | Grades 1 < 2, 3 | The 3 proteins may constitute a panel that correlates with meningioma progression | [82], 2019 | |
| EFEMP1 | A study of CSF and serum of 45 meningioma patients and 30 healthy controls | Diagnosis | CSF and serum EFEMP1 levels significantly higher meningioma patients | [83], 2017 | ||
| Histological | SSTR2A and Claudin-1 | 35 meningiomas, 10 intracranial schwannoma, and 10f hemangiopericytoma cases | Diagnosis | SSTRA: Grades 1, 2 > 3 Claudin-1: Grades 1, 2 < 3 | Distinguishes meningiomas from schwannoma and hemangiopericytoma | [84], 2018 |
| CA9 | Immunohistochemistry of paraffin-embedded sections of 25 Grade 1, 17 Grade 2, and 20 Grade 3 meningiomas | Prognosis | Grade 3 | Associated with higher grade histology and common in recurrent tumors | [85], 2007 | |
| Metabolomic | Alanine and Glutamine/Glutamate | 1H NMR of 23 Grade 1 and 10 Grade 2 meningioma tissues | Diagnosis/Prognosis | Glutamine metabolism: Grades 1 > 2 | Predominantly elevated in Grade 2 meningiomas | [86], 2022 |
| Glycine/Serine | Validation of 43 meningioma patients | Diagnosis/Prognosis | Grades 1 > 2 > 3 | Grade 1 associated with lower proliferation and longer progression-free survival | [87], 2021 | |
| Choline/Tryptophan | Validation of 43 meningioma patients | Diagnosis/Prognosis | Grades 2, 3 > 1 | Higher tryptophan/choline associated with shorter progression-free survival Similar incidence of Grades 1, 2, and 3 | [87], 2021, | |
| Sphingolipid Galactosyl Ceramide | Discovery using LC-MS/MS and validation in 85 meningioma biopsies of different grades | Diagnosis/Prognosis | Grades 2, 3 > Grade 1 | Higher levels in WHO Grades 2 and 3 than Grade 1 | [88], 2023 | |
| High acetate, threonine, N-acetyl-lysine, hydroxybutyrate, myoinositol, ascorbate, and total choline and low aspartate, glucose, isoleucine, valine, adenosine, arginine, and alanine | Metabolomics analysis using HRMAS NMR of 62 human meningioma samples | Diagnosis/Prognosis | Aggressive Grade 1 and Grade 2 have similar metabolic signature to Grade 3 | Poor prognosis and high proliferation and histological grade | [89], 2020 | |
| Integrated systems of molecular/histological biomarkers | WHO grade, methylation class, and absence of chromosomes 1p, 6q, and 14q | Retrospective and prospective multi-center clinical study of 514 meningiomas and validation in 471 samples | Diagnosis/Prognosis | Nine-point scoring system Final scores of 3–5 → low risk of recurrence; 3–5 → intermediate risk; and score of 6–9 → high risk of recurrence | [90], 2021 | |
| Mitotic index, CDKN2A/B homologous deletion, and alterations of copy number of specific chromosomes | Discovery cohort of 527 meningiomas and a validation set of 172 meningiomas | Diagnosis/Prognosis | Points scoring system Final scores of 0–1 → low risk of recurrence; 2–3 → intermediate risk; and score of 4 or more → high risk of recurrence | [91], 2022 |
| Mouse Model | Advantages | Limitations |
|---|---|---|
| Heterotopic xenograft model | Very reliable in terms of tumor take rates. | Lacks the key components of the meningioma’s specific microenvironment. |
| Orthotopic xenograft model |
|
|
| Genetically Engineered Mouse Models (GEMMs) |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halabi, R.; Dakroub, F.; Haider, M.Z.; Patel, S.; Amhaz, N.A.; Reslan, M.A.; Eid, A.H.; Mechref, Y.; Darwiche, N.; Kobeissy, F.; et al. Unveiling a Biomarker Signature of Meningioma: The Need for a Panel of Genomic, Epigenetic, Proteomic, and RNA Biomarkers to Advance Diagnosis and Prognosis. Cancers 2023, 15, 5339. https://doi.org/10.3390/cancers15225339
Halabi R, Dakroub F, Haider MZ, Patel S, Amhaz NA, Reslan MA, Eid AH, Mechref Y, Darwiche N, Kobeissy F, et al. Unveiling a Biomarker Signature of Meningioma: The Need for a Panel of Genomic, Epigenetic, Proteomic, and RNA Biomarkers to Advance Diagnosis and Prognosis. Cancers. 2023; 15(22):5339. https://doi.org/10.3390/cancers15225339
Chicago/Turabian StyleHalabi, Reem, Fatima Dakroub, Mohammad Z. Haider, Stuti Patel, Nayef A. Amhaz, Mohammad A. Reslan, Ali H. Eid, Yehia Mechref, Nadine Darwiche, Firas Kobeissy, and et al. 2023. "Unveiling a Biomarker Signature of Meningioma: The Need for a Panel of Genomic, Epigenetic, Proteomic, and RNA Biomarkers to Advance Diagnosis and Prognosis" Cancers 15, no. 22: 5339. https://doi.org/10.3390/cancers15225339
APA StyleHalabi, R., Dakroub, F., Haider, M. Z., Patel, S., Amhaz, N. A., Reslan, M. A., Eid, A. H., Mechref, Y., Darwiche, N., Kobeissy, F., Omeis, I., & Shaito, A. A. (2023). Unveiling a Biomarker Signature of Meningioma: The Need for a Panel of Genomic, Epigenetic, Proteomic, and RNA Biomarkers to Advance Diagnosis and Prognosis. Cancers, 15(22), 5339. https://doi.org/10.3390/cancers15225339












