Simple Summary
Despite recent improvement in chemotherapy regimens for pancreatic adenocarcinoma (PDAC), the clinical outcomes are still unsatisfactory compared to other solid tumors. Radiotherapy was demonstrated to improve locoregional control of PDAC; however, the survival benefit of radiotherapy in localized PDAC is undefined due to early distant progression in the majority of patients. Upfront chemotherapy for localized PDAC was suggested recently to avoid radical local therapy for patients of localized PDAC high risk of distant metastasis. Potential tissue biomarkers were developed to select PDAC patients who will benefit from local radiotherapy. This review summarizes potential tissue biomarkers reported to predict the efficacy and survival benefits of radiotherapy for localized PDAC including SMAD4, a biomarker validated in a prospective clinical trial to correlate with failure pattern of localized PDAC after radiotherapy. In particular, we describe Krüppel-like factor 10 (KLF10), lost in two thirds of PDAC patients, in association with distant metastasis and radio-resistance of PDAC. From tumor tissues of patients with resectable PDAC enrolled to a clinical trial, we demonstrated that the combination of KLF10 and SMAD4 expression in tumor tissues may help select those who may benefit the most from additional radiotherapy. Though promising, these potential biomarkers should be validated in prospective clinical trials.
Abstract
The prognosis of pancreatic adenocarcinoma (PDAC) remains poor, with a 5-year survival rate of 12%. Although radiotherapy is effective for the locoregional control of PDAC, it does not have survival benefits compared with systemic chemotherapy. Most patients with localized PDAC develop distant metastasis shortly after diagnosis. Upfront chemotherapy has been suggested so that patients with localized PDAC with early distant metastasis do not have to undergo radical local therapy. Several potential tissue markers have been identified for selecting patients who may benefit from local radiotherapy, thereby prolonging their survival. This review summarizes these biomarkers including SMAD4, which is significantly associated with PDAC failure patterns and survival. In particular, Krüppel-like factor 10 (KLF10) is an early response transcription factor of transforming growth factor (TGF)-β. Unlike TGF-β in advanced cancers, KLF10 loss in two-thirds of patients with PDAC was associated with rapid distant metastasis and radioresistance; thus, KLF10 can serve as a predictive and therapeutic marker for PDAC. For patients with resectable PDAC, a combination of KLF10 and SMAD4 expression in tumor tissues may help select those who may benefit the most from additional radiotherapy. Future trials should consider upfront systemic therapy or include molecular biomarker-enriched patients without early distant metastasis.
1. Introduction
1.1. Controversies Regarding Radiotherapy for Pancreatic Adenocarcinoma
Pancreatic adenocarcinoma (PDAC) is notoriously well known for its dismal survival outcomes. It is characterized by rapid distant metastasis or local destructive progression with a 5-year survival rate of 12% [1]. In patients with metastatic or unresectable PDAC, combination chemotherapy regimens consisting of (modified) FOLFIRINOX [2,3] and gemcitabine (GEM) plus nab-paclitaxel [4] have achieved better tumor responses (31.6% vs. 9.4% and 23% vs. 7%, both p < 0.001) and overall survival (OS) than single-agent GEM or 5-fluorouracil (11.1 vs. 6.8 months and 8.5 vs. 6.7 months, both p < 0.001). Prospective randomized trials revealed the survival benefit of adjuvant chemotherapy with FOLFIRINOX (p = 0.003) [5], GEM plus capecitabine (p = 0.032) [6], or GEM plus nab-paclitaxel (p = 0.009) [7] over single-agent GEM after PDAC resection. For borderline resectable PDAC, neoadjuvant chemotherapy achieved a better R0 resection rate (71% vs. 40%, p < 0.01) and survival (15.7 vs. 14.3 months, p = 0.025) than upfront surgery [8,9,10].
Unlike chemotherapy, the efficacy of radiotherapy as an adjuvant or curative treatment for PDAC remains controversial. The European Study Group for Pancreatic Cancer (ESPAC)-1 trial indicated no benefit of radiotherapy for resectable PDAC [11]. Our prospective randomized study to revealed that chemoradiotherapy (CRT) with adjuvant GEM for six months improved local control (GEM-CRT arm vs. GEM arm, locoregional recurrence rate: 41.4% vs. 58.1%, p = 0.039) but additional CRT had no survival benefit (GEM-CRT arm vs. GEM arm; OS: 21.5 vs. 23.5 months, p = 0.82) for patients with resescted PDAC [12]. However, long-term outcomes from the Dutch Pancreatic Cancer Group-initiated PREOPANC study revealed that in patients with (borderline) resectable PDAC receiving adjuvant GEM, neoadjuvant GEM-based CRT had a substantial advantage (5-year OS rate: 20.5% vs. 6.5%, p = 0.025) and improved locoregional control (p = 0.004) compared with upfront surgery [9,10]. The authors suggested that CRT might benefit patients with PDAC who do not have early distant metastasis. The survival benefit of neoadjuvant therapy for localized PDAC was demonstrated from the ESPAC-5 [13] and the National Clinical Trials Network cooperative groups initiated A021501 [14] trials, especially chemotherapy with FOLFIRINOX in ESPAC-5 showing the 1-year OS rate: 84% vs. 39% (p = 0.0028). Neoadjuvant capecitabine-based CRT provided a moderate survival benefit (60% vs. 39%) compared with immediate surgery despite the improved R0 resection and pathologic complete remission rates. The efficacy of neoadjuvant CRT could not be determined in the A021501 trial due to the insufficient accrual of patients after early termination due to the low R0 resection rate in the neoadjuvant CRT arm. The authors concluded that preoperative radiotherapy using other delivery approaches may benefit a subpopulation of patients. Regarding locally advanced PDAC (LAPC), the international LAP07 study identified that the addition of CRT after GEM induction therapy improved local control from 32% to 46% (p = 0.03) without survival benefit (p = 0.09), partly due to rapid distant metastasis [15]. Conversely, the Eastern Cooperative Oncology Group trial disclosed that upfront GEM-based CRT prolonged median survival duration compared with GEM alone (p = 0.017) in LAPC [16]. The conflicting results of randomized studies on localized PDAC imply a narrow therapeutic window for local radiotherapy.
Despite improved clinical outcomes with the combination chemotherapy and neoadjuvant strategy, the survival of patients with PDAC remains inferior to that of patients with other solid tumors [1]. Local recurrence remains one of the essential issues for survival and life quality of PDAC patients. One third of patients with PDAC, disclosed from rapid autopsy, died from local destructive progression without prominent distant metastasis [17]. Despite significant amelioration in recurrence with the mFOLFIRINOX regimen compared with GEM alone in patients with resectable PDAC [5], the pattern of recurrence remained unaffected with isolated locoregional recurrence accounting for 24.6% and 24% of all recurrences in patients who had undergone mFOLFIRINOX and GEM, respectively. A similar observation was noted in the ESPAC-4 study, with local recurrence rates of 53% and 46% in the GEM arm and Gem-CRT arm, respectively [6].
Using personalized radiotherapy, in 49 patients with LAPC, according to the response to induction chemotherapy of eight cycles of FOLFIRINOX and losartan, an inhibitor of thrombospondin-1-mediated activation of latent TGF-β, a phase II trial demonstrated a prominent down-staging and R0 resection rate of 61% with significantly prolonged median progression-free survival (PFS) and OS (17.5 and 31.4 months, respectively) [18]. Ablative radiotherapy following induction chemotherapy with a combination regimen revealed, in 119 patients with inoperable PDAC, safe and prolonged local control, with a median OS of 26.8 months [19]. Advancements in treatment techniques and radiotherapy strategies can be applied to current standard approaches for improving the currently unsatisfactory clinical outcomes of patients with PDAC. Modern radiotherapy provides excellent locoregional control and it should therefore be incorporated into the multimodal treatment of PDAC. Recent clinical trials, especially the PREOPAC study, have implied that administering CRT to patients with a low risk of early distant metastasis can translate local control into survival benefit [4]. Research must be conducted to find patients with PDAC who will benefit the most from CRT by using molecular biomarkers related to PDAC tumorigenesis and progression.
1.2. Tissue Biomarkers of Radiotherapy Responses in PDAC
The heterogeneity and aggressive biology of PDAC are classified based on epigenetic, genomic, transcriptomic, and proteomic data [20]. Several potential tissue biomarkers were identified for differentiating progression patterns in patients with PDAC (Table 1). A radiosensitivity index (RSI) for intrinsic radiosensitivity of tumors was developed from a linear regression algorithm of the surviving fraction of 48 cancer cells after 2 Gy and the expression levels of 10 genes including HDAC1, SUMO1, PKCb, c-Abl, STAT1, AR, Cdk1, c-Jun, RelA, and IRF1. In 73 patients with PDAC receiving surgery with or without radiotherapy, patients with RSI-high radioresistant tumors tended to have shorter survival (hazard ratio [HR]: 2.1, 95% confidence interval [CI]: 1.0–4.3, p = 0.054). For the 31 high-risk patients (positive lymph nodes, positive margins, or postoperative CA19-9 levels > 90 U/mL) who underwent radiotherapy, radio-sensitive patients (i.e., with a low RSI) had significantly improved survival compared with radioresistant patients (i.e., with a high RSI) (OS: 31.2 vs. 13.2 months, p = 0.04) [21]. The authors concluded that integrating the RSI with high-risk variables can refine the prognosis of patients with pancreatic cancer treated with radiotherapy. An optimal radiotherapy dose at the individual specific molecular signature level genomic-adjusted radiation dose (GARD) was obtained by combing RSI with a linear-quadratic model. Using data from the total cancer care (TCC) study, the GARD was calculated for 20 primary tumors from various sites treated with the corresponding conventional radiotherapy doses. Despite this uniformity of the radiation dose for a specific tumor type, GARD varied widely across the TCC cohort, implying that a high dose does not always result in a high therapeutic effect. The median GARD was higher in patients with oropharyngeal cancer than in those with non-oropharyngeal head and neck cancer (39.71 vs. 32.56, p = 0.042) after 70 Gy; this finding is in concordance with the observation of better efficacy of radiotherapy in patients with oropharyngeal cancer [22,23]. Among the 40 patients in the Moffitt pancreas cancer cohort, the GARD ranged between 16 and 40 and predicted OS independently to a statistically significant level (HR: 2.6, 95% CI: 1.1–6.0; p = 0.029). Higher GARDs predicted a better radiotherapeutic effect, longer time to recurrence, and longer survival; moreover, GARD enabled the individualization of the radiation dosage according to tumor radiosensitivity [22,23]. Preclinical studies have demonstrated that indoleamine 2,3 dioxygenase-2 (IDO2), a tryptophan catabolic enzyme, promotes pancreatic tumorigenesis. PDAC development reduced in IDO2−/− mice (30% vs. 10%, p < 0.05) [24]. In humans, the high prevalence of two inactivating single-nucleotide variations, rs4503083 [Exon 11] and rs10109853 [Exon 9], of IDO2 was noted. A DNA analysis of 200 patients from two pancreatic cancer cohorts (The cancer genome atlas and the Thomas Jefferson University Hospital dataset) indicated that an IDO2-deficient genotype was correlated with longer PFS in PDAC patients receiving adjuvant radiotherapy (p = 0.023). The choline phosphorylation pathway is upregulated in PDAC. From 88 patients with resectable PDAC, metabolic profile analysis demonstrated a prominent difference between good and poor responders in tumors’ choline metabolites (including N-acetylglucosamine-1-phosphate, 1-methylnicotinamide, carnitine, glucose, glutathione, N-acetylglucosamine-6-phosphate, and uridine-5″-monophosphate) regardless of whether they received neoadjuvant CRT. In patients receiving neoadjuvant CRT (n = 62), the levels of carnitine (≤130 nmol/mg), choline (≤283 nmol/mg), phosphocholine (≤749 nmol/mg), and glutathione (≤373 nmol/mg) predicted better PFS (all p < 0.05). Multivariate analysis revealed that choline levels of > 284 nmol/mg were significantly associated with recurrence. Microarray analysis confirmed significant suppression of the gene expression levels of the choline transporter CTL1-4 (SLC44A1-44A4) in pancreatic tumor tissues after neoadjuvant CRT. Thus, choline metabolism was suggested as a target and biomarker of neoadjuvant CRT for localized PDAC [25]. Another study integrated genomic profiling and clinical information to predict the radiotherapy response and noted that among 88 patients with cancer receiving radiotherapy, mutations of CHEK2 (p = 0.049), MSH2 (p = 0.014), and NOTCH1 (p = 0.031) were more frequently found in patients with a durable local control of ≥6 months (n = 47). Derangements of DNA repair pathways were associated with better local control (p = 0.014). The somatic mutation signature of smoking was observed more often in the durable local control group with a prediction probability of 0.83 for the 6-month local control [26].

Table 1.
Tissue biomarkers to predict radiotherapy responses in pancreatic adenocarcinoma. Representative clinical studies evaluating potential tissue biomarkers in correlation with survival or failure pattern of radiotherapy to PDAC.
SMAD4 is the only tissue biomarker validated by a prospective trial for predicting failure patterns in PDAC. Among LAPC patients, a local dominant failure pattern was noted in patients with SMAD4 expression compared to those with SMAD4 loss (73% vs. 28%, p = 0.016) [27]. A retrospective analysis of more than 600 patients with resected PDAC also demonstrated improved survival with adjuvant radiotherapy in only SMAD4-positive patients (p = 0.002). SMAD4 loss was significantly associated with metastatic recurrence (HR: 4.28, 95% CI: 2.75–6.68) [28]. SMAD4 status and expression were correlated with radiosensitivity and PDAC failure patterns in clinical and preclinical studies [17,31,32]. Further studies have demonstrated that the SMAD4 heterozygous mutation ameliorated PDAC metastatic yet increased its proliferation ability. Loss of SMAD4 heterozygosity regained PDAC metastatic competency in addition to increased proliferation. Further studies revealed that RUNX3 interacted with SMAD4 to modulate cancer cell division and dissemination. This observation implies that a combination of RUNX3 and SMAD4 levels can help clinical decision making for resectable PDAC [29].
These tissue biomarkers may optimize the integration of radiotherapy in multimodality treatment for patients with PDAC. Advances in tissue biomarkers facilitate the stratification of patients with PDAC with various potential for distant metastasis and the prediction of those who would benefit the most from additional radiotherapy. With the increasing use of neoadjuvant CRT, especially in borderline resectable PDAC, the value of potential biomarkers in specimens of biopsy, cytology, or peripheral blood should be developed in the future.
Several studies including ours have demonstrated that Krüppel-like factor (KLF) 10, a TGF-β early-response gene, contributes to radiosensitivity and cancer progression [33,34,35,36]. In the current review, we summarize recent progress in clinical studies of molecular mechanisms of KLF10 as a predictor of radiotherapy in patients with PDAC.
2. Main Text
2.1. KLFs
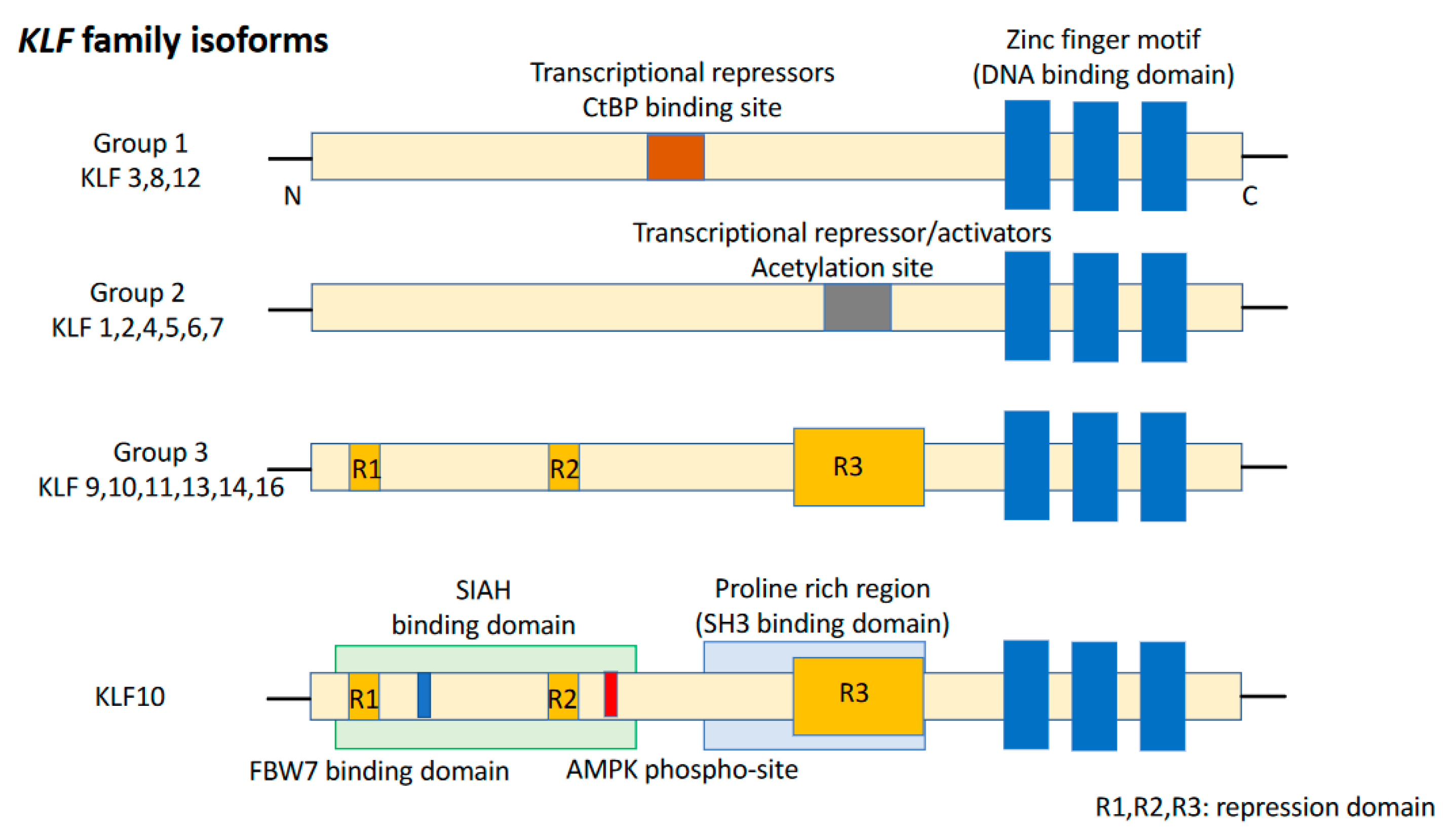
KLFs are of the specificity protein 1 (SP1)-like/KLF transcription factor superfamily and are characterized by the absence of a Buttonhead box, namely CXCPXC [37]. The DNA-binding domain of KLFs, located at the carboxyl terminus, contains three conserved C2H2 zinc finger structures. It enables KLFs to recognize CACCC elements or GC-boxes and to bind to regulatory regions of the target genes [38]. Eighteen unique members of the KLF family were identified, with a >65% sequence similarity for zinc finger motifs, resulting in competition for binding to promoters of target genes (Figure 1). Group 1 consists of KLF3, KLF8, and KLF12 which behave as transcriptional repressors by interacting with proteins binding to the carboxyl terminus. Group 2 includes KLF1, KLF2, KLF4, KLF5, KLF6, and KLF7 which bind to acetyltransferases and function as transcriptional activators. Group 3 comprises KLF9, KLF10, KLF11, KLF13, KLF14, and KLF16 which are transcriptional repressors and interact with switch-independent-3 family member A (Sin3A), a common transcriptional corepressor. Nowadays, KLF15, KLF17, and KLF18 remain unclassified [39]. KLFs are known to be critical regulators of many important biological processes, such as cell proliferation, differentiation, survival, cell cycle, epithelial–mesenchymal transition (EMT), invasion, metastasis, cell maturation, and organogenesis. Dysregulation of KLF function can lead to the development of cancer and other disorders [40].
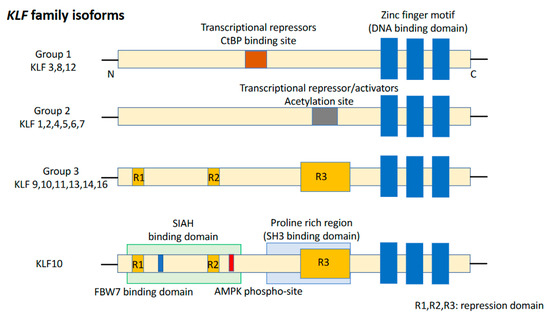
Figure 1.
Structure of the KLF family and KLF10. KLF isoforms can be divided into three groups. KLF10 belongs to Group 3. KLF15, 17, and 18 are not included in any of these groups.
2.2. KLF10
KLF10 was identified in human fetal osteoblasts as a positive regulator of bone growth [41]. The protein homology of KLF10 among humans, Mus musculus, Bos taurus, and Liacerta agilis is as high as 81.28%, suggesting its critical role in biological processes [42]. KLF10 is an early-response mediator of TGFβ/SMAD signaling. It forms a positive feedback loop with TGF-β signaling by transcriptionally regulating SMAD2 and SMAD7 [43]. Estrogen stimulates KLF10 expression, which inhibits BAX inhibitor-1 transcription and enhances breast cancer cell apoptosis [44]. Jun B and lysine demethylase 6A may facilitate KLF10 transcription to exacerbate diabetic nephropathy [45]. Multiple long noncoding RNAs (lncRNA) and microRNAs (miRNA) were identified as upstream regulators of KLFs, thus providing essential pathways for targeting KLFs. E3 ubiquitin ligases, including seven in absentia homolog-1 (SIAH1) and FBW7, interact with KLF10 through conserved binding motifs to promote the proteasomal degradation of KLF10. The binding of KLF10 to itchy E3 ubiquitin ligase (ITCH) increases KLF10 levels and activates Foxp3 transcription in regulatory T cells [46,47]. We previously reported that KLF10 is a phosphorylated protein at Thr-93 in the N-terminal region. RAF-1 phosphorylation and PIN1 isomerization coordinately regulate KLF10 stability and tumor progression [48].
2.3. Involvement of KFL10 in Multiple Diseases
KLF10 is involved in glucose and lipid metabolism, mitochondrial structure and function, cell proliferation, and apoptosis and it plays critical roles in multiple diseases [49]. It is a clock-controlled gene that maintains the hepatic circadian rhythm which is essential for regulating hepatic glucose and lipid homeostasis [42]. Sex-dependent differences were found in the metabolic phenotypes of KLF10-knockout mice. Male mice exhibited post-prandial and fasting hyperglycaemia whereas female mice exhibited increased plasma triglyceride levels. As a circadian-clock-controlled transcription factor, KLF10 suppresses lipogenic genes of glucose and lipid metabolism in the liver and it affects gluconeogenesis, contributing to diabetes [50,51]. KLF10 alleviates hepatic steatosis and nonalcoholic-steatohepatitis by downregulating SREBP-1c involving lipogenesis [52,53]. KLF10-deficient mice exhibit reduced receptor activator of nuclear factor kappa-B ligand, increased osteoprotegerin, and delayed8 osteoclast differentiation which led to reduced bone turnover and osteopenia [49,51,54]. A study reported that male KLF-knockout mice developed cardiac hypertrophy after approximately 16 months due to the angiotensin II-induced cardiac transcription factor, GATA4, and the atrial natriuretic factor, brain natriuretic peptide [55]. KLF10 can transactivate Foxp3 promoters in regulatory T cells in response to TG-β1 to promote atherosclerosis [56,57].
2.4. KLF10 in Cancer
Many studies have demonstrated the tumor suppressor function of KLF10 in terms of cell proliferation inhibition and apoptosis induction [58,59]. KLF10 loss activates PTEN/PI3K/AKT activity in multiple myeloma and bladder cancer [60,61]. KLF10 overexpression can suppress Wnt signaling and GSK3β phosphorylation to inhibit the proliferation, migration, and drug resistance of multiple myeloma cells. Knock-down of securin, the downstream target of KLF10, can mimic the tumor suppressor role of KLF10 in multiple myeloma [62]. In advanced-stage cancer, TGF-β signaling enhances the EMT whereas KLF10 inhibits TGF-β-induced EMT. KLF10 can suppress lung and pancreatic cancer EMT and invasion by recruiting HDAC1 to suppress the SNAI2 promoter for the removal of histone acetylation (H3K9ac and H3K27ac) [63]. In oral squamous cell carcinoma, KLF10 was identified as a differentially expressed circadian-related gene that was correlated with OS (p < 0.05) and the drug response (p = 0.0014) [64]. By directly binding to the LINC00629 promoter to induce Mcl1 degradation, KLF10 exerts antitumor activity in oral squamous cell carcinoma treated with apigenin, a flavonoid [65]. KLF10 is involved in cervical cancer immunoediting by transcriptionally regulating IL6, IL25, and pregnancy-specific beta-1 glycoproteins 2 and 5 [66]. Conversely, the tumor suppressive role of KLF10 may vary depending on the tumor cells types and the microenvironments. In KLF10-knockout mice, the TGF-β-SMAD signaling pathway was activated to suppress diethylnitrosamine-induced hepatocyte proliferation in the liver cancer [67].
2.5. Role of KLF10 in PDAC Progression
Studies have revealed associations between PDAC and alterations in TGF-β receptor genes and SMAD [68,69]. However, no alterations in KLF10 expression were found in a mutation screening study of 22 human pancreatic cancer cell lines [70]. KLF10 expression in various cancer tissues has been reported to be significantly lower than that in normal tissues [63,71]. In PDAC, KLF10 expression was low in two thirds of patients and was inversely correlated with the cancer stage [36,53]. Despite alterations in the TGF-β signaling pathway components in patients with PDAC, KLF 10 could regulate TGF-β signaling and inhibit epithelial cell proliferation in pancreatic cancer cells [72]. KLF10 expression can be increased by a noncoding RNA, lncRNA FLVCR1-AS1, by acting as a competitive endogenous RNA to sequester the inhibitory effects of miR-513c-5p or miR-514b-5p. Since lncRNA FLVCR1-AS1 is a direct transcriptional target of KLF10, this FLVCR1-AS1/KLF10 positive feedback loop can suppress PDAC progression [73].
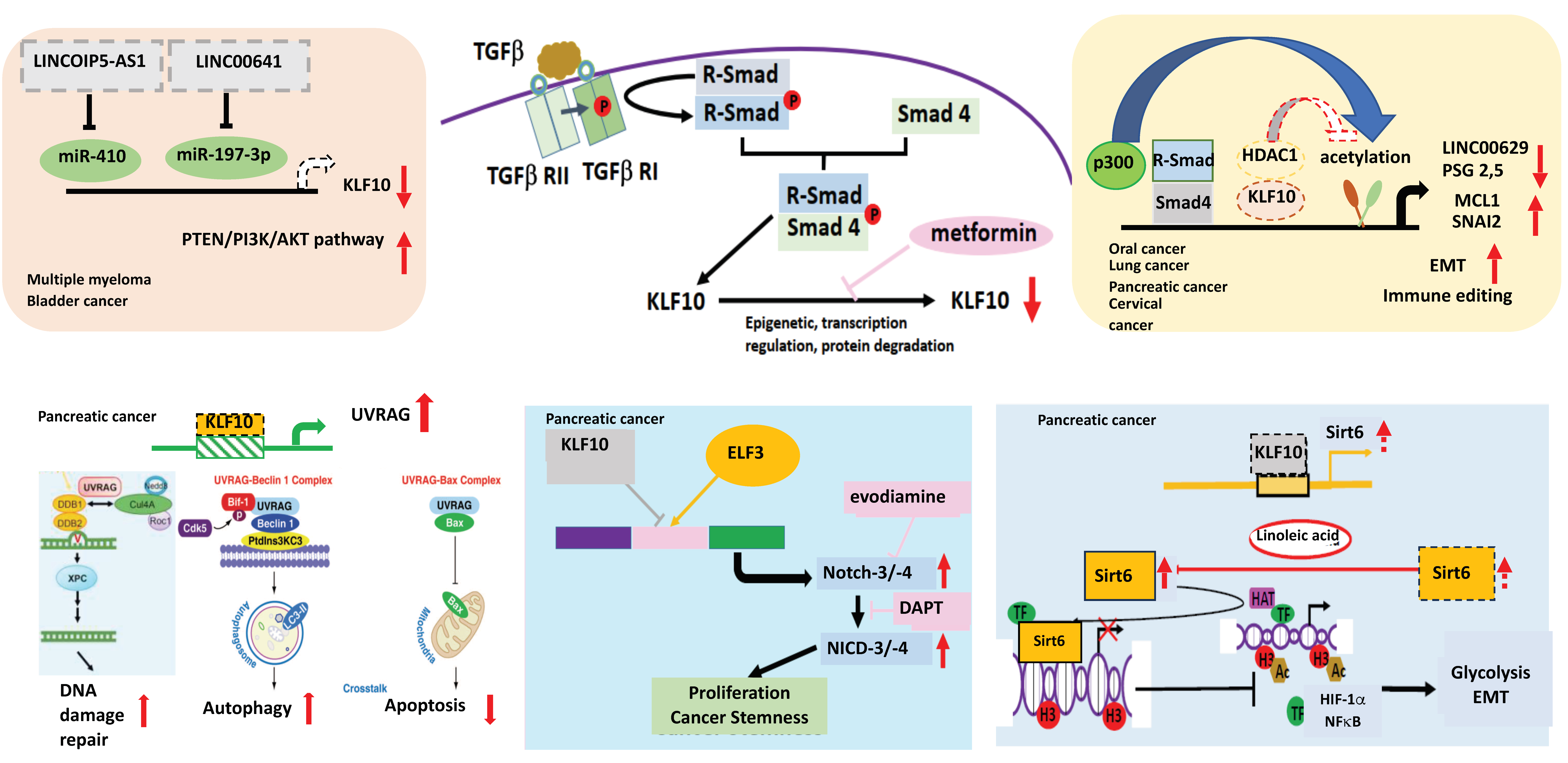
In the murine model of pancreas-specific KLF10 deletion (Pdx-1Cre KLF10L/L), no evidence of abnormal pancreas development or neoplastic lesions was noted. The synergistic effects of KLF10 inactivation-activated mutant KrasG12D in cross-breed mice led to the rapid onset of advanced PDAC with 50% penetrance. The upregulation of c-Jun and SDF-1/CXCR4 signaling after KLF10 deletion was responsible for accelerated PDAC cell growth and distant metastasis [74]. Since KLF10-knockout mice exhibited a high incidence of metabolic disorders, we previously explored sirtuin6, an NAD+-dependent deacetylase downstream of KLF10, as a key regulator of glucose homeostasis and a tumor suppressor. Our findings indicated that KLF10 transcriptionally activated sirtuin6 to modulate the EMT and glycolysis of PDAC coordinately through NFκB and HIF1α [75]. In addition to the Wnt/β-catenin signaling pathway, we demonstrated that KLF10 contributed to the cancer stemness phenotype by transcriptionally regulating Notch-3 and Notch-4 and competing with E74-like ETS transcription factor 3 (ELF3) for promoter binding. A combination of metformin, which upregulates KLF10 by phosphorylating AMP-activated protein kinase, and evodiamine, a nontoxic Notch-3 methylation stimulator, ameliorated PDAC growth through KLF10 downregulation [76] (Figure 2).
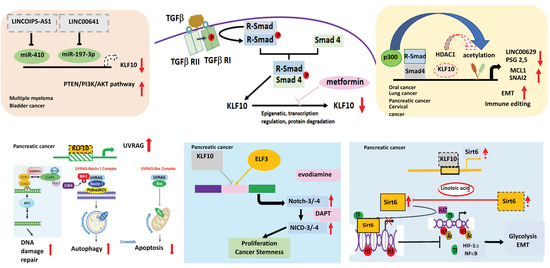
Figure 2.
The role of KLF10 in cancers. KLF10 can transcriptionally regulate lnc00629 to modulate MCL1 expression in oral cancer. KLF10 inhibits multiple myeloma (MM) and bladder cancer by increasing the expression of PTEN to reduce AKT activity. In MM, miR410, which can be suppressed by lncRNAOIP5-AS1 inhibits the expression of KLF10. In bladder cancer, lncRNA00641 can suppress miR-197-3p to modulate KLF10 transcription. In cervical cancer, KLF10 regulates PSG2,5 to modulate the tumor immune environment. KLF10 can inhibit the TGFβ-induced epithelial–mesenchymal transition (EMT) to suppress cancer invasion in the lung and pancreas by recruiting HDAC1 to block the expression of SNAI2. In pancreatic adenocarcinoma, KLF10 transcriptionally suppresses the UV radiation-associated gene (UVRAG) to modulate DNA damage repair, autophagy, and the apoptosis of cancer cells. KLF10 competes with E74-like ETS transcription factor 3 (ELF3) in binding to Notch-3 and -4 promoters to suppress cancer proliferation and the stemness phenotype. KLF10 can transcriptionally activate sirtuin6 to coordinate glycolysis and EMT of pancreatic cancer via HIF1α and NFkB. Upward arrows denotes increase; downward arrows represent decrease. Lines with blunt end means inhibit. Thicker lines represent prominent increase (or decrease) and vice versa.
2.6. Role of KLF10 in PDAC Resistance to Radiotherapy
The KLF family regulate radiosensitivity in various cancers (Table 2). KLF2 and KLF4 are positive regulators of endothelial-protective molecules such as nitric oxide and thrombomodulin. Compared with single-dose radiation, fractionated radiation markedly reduced the ERK5/KLF2 pathway and enhanced ICAM-1 expression, leading to endothelial dysfunction [77]. KLF4 and KLF5 may prevent radiation-induced intestinal injury by inhibiting apoptosis and modulating DNA repair pathways [78,79]. KLF4 expression can predict radiotherapy resistance and poor clinical outcomes for cervical cancer. From tumor tissues of 117 patients with locally advanced cervical cancer, KLF4 was disclosed as a risk factor for radioresistance (p = 0.032), poor PFS (p = 0.001), and OS (p < 0.001) [80]. KLF5 was the predictor of poor response to CRT in rectal cancer [81]. In colon cancer cells, radiation time-dependently and dose-dependently stabilized KLF5 levels. KLF5 increased cyclin D1 and β-catenin levels to mediate cell survival. A study assessing 60 colorectal tumor tissues before radiotherapy indicated that high KLF5 expression was correlated with pathologic complete remission (p = 0.023) and radioresistance in colorectal cancer [81]. High KLF6 expression level was associated with a nearly four times higher risk of local recurrence in head and neck cancer patients after radiotherapy (p = 0.008) [82].

Table 2.
KLFs in regulating the radiation sensitivity of cancers. Representative studies evaluating KLF family members in correlation with clinical outcomes of radiotherapy in various cancers.
KLF10 gene expression can be used to discriminate between γ-radiation and α-radiation quality [83]. Radiation-induced delayed neuropsychiatric disorders was associated with biological processes, such as protein kinase activity, circadian behavior, and cell differentiation. The alteration of expression levels of six genes, including KLF10, in the chronic phase of radiation increased anxiety-like behaviors in mice [84]. Radiation-induced KLF10 upregulation was noted in many cancer cell lines and murine models [83,84,85]. KLF10 transcriptionally downregulated EGFR and modulated gemcitabine-resistance in cholangiocarcinoma [86]. In esophageal squamous cell carcinoma, exosomes secreted from hypoxic tumors after radiation expressed high levels of miR-340-5p, which suppressed KLF10 transcription. Higher miR-340-5p expression and lower KLF10 expression in plasma exosomes from patients with esophageal cancer patients were associated with poorer radiation responses and prognosis [33]. Several studies, including ours, have demonstrated that KLF10 transcriptionally suppresses the UV radiation resistance-associated gene (UVRAG) and modulates apoptosis, DNA repair, and autophagy in cancer cells. Metformin might decrease radioresistance in pancreatic and esophageal cancers by elevating KLF10 expression [33,35]. Furthermore, EMT and cancer stem cell phenotypes also contribute to radioresistance [87,88]. KLF10 modulates EMT and can lead to cancer stemness phenotypes by transcriptionally regulating sirtuin6, Notch-3, and Notch-4, respectively, and thus may cause radioresistance in PDAC [74,75,76]. Whether KLF family members share promoter binding sites on UVRAG or other signal targets and regulate the balance between radiosensitivity and radioresistance warrants further exploration.
2.7. Selection of Patients with Resectable PDAC for Radiotherapy Using KLF10 and SMAD4
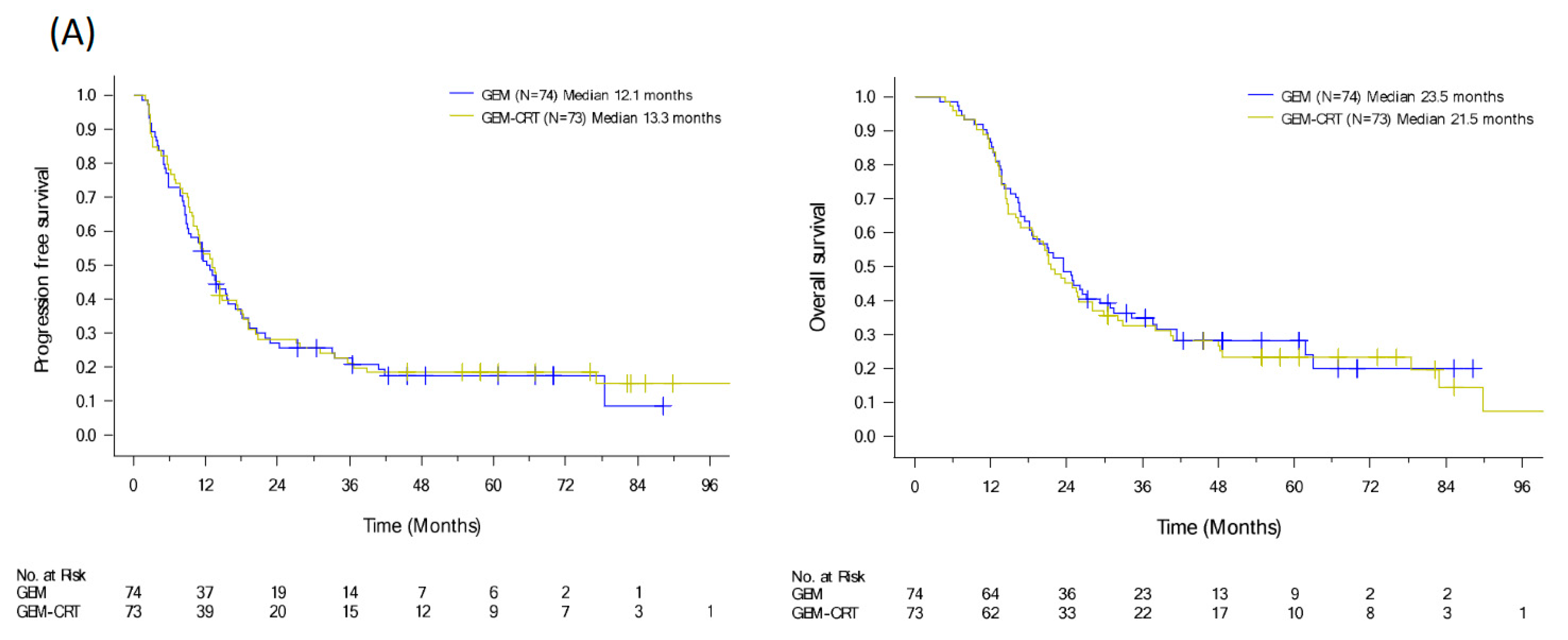
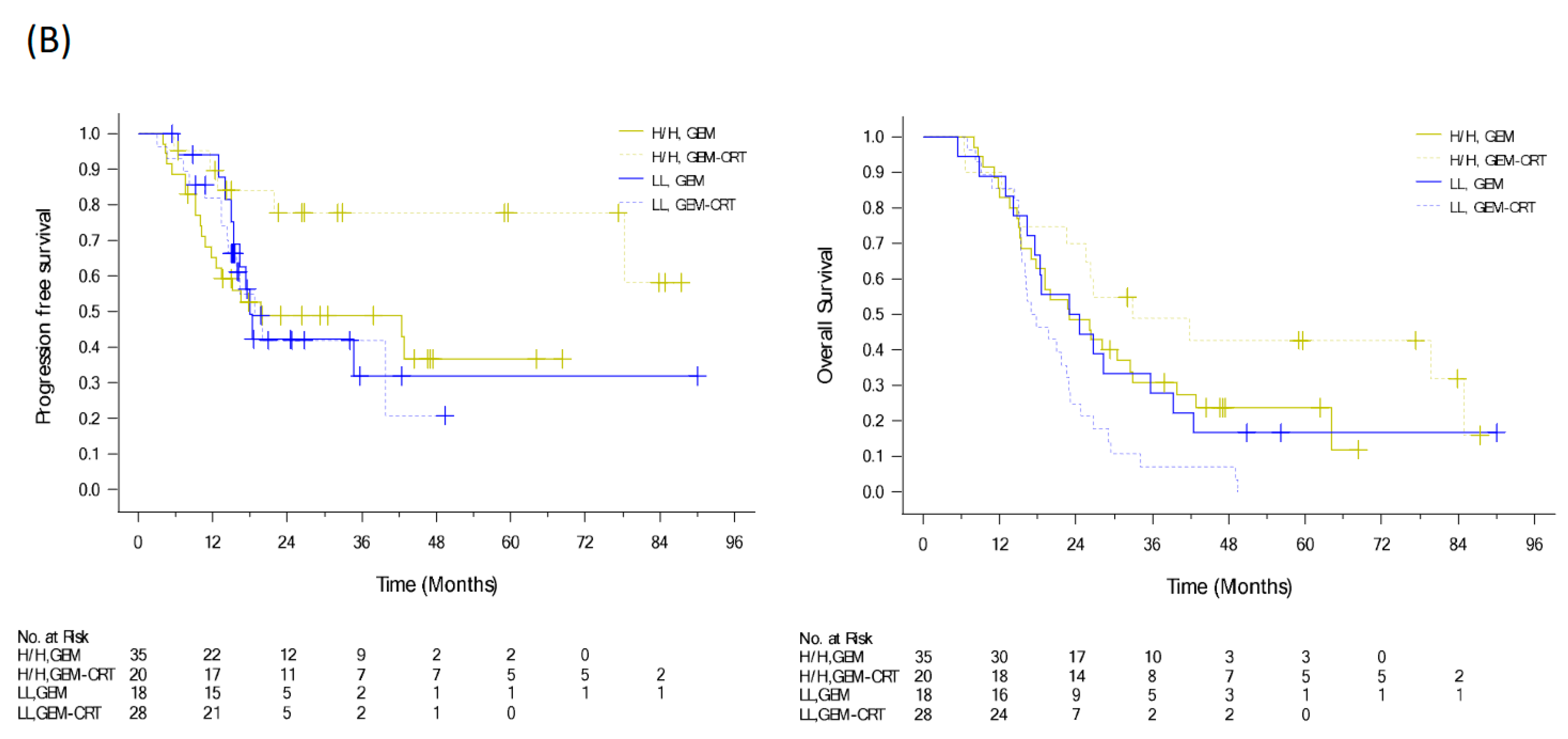
To evaluate the benefits of additional CRT to standard adjuvant chemotherapy in patients with resected PDAC, we conducted a randomized clinical trial from 2009 to 2015 [12]. We enrolled 147 patients with PDAC after curative resection and randomized them to either adjuvant GEM 1000 mg/m2 infusion weekly for six cycles or adjuvant GEM for three cycles and GEM (400 mg/m2 weekly)-based CRT and another three cycles of GEM. Despite the significant locoregional benefit (p = 0.039) of additional CRT, the median recurrence-free survival and OS were of no significant difference in the two arms (HR: 0.98, p = 0.89 and HR: 1.04, p = 0.82), respectively (Figure 3A) [12]. Tumor specimens were collected from 111 patients. Immunohistochemical expression of biomarkers including KLF10, SMAD4, and RUNX3 was evaluated by pathologists using a visual grading system based on staining intensity and extent. The postoperative CA19-9 level and protein expression of KLF10 and SAMD4, were significantly associated with OS (p = 0.047, 0.013, and 0.045, respectively). High KLF10 or SMAD4 expression in patients (n = 55) receiving additional adjuvant CRT had a significantly prolonged local control time (ꚙ vs. 19.8 months, p = 0.026) and a better OS (33.0 vs. 23.0 months, p = 0.12) than those receiving GEM alone. In resected PDAC patients who had a loss of both SMAD4 and KLF10, additional adjuvant CRT caused the rapid development of distant metastasis and worse clinical outcomes (Figure 3B) [30]. The combination of KLF10 and CA19-9 levels did not reveal significant differences in survival outcomes between the treatment arms [30]. On the basis of these findings, we concluded that the chances of translating locoregional control of CRT into prolonged survival were high in patients with KLF10- or SMAD4-expressing tumors. Although these findings are promising, a prospective study is warranted to validate the results.
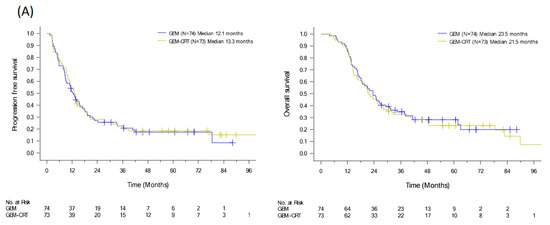
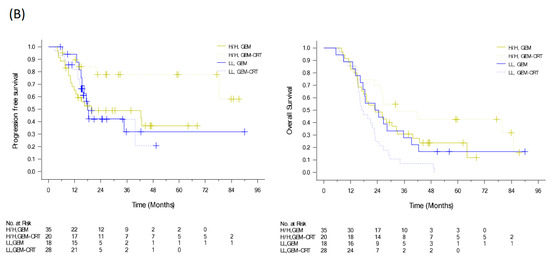
Figure 3.
PFS and OS curves of (A) 147 resectable pancreatic cancer patients randomized to adjuvant gemcitabine (GEM, n = 74) with or without additional adjuvant gemcitabine-based chemoradiotherapy (GEM-CRT, n = 73). (B) In total, 101 patients enrolled to the above mentioned clinical trial with qualified tumor tissues. The levels of KLF10 and SMAD4 were evaluated by two pathologists blinded to the clinical information. Patients with high expression of KLF10 or SMAD4 (H/H, n = 55) should receive additional adjuvant CRT than GEM only due to a significantly better PFS (NA vs. 19.8 months; p = 0.026) and a longer OS (33.0 vs. 23.0 months; p = 0.12). Conversely, adjuvant CRT after curative resection may not be suitable in those with low expression of both KLF10 and SMAD4 (LL) who develop distant metastasis rapidly.
3. Conclusions
The value of radiotherapy in PDAC remains unclear due to conflicting results of clinical trials [89,90,91]. Modern radiotherapy is efficacious, resulting in a satisfactory safety profile and local control for patients with localized PDAC [18,19]. Locoregional control of the primary tumor is crucial for patients with PDAC and is increasingly possible with advancements in chemotherapy [92]. Retrospective studies have identified potential tissue biomarkers for predicting the benefits of enhanced locoregional therapy. However, most candidate biomarkers were only correlated with survival but not with failure patterns. Thus, prospective clinical trials in patients with PDAC receiving modern chemotherapy with or without up-to-date radiotherapy are required to validate the efficacy of biomarkers in selecting optimal patients for radiotherapy.
In preclinical studies, KLF10 was demonstrated to be correlated with PDAC progression and resistance and it was reported to modulate distant metastasis, cancer stemness, and radio-sensitivity. A retrospective analysis of prospective randomized trials concluded that the combination of KLF10 and SMAD4 expression can help select patients with resected PDAC who may be suitable for local radiotherapy. Current enthusiasm of upfront systemic chemotherapy for localized PDAC aims to prevent patients with rapid distant metastasis from radical local therapy including radiotherapy. Future trials evaluating the efficacy of radiotherapy in PDAC should focus on molecular biomarker-enriched patients who carry a low risk of early distant metastasis.
Author Contributions
Conceptualization, H.-J.C. and Y.-C.T.; methodology, Y.-C.T. and R.-J.L.; software, M.-C.H. and T.-W.L.; validation, Y.-C.T. and M.-C.H.; formal analysis, R.-J.L. and T.-W.L.; investigation, H.-J.C. and T.-W.L.; resources, H.-J.C.; data curation, R.-J.L. and T.-W.L.; writing—original draft preparation, Y.-C.T.; writing—review and editing, H.-J.C.; visualization, R.-J.L. and H.-J.C.; supervision, H.-J.C.; project administration, R.-J.L.; funding acquisition, H.-J.C. All authors have read and agreed to the published version of the manuscript.
Funding
National Health Research Institutes, Taiwan (CA-112-PP 14).
Conflicts of Interest
The authors declare no conflict of interest.
References
- American Cancer Society. Survival Rates for Pancreatic Cancer. 2023. Available online: https://www.cancer.org/cancer/types/pancreatic-cancer/detection-diagnosis-staging/survival-rates.html (accessed on 12 June 2023).
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Stein, S.M.; James, E.S.; Deng, Y.; Cong, X.; Kortmansky, J.S.; Li, J.; Staugaard, C.; Indukala, D.; Boustani, A.M.; Patel, V.; et al. Final analysis of a phase II study of modified FOLFIRINOX in locally advanced and metastatic pancreatic cancer. Br. J. Cancer 2016, 114, 737–743. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Chone, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Palmer, D.H.; Ghaneh, P.; Psarelli, E.E.; Valle, J.W.; Halloran, C.M.; Faluyi, O.; O’Reilly, D.A.; Cunningham, D.; Wadsley, J.; et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): A multicentre, open-label, randomised, phase 3 trial. Lancet 2017, 389, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Tempero, M.A.; Pelzer, U.; O’Reilly, E.M.; Winter, J.; Oh, D.Y.; Li, C.P.; Tortora, G.; Chang, H.M.; Lopez, C.D.; Bekaii-Saab, T.; et al. Adjuvant nab-Paclitaxel + Gemcitabine in Resected Pancreatic Ductal Adenocarcinoma: Results From a Randomized, Open-Label, Phase III Trial. J. Clin. Oncol. 2023, 41, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.A.; Duong, M.; Sohal, D.P.S.; Gandhi, N.S.; Beg, M.S.; Wang-Gillam, A.; Wade, J.L., 3rd; Chiorean, E.G.; Guthrie, K.A.; Lowy, A.M.; et al. Surgical Outcome Results From SWOG S1505: A Randomized Clinical Trial of mFOLFIRINOX Versus Gemcitabine/Nab-paclitaxel for Perioperative Treatment of Resectable Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2020, 272, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Versteijne, E.; Suker, M.; Groothuis, K.; Akkermans-Vogelaar, J.M.; Besselink, M.G.; Bonsing, B.A.; Buijsen, J.; Busch, O.R.; Creemers, G.M.; van Dam, R.M.; et al. Preoperative Chemoradiotherapy Versus Immediate Surgery for Resectable and Borderline Resectable Pancreatic Cancer: Results of the Dutch Randomized Phase III PREOPANC Trial. J. Clin. Oncol. 2020, 38, 1763–1773. [Google Scholar] [CrossRef]
- Versteijne, E.; van Dam, J.L.; Suker, M.; Janssen, Q.P.; Groothuis, K.; Akkermans-Vogelaar, J.M.; Besselink, M.G.; Bonsing, B.A.; Buijsen, J.; Busch, O.R.; et al. Neoadjuvant Chemoradiotherapy Versus Upfront Surgery for Resectable and Borderline Resectable Pancreatic Cancer: Long-Term Results of the Dutch Randomized PREOPANC Trial. J. Clin. Oncol. 2022, 40, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.A.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-J.; Chiu, Y.-F.; Chen, J.-S.; Li, C.; Ho, C.; Shyr, Y.; Chiou, W.; Yeh, C.; Hsieh, R.; Lin, Y.; et al. Randomized, phase III trial comparing adjuvant gemcitabine (Gem) versus Gem plus chemoradiation (CCRT) in curatively resected pancreatic ductal adenocarcinoma (PDAC): A Taiwan cooperative oncology group study. Ann. Oncol. 2018, 29 (Suppl. S8), viii210. [Google Scholar] [CrossRef]
- Ghaneh, P.; Palmer, D.; Cicconi, S.; Jackson, R.; Halloran, C.M.; Rawcliffe, C.; Sripadam, R.; Mukherjee, S.; Soonawalla, Z.; Wadsley, J.; et al. Immediate surgery compared with short-course neoadjuvant gemcitabine plus capecitabine, FOLFIRINOX, or chemoradiotherapy in patients with borderline resectable pancreatic cancer (ESPAC5): A four-arm, multicentre, randomised, phase 2 trial. Lancet Gastroenterol. Hepatol. 2023, 8, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.H.G.; Shi, Q.; Meyers, J.; Herman, J.M.; Chuong, M.; Wolpin, B.M.; Ahmad, S.; Marsh, R.; Schwartz, L.; Behr, S.; et al. Efficacy of Preoperative mFOLFIRINOX vs mFOLFIRINOX Plus Hypofractionated Radiotherapy for Borderline Resectable Adenocarcinoma of the Pancreas: The A021501 Phase 2 Randomized Clinical Trial. JAMA Oncol. 2022, 8, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Hammel, P.; Huguet, F.; van Laethem, J.L.; Goldstein, D.; Glimelius, B.; Artru, P.; Borbath, I.; Bouche, O.; Shannon, J.; Andre, T.; et al. Effect of Chemoradiotherapy vs Chemotherapy on Survival in Patients With Locally Advanced Pancreatic Cancer Controlled After 4 Months of Gemcitabine With or Without Erlotinib: The LAP07 Randomized Clinical Trial. JAMA 2016, 315, 1844–1853. [Google Scholar] [CrossRef]
- Loehrer, P.J., Sr.; Feng, Y.; Cardenes, H.; Wagner, L.; Brell, J.M.; Cella, D.; Flynn, P.; Ramanathan, R.K.; Crane, C.H.; Alberts, S.R.; et al. Gemcitabine alone versus gemcitabine plus radiotherapy in patients with locally advanced pancreatic cancer: An Eastern Cooperative Oncology Group trial. J. Clin. Oncol. 2011, 29, 4105–4112. [Google Scholar] [CrossRef] [PubMed]
- Iacobuzio-Donahue, C.A.; Fu, B.; Yachida, S.; Luo, M.; Abe, H.; Henderson, C.M.; Vilardell, F.; Wang, Z.; Keller, J.W.; Banerjee, P.; et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J. Clin. Oncol. 2009, 27, 1806–1813. [Google Scholar] [CrossRef]
- Murphy, J.E.; Wo, J.Y.; Ryan, D.P.; Clark, J.W.; Jiang, W.; Yeap, B.Y.; Drapek, L.C.; Ly, L.; Baglini, C.V.; Blaszkowsky, L.S.; et al. Total Neoadjuvant Therapy With FOLFIRINOX in Combination With Losartan Followed by Chemoradiotherapy for Locally Advanced Pancreatic Cancer: A Phase 2 Clinical Trial. JAMA Oncol. 2019, 5, 1020–1027. [Google Scholar] [CrossRef]
- Reyngold, M.; O’Reilly, E.M.; Varghese, A.M.; Fiasconaro, M.; Zinovoy, M.; Romesser, P.B.; Wu, A.; Hajj, C.; Cuaron, J.J.; Tuli, R.; et al. Association of Ablative Radiation Therapy with Survival Among Patients With Inoperable Pancreatic Cancer. JAMA Oncol. 2021, 7, 735–738. [Google Scholar] [CrossRef]
- Taherian, M.; Wang, H.; Wang, H. Pancreatic Ductal Adenocarcinoma: Molecular Pathology and Predictive Biomarkers. Cells 2022, 11, 3068. [Google Scholar] [CrossRef] [PubMed]
- Strom, T.; Hoffe, S.E.; Fulp, W.; Frakes, J.; Coppola, D.; Springett, G.M.; Malafa, M.P.; Harris, C.L.; Eschrich, S.A.; Torres-Roca, J.F.; et al. Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother. Oncol. 2015, 117, 159–164. [Google Scholar] [CrossRef]
- Scott, J.G.; Sedor, G.; Ellsworth, P.; Scarborough, J.A.; Ahmed, K.A.; Oliver, D.E.; Eschrich, S.A.; Kattan, M.W.; Torres-Roca, J.F. Pan-cancer prediction of radiotherapy benefit using genomic-adjusted radiation dose (GARD): A cohort-based pooled analysis. Lancet Oncol. 2021, 22, 1221–1229. [Google Scholar] [CrossRef]
- Scott, J.G.; Berglund, A.; Schell, M.J.; Mihaylov, I.; Fulp, W.J.; Yue, B.; Welsh, E.; Caudell, J.J.; Ahmed, K.; Strom, T.S.; et al. A genome-based model for adjusting radiotherapy dose (GARD): A retrospective, cohort-based study. Lancet Oncol. 2017, 18, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Nevler, A.; Muller, A.J.; Sutanto-Ward, E.; DuHadaway, J.B.; Nagatomo, K.; Londin, E.; O’Hayer, K.; Cozzitorto, J.A.; Lavu, H.; Yeo, T.P.; et al. Host IDO2 Gene Status Influences Tumor Progression and Radiotherapy Response in KRAS-Driven Sporadic Pancreatic Cancers. Clin. Cancer Res. 2019, 25, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Okano, K.; Sato, K.; Sugimoto, M.; Shimomura, A.; Nagao, M.; Matsukawa, H.; Ando, Y.; Suto, H.; Oshima, M.; et al. Tumor metabolic alterations after neoadjuvant chemoradiotherapy predict postoperative recurrence in patients with pancreatic cancer. Jpn. J. Clin. Oncol. 2022, 52, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.S.; Chang, J.H.; Jeon, S.H.; Song, M.G.; Lee, K.H.; Im, S.A.; Kim, J.I.; Kim, T.Y.; Chie, E.K. Radiation Response Prediction Model Based on Integrated Clinical and Genomic Data Analysis. Cancer Res. Treat. 2022, 54, 383–395. [Google Scholar] [CrossRef]
- Crane, C.H.; Varadhachary, G.R.; Yordy, J.S.; Staerkel, G.A.; Javle, M.M.; Safran, H.; Haque, W.; Hobbs, B.D.; Krishnan, S.; Fleming, J.B.; et al. Phase II trial of cetuximab, gemcitabine, and oxaliplatin followed by chemoradiation with cetuximab for locally advanced (T4) pancreatic adenocarcinoma: Correlation of Smad4(Dpc4) immunostaining with pattern of disease progression. J. Clin. Oncol. 2011, 29, 3037–3043. [Google Scholar] [CrossRef]
- Shin, S.H.; Kim, H.J.; Hwang, D.W.; Lee, J.H.; Song, K.B.; Jun, E.; Shim, I.K.; Hong, S.M.; Kim, H.J.; Park, K.M.; et al. The DPC4/SMAD4 genetic status determines recurrence patterns and treatment outcomes in resected pancreatic ductal adenocarcinoma: A prospective cohort study. Oncotarget 2017, 8, 17945–17959. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.C.; Izeradjene, K.; Rani, P.G.; Feng, L.; Carlson, M.A.; DelGiorno, K.E.; Wood, L.D.; Goggins, M.; Hruban, R.H.; Chang, A.E.; et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell 2015, 161, 1345–1360. [Google Scholar] [CrossRef]
- Pen, S.L.; Shan, Y.S.; Hsiao, C.F.; Liu, T.W.; Chen, J.S.; Ho, C.L.; Chou, W.C.; Hsieh, R.K.; Chen, L.T.; Ch’ang, H.J. High expression of kruppel-like factor 10 or Smad4 predicts clinical benefit of adjuvant chemoradiotherapy in curatively resected pancreatic adenocarcinoma: From a randomized phase III trial. Radiother. Oncol. 2021, 158, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xia, X.; Yang, C.; Shen, J.; Mai, J.; Kim, H.C.; Kirui, D.; Kang, Y.; Fleming, J.B.; Koay, E.J.; et al. SMAD4 Gene Mutation Renders Pancreatic Cancer Resistance to Radiotherapy through Promotion of Autophagy. Clin. Cancer Res. 2018, 24, 3176–3185. [Google Scholar] [CrossRef]
- Hu, B.; Ma, X.; Huang, R.; Wu, Z.; Lu, J.; Guo, Y.; Tang, J.; Ma, C.; Ma, J.; Zhang, L.; et al. Identification of Key Genes Mutations Associated With the Radiosensitivity by Whole Exome Sequencing in Pancreatic Cancer. Front. Oncol. 2021, 11, 697308. [Google Scholar] [CrossRef]
- Chen, F.; Xu, B.; Li, J.; Yang, X.; Gu, J.; Yao, X.; Sun, X. Hypoxic tumour cell-derived exosomal miR-340-5p promotes radioresistance of oesophageal squamous cell carcinoma via KLF10. J. Exp. Clin. Cancer Res. 2021, 40, 38. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Yeh, K.T.; Yeh, C.M.; Soon, M.S.; Hsu, L.S. KLF10 Functions as an Independent Prognosis Factor for Gastric Cancer. Medicina 2022, 58, 711. [Google Scholar] [CrossRef]
- Chang, V.H.; Tsai, Y.C.; Tsai, Y.L.; Peng, S.L.; Chen, S.L.; Chang, T.M.; Yu, W.C.; Ch’ang, H.J. Krupple-like factor 10 regulates radio-sensitivity of pancreatic cancer via UV radiation resistance-associated gene. Radiother. Oncol. 2017, 122, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.H.; Chu, P.Y.; Peng, S.L.; Mao, T.L.; Shan, Y.S.; Hsu, C.F.; Lin, C.Y.; Tsai, K.K.; Yu, W.C.; Ch’ang, H.J. Kruppel-like factor 10 expression as a prognostic indicator for pancreatic adenocarcinoma. Am. J. Pathol. 2012, 181, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Suske, G.; Bruford, E.; Philipsen, S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics 2005, 85, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Lomberk, G.; Urrutia, R. The family feud: Turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 2005, 392, 1–11. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Do, D.T.; Nguyen, T.T.; Le, Q.A. A sequence-based prediction of Kruppel-like factors proteins using XGBoost and optimized features. Gene 2021, 787, 145643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, C.; Ju, Z.; Jiao, D.; Hu, D.; Qi, L.; Liu, S.; Wu, X.; Zhao, C. Kruppel-like factors in tumors: Key regulators and therapeutic avenues. Front. Oncol. 2023, 13, 1080720. [Google Scholar] [CrossRef]
- Subramaniam, M.; Harris, S.A.; Oursler, M.J.; Rasmussen, K.; Riggs, B.L.; Spelsberg, T.C. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995, 23, 4907–4912. [Google Scholar] [CrossRef]
- Luo, H.Y.; Zhu, J.Y.; Chen, M.; Mu, W.J.; Guo, L. Kruppel-like factor 10 (KLF10) as a critical signaling mediator: Versatile functions in physiological and pathophysiological processes. Genes Dis. 2023, 10, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, S.A.; Subramaniam, M.; Monroe, D.G.; Janknecht, R.; Spelsberg, T.C. Modulation of transforming growth factor beta (TGFbeta)/Smad transcriptional responses through targeted degradation of TGFbeta-inducible early gene-1 by human seven in absentia homologue. J. Biol. Chem. 2002, 277, 30754–30759. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.F.; Sui, C.L.; Wu, W.C.; Wang, J.J.; Yang, D.H.; Chen, Y.C.; Yu, W.C.; Chang, H.S. Klf10 induces cell apoptosis through modulation of BI-1 expression and Ca2+ homeostasis in estrogen-responding adenocarcinoma cells. Int. J. Biochem. Cell Biol. 2011, 43, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Hsu, Y.C.; Huang, Y.T.; Shih, Y.H.; Wang, C.J.; Chiang, W.C.; Chang, P.J. A KDM6A-KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction. EMBO Mol. Med. 2019, 11, e9828. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef]
- Hwang, Y.C.; Yang, C.H.; Lin, C.H.; Ch’ang, H.J.; Chang, V.H.S.; Yu, W.C.Y. Destabilization of KLF10, a tumor suppressor, relies on thr93 phosphorylation and isomerase association. Biochim. Biophys. Acta 2013, 1833, 3035–3045. [Google Scholar] [CrossRef]
- Subramaniam, M.; Hawse, J.R.; Johnsen, S.A.; Spelsberg, T.C. Role of TIEG1 in biological processes and disease states. J. Cell. Biochem. 2007, 102, 539–548. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Ho, C.; Shih, Y.H.; Ni, W.C.; Li, Y.C.; Chang, H.C.; Lin, C.L. Knockout of KLF10 Ameliorated Diabetic Renal Fibrosis via Downregulation of DKK-1. Molecules 2022, 27, 2644. [Google Scholar] [CrossRef]
- Gutierrez-Aguilar, R.; Benmezroua, Y.; Balkau, B.; Marre, M.; Helbecque, N.; Charpentier, G.; Polychronakos, C.; Sladek, R.; Froguel, P.; Neve, B. Minor contribution of SMAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab. 2007, 33, 372–378. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, R.J.; Peng, S.Y.; Yu, W.C.Y.; Chang, V.H. Therapeutic Targeting of Nonalcoholic Fatty Liver Disease by Downregulating SREBP-1C Expression via AMPK-KLF10 Axis. Front. Mol. Biosci. 2021, 8, 751938. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Jia, L.; Xiang, J.; Yang, G.; Qiu, S.; Kang, L.; Zheng, P.; Liang, Z.; Lu, Y. KLF10 promotes nonalcoholic steatohepatitis progression through transcriptional activation of zDHHC7. EMBO Rep. 2022, 23, e54229. [Google Scholar] [CrossRef]
- Hawse, J.R.; Cicek, M.; Grygo, S.B.; Bruinsma, E.S.; Rajamannan, N.M.; van Wijnen, A.J.; Lian, J.B.; Stein, G.S.; Oursler, M.J.; Subramaniam, M.; et al. TIEG1/KLF10 modulates Runx2 expression and activity in osteoblasts. PLoS ONE 2011, 6, e19429. [Google Scholar] [CrossRef]
- Li, Q.; Shen, P.; Zeng, S.; Liu, P. TIEG1 Inhibits Angiotensin II-induced Cardiomyocyte Hypertrophy by Inhibiting Transcription Factor GATA4. J. Cardiovasc. Pharmacol. 2015, 66, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Wara, A.K.; Rawal, S.; Yang, X.; Perez-Cremades, D.; Sachan, M.; Chen, J.; Feinberg, M.W. KLF10 deficiency in CD4(+) T cells promotes atherosclerosis progression by altering macrophage dynamics. Atherosclerosis 2022, 359, 27–41. [Google Scholar] [CrossRef]
- Cao, Z.; Wara, A.K.; Icli, B.; Sun, X.; Packard, R.R.; Esen, F.; Stapleton, C.J.; Subramaniam, M.; Kretschmer, K.; Apostolou, I.; et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(−) T cells and T regulatory cells. J. Biol. Chem. 2009, 284, 24914–24924. [Google Scholar] [CrossRef]
- Memon, A.; Lee, W.K. KLF10 as a Tumor Suppressor Gene and Its TGF-beta Signaling. Cancers 2018, 10, 161. [Google Scholar] [CrossRef]
- Engelmann, D.; Knoll, S.; Ewerth, D.; Steder, M.; Stoll, A.; Putzer, B.M. Functional interplay between E2F1 and chemotherapeutic drugs defines immediate E2F1 target genes crucial for cancer cell death. Cell. Mol. Life Sci. 2010, 67, 931–948. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chen, J.; Zhang, H.; Wang, X.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef]
- Li, Z.; Hong, S.; Liu, Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem. Biophys. Res. Commun. 2018, 503, 1825–1829. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.; Zhang, H.; Liu, H.; Yao, H.; Wang, X.; Zhang, W.; Zhao, Y.; Yang, N. KLF10 inhibits cell growth by regulating PTTG1 in multiple myeloma under the regulation of microRNA-106b-5p. Int. J. Biol. Sci. 2020, 16, 2063–2071. [Google Scholar] [CrossRef]
- Mishra, V.K.; Subramaniam, M.; Kari, V.; Pitel, K.S.; Baumgart, S.J.; Naylor, R.M.; Nagarajan, S.; Wegwitz, F.; Ellenrieder, V.; Hawse, J.R.; et al. Kruppel-like Transcription Factor KLF10 Suppresses TGFbeta-Induced Epithelial-to-Mesenchymal Transition via a Negative Feedback Mechanism. Cancer Res. 2017, 77, 2387–2400. [Google Scholar] [CrossRef]
- Zhang, C.; Dang, D.; Wang, H.; Shi, S.; Dai, J.; Yang, M. Acircadian rhythm-related gene signature for predicting survival and drug response in HNSC. Front. Immunol. 2022, 13, 1029676. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Ma, C.; Ren, C.; Li, N.; Liu, X.; Zhang, Y.; Wang, Y.; Li, X.; Lv, P.; Han, C.; et al. LINC00629, a KLF10-responsive lncRNA, promotes the anticancer effects of apigenin by decreasing Mcl1 stability in oral squamous cell carcinoma. Aging 2022, 14, 9149–9166. [Google Scholar] [CrossRef] [PubMed]
- Marrero-Rodriguez, D.; Taniguchi-Ponciano, K.; Subramaniam, M.; Hawse, J.R.; Pitel, K.S.; Arreola-De la Cruz, H.; Huerta-Padilla, V.; Ponce-Navarrete, G.; Figueroa-Corona, M.D.P.; Gomez-Virgilio, L.; et al. Kruppel-Like Factor 10 participates in cervical cancer immunoediting through transcriptional regulation of Pregnancy-Specific Beta-1 Glycoproteins. Sci. Rep. 2018, 8, 9445. [Google Scholar] [CrossRef]
- Heo, S.H.; Jeong, E.S.; Lee, K.S.; Seo, J.H.; Lee, W.K.; Choi, Y.K. Kruppel-like factor 10 null mice exhibit lower tumor incidence and suppressed cellular proliferation activity following chemically induced liver tumorigenesis. Oncol. Rep. 2015, 33, 2037–2044. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Padua, D.; Massague, J. Roles of TGFbeta in metastasis. Cell Res. 2009, 19, 89–102. [Google Scholar] [CrossRef]
- Antonello, D.; Moore, P.S.; Zamboni, G.; Falconi, M.; Scarpa, A. Absence of mutations in the transforming growth factor-beta inducible early gene 1, TIEG1, in pancreatic cancer. Cancer Lett. 2002, 183, 179–183. [Google Scholar] [CrossRef]
- Subramaniam, M.; Hefferan, T.E.; Tau, K.; Peus, D.; Pittelkow, M.; Jalal, S.; Riggs, B.L.; Roche, P.; Spelsberg, T.C. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J. Cell. Biochem. 1998, 68, 226–236. [Google Scholar] [CrossRef]
- Cook, T.; Urrutia, R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 278, G513–G521. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhai, S.; Zou, S.; Xu, Z.; Zhang, J.; Jiang, L.; Deng, X.; Chen, H.; Peng, C.; Zhang, J.; et al. Positive feedback between lncRNA FLVCR1-AS1 and KLF10 may inhibit pancreatic cancer progression via the PTEN/AKT pathway. J. Exp. Clin. Cancer Res. 2021, 40, 316. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.C.; Hawse, J.R.; Subramaniam, M.; Chang, V.H.S.; Yu, W.C.Y.; Hung, W.C.; Chen, L.T.; Cheng, K.H. KLF10 loss in the pancreas provokes activation of SDF-1 and induces distant metastases of pancreatic ductal adenocarcinoma in the Kras(G12D) p53(flox/flox) model. Oncogene 2017, 36, 5532–5543. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Chen, S.L.; Peng, S.L.; Tsai, Y.L.; Chang, Z.M.; Chang, V.H.; Ch’ang, H.J. Upregulating sirtuin 6 ameliorates glycolysis, EMT and distant metastasis of pancreatic adenocarcinoma with kruppel-like factor 10 deficiency. Exp. Mol. Med. 2021, 53, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Cheng, K.H.; Jiang, S.S.; Hawse, J.R.; Chuang, S.E.; Chen, S.L.; Huang, T.S.; Ch’ang, H.J. Kruppel-like factor 10 modulates stem cell phenotypes of pancreatic adenocarcinoma by transcriptionally regulating notch receptors. J. Biomed. Sci. 2023, 30, 39. [Google Scholar] [CrossRef]
- Sadhukhan, R.; Leung, J.W.C.; Garg, S.; Krager, K.J.; Savenka, A.V.; Basnakian, A.G.; Pathak, R. Fractionated radiation suppresses Kruppel-like factor 2 pathway to a greater extent than by single exposure to the same total dose. Sci. Rep. 2020, 10, 7734. [Google Scholar] [CrossRef] [PubMed]
- Talmasov, D.; Xinjun, Z.; Yu, B.; Nandan, M.O.; Bialkowska, A.B.; Elkarim, E.; Kuruvilla, J.; Yang, V.W.; Ghaleb, A.M. Kruppel-like factor 4 is a radioprotective factor for the intestine following gamma-radiation-induced gut injury in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G121–G138. [Google Scholar] [CrossRef]
- Li, M.; Gu, Y.; Ma, Y.C.; Shang, Z.F.; Wang, C.; Liu, F.J.; Cao, J.P.; Wan, H.J.; Zhang, X.G. Kruppel-Like Factor 5 Promotes Epithelial Proliferation and DNA Damage Repair in the Intestine of Irradiated Mice. Int. J. Biol. Sci. 2015, 11, 1458–1468. [Google Scholar] [CrossRef]
- Liu, H.X.; Li, N.; Wei, L.; Zhou, F.X.; Ma, R.; Xiao, F.; Zhang, W.; Zhang, Y.; Hui, Y.P.; Song, H.; et al. High expression of Kruppel-like factor 4 as a predictor of poor prognosis for cervical cancer patient response to radiotherapy. Tumour. Biol. 2017, 39, 1010428317710225. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.G.; Kim, K.S.; Choi, Y.H.; Kim, N.K. The Kruppel-like factor (KLF5) as a predictive biomarker in preoperative chemoradiation therapy for rectal cancer. Ann. Surg. Treat. Res. 2019, 97, 83–92. [Google Scholar] [CrossRef]
- Leon, X.; Venegas, M.; Pujol, A.; Bulboa, C.; Llansana, A.; Casasayas, M.; Quer, M.; Camacho, M. Predictive value of transcriptional expression of Kruppel-like factor-6 (KLF6) in head and neck carcinoma patients treated with radiotherapy. Clin. Transl. Oncol. 2021, 23, 2507–2512. [Google Scholar] [CrossRef] [PubMed]
- Unverricht-Yeboah, M.; Giesen, U.; Kriehuber, R. Comparative gene expression analysis after exposure to 123I-iododeoxyuridine, gamma- and alpha-radiation-potential biomarkers for the discrimination of radiation qualities. J. Radiat. Res. 2018, 59, 411–429. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, J.S.; Moon, C.; Son, Y. Profiling of gene expression in the brain associated with anxiety-related behaviors in the chronic phase following cranial irradiation. Sci. Rep. 2022, 12, 13162. [Google Scholar] [CrossRef]
- Udayakumar, T.S.; Betancourt, D.M.; Ahmad, A.; Tao, W.; Totiger, T.M.; Patel, M.; Marples, B.; Barber, G.; Pollack, A. Radiation Attenuates Prostate Tumor Antiviral Responses to Vesicular Stomatitis Virus Containing IFNbeta, Resulting in Pronounced Antitumor Systemic Immune Responses. Mol. Cancer Res. 2020, 18, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J.; Yao, L.; Wu, L. Effect of Photodynamic Therapy on Gemcitabine-Resistant Cholangiocarcinoma in vitro and in vivo Through KLF10 and EGFR. Front. Cell Dev. Biol. 2021, 9, 710721. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Graham, P.H.; Hao, J.; Ni, J.; Bucci, J.; Cozzi, P.J.; Kearsley, J.H.; Li, Y. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013, 4, e875. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef]
- Choti, M.A. Adjuvant therapy for pancreatic cancer—The debate continues. N. Engl. J. Med. 2004, 350, 1249–1251. [Google Scholar] [CrossRef]
- Koshy, M.C.; Landry, J.C.; Cavanaugh, S.X.; Fuller, C.D.; Willett, C.G.; Abrams, R.A.; Hoffman, J.P.; Thomas, C.R., Jr. A challenge to the therapeutic nihilism of ESPAC-1. Int. J. Radiat. Oncol. Biol. Phys. 2005, 61, 965–966. [Google Scholar] [CrossRef]
- Abrams, R.A.; Winter, K.A.; Regine, W.F.; Safran, H.; Hoffman, J.P.; Lustig, R.; Konski, A.A.; Benson, A.B.; Macdonald, J.S.; Rich, T.A.; et al. Failure to adhere to protocol specified radiation therapy guidelines was associated with decreased survival in RTOG 9704—A phase III trial of adjuvant chemotherapy and chemoradiotherapy for patients with resected adenocarcinoma of the pancreas. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 809–816. [Google Scholar] [CrossRef]
- Hall, W.A.; Goodman, K.A. Radiation therapy for pancreatic adenocarcinoma, a treatment option that must be considered in the management of a devastating malignancy. Radiat. Oncol. 2019, 14, 114. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).