The Clinical Impact of Death Domain-Associated Protein and Holliday Junction Recognition Protein Expression in Cancer: Unmasking the Driving Forces of Neoplasia
Abstract
Simple Summary
Abstract
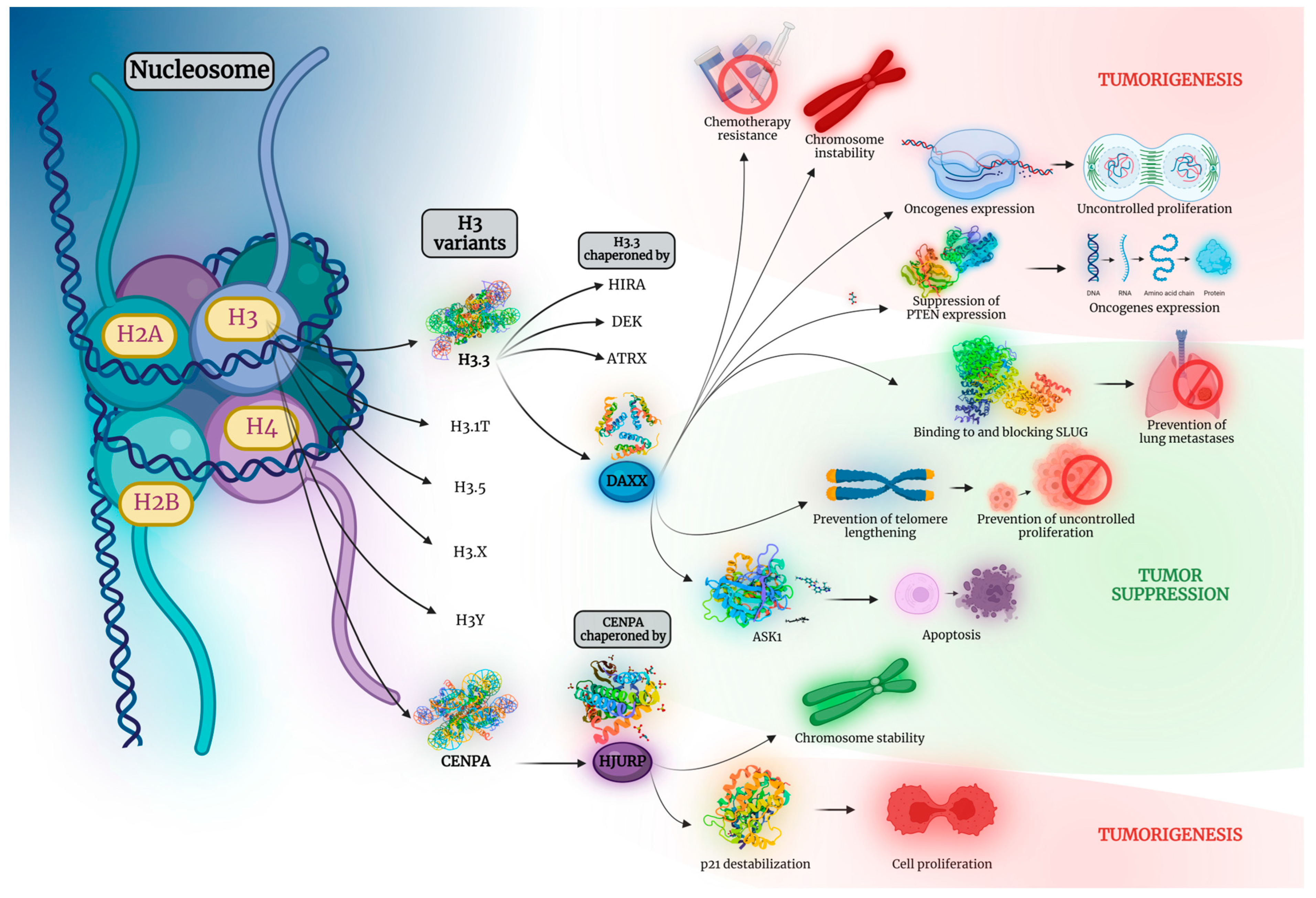
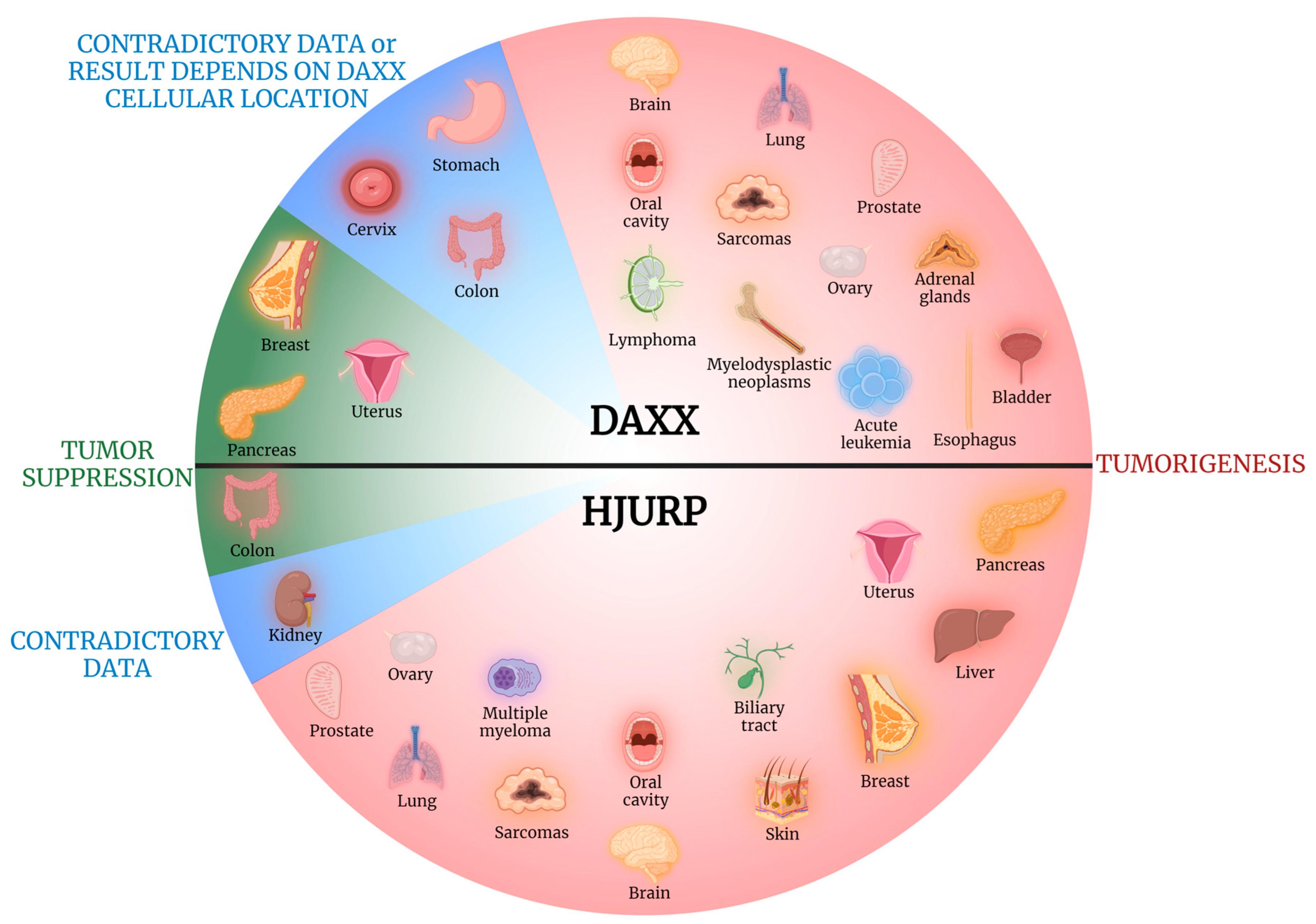
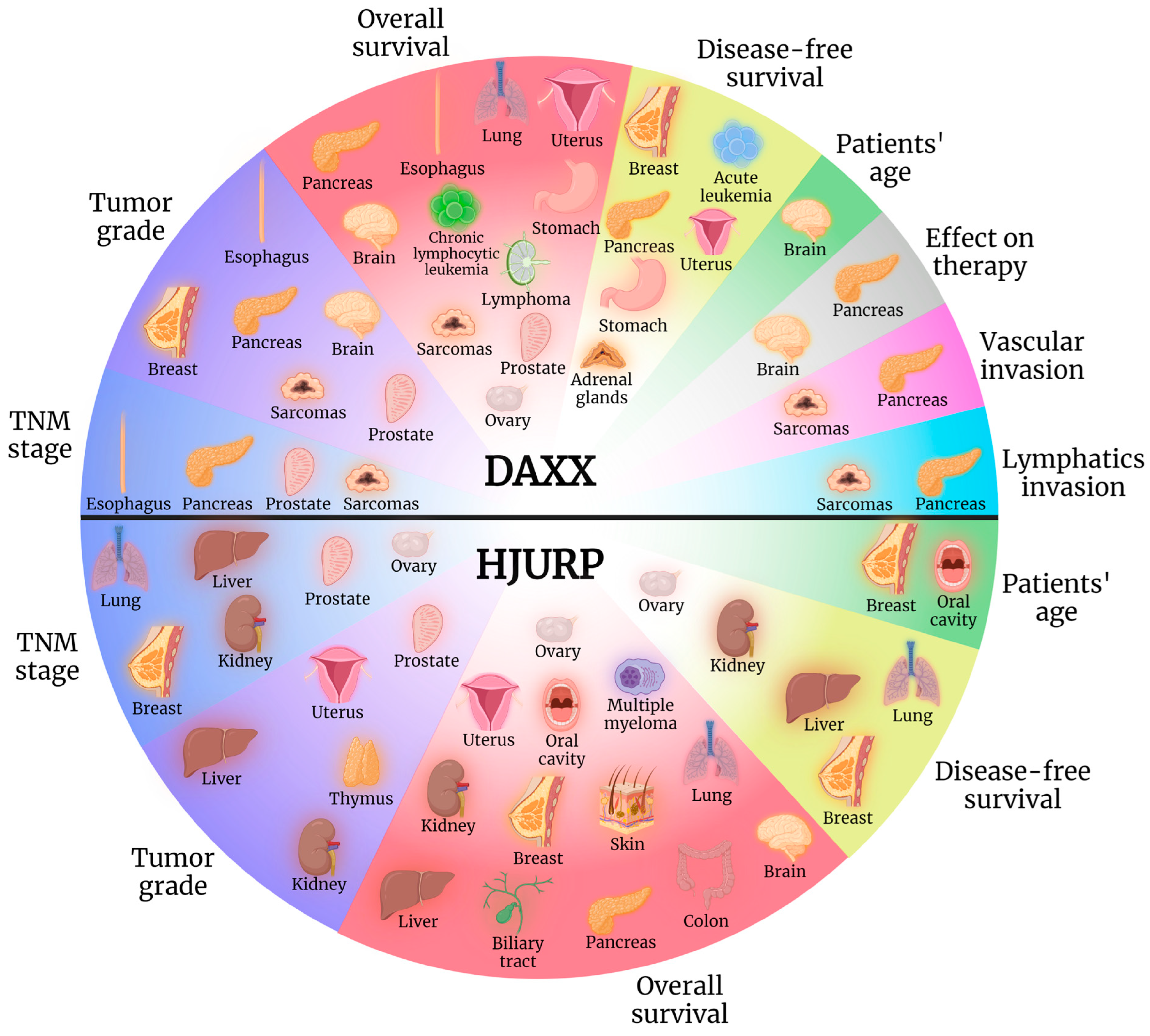
1. Introduction
2. Head and Neck, Thymus, and Skin Tumors
2.1. Brain
2.2. Oral Cavity
2.3. Thymus
2.4. Skin
3. Lung, Breast, and Sarcomas
3.1. Lung
3.2. Breast
3.3. Sarcomas
4. Gastrointestinal Tract, Liver, Biliary Tract, and Pancreas
4.1. Esophagus
4.2. Stomach
4.3. Colon
4.4. Liver
4.5. Biliary Tract
4.6. Pancreas
5. Urinary Tract, Prostate, Testis, and Adrenal Glands
5.1. Kidney
5.2. Bladder
5.3. Prostate
5.4. Adrenal Glands
6. Gynecologic Tumors
6.1. Ovary
6.2. Uterus
6.3. Cervix
7. Hematological Malignancies
7.1. Acute Leukemias
7.2. Myelodysplastic Neoplasms
7.3. Chronic Lymphocytic Leukemia
7.4. Lymphomas
7.5. Multiple Myeloma
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Cancer Observatory. 2020. Available online: https://gco.iarc.fr/ (accessed on 1 May 2023).
- Mahmud, I.; Liao, D. DAXX in cancer: Phenomena, processes, mechanisms and regulation. Nucleic Acids Res. 2019, 47, 7734–7752. [Google Scholar] [CrossRef]
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.-M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 2010, 107, 14075–14080. [Google Scholar] [CrossRef]
- Yang, X.; Khosravi-Far, R.; Chang, H.Y.; Baltimore, D. Daxx, a Novel Fas-Binding Protein That Activates JNK and Apoptosis. Cell 1997, 89, 1067–1076. [Google Scholar] [CrossRef]
- Shiizaki, S.; Naguro, I.; Ichijo, H. Activation mechanisms of ASK1 in response to various stresses and its significance in intracellular signaling. Adv. Biol. Regul. 2013, 53, 135–144. [Google Scholar] [CrossRef]
- Obsilova, V.; Honzejkova, K.; Obsil, T. Structural Insights Support Targeting ASK1 Kinase for Therapeutic Interventions. Int. J. Mol. Sci. 2021, 22, 13395. [Google Scholar] [CrossRef]
- Nye, J.; Melters, D.P.; Dalal, Y. The Art of War: Harnessing the epigenome against cancer. F1000Research 2018, 7, 141. [Google Scholar] [CrossRef]
- Pan, W.-W.; Zhou, J.-J.; Liu, X.-M.; Xu, Y.; Guo, L.-J.; Yu, C.; Shi, Q.-H.; Fan, H.-Y. Death Domain-associated Protein DAXX Promotes Ovarian Cancer Development and Chemoresistance. J. Biol. Chem. 2013, 288, 13620–13630. [Google Scholar] [CrossRef]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 1 October 2023).
- Cavalcante, S.G.; Pereira, B.J.; Lerario, A.M.; Sola, P.R.; Oba-Shinjo, S.M.; Marie, S.K. The chromatin remodeler complex ATRX-DAXX-H3.3 and telomere length in meningiomas. Clin. Neurol. Neurosurg. 2021, 210, 106962. [Google Scholar] [CrossRef]
- Cantero, D.; de Lope, R.; de la Presa, R.M.; Sepúlveda, J.M.; Borrás, J.M.; Castresana, J.S.; D’haene, N.; García, J.F.; Salmon, I.; Mollejo, M.; et al. Molecular Study of Long-Term Survivors of Glioblastoma by Gene-Targeted Next-Generation Sequencing. J. Neuropathol. Exp. Neurol. 2018, 77, 710–716. [Google Scholar] [CrossRef]
- Aksoy, S.A.; Mutlu, M.; Tunca, B.; Kocaeli, H.; Taskapilioglu, M.O.; Bekar, A.; Tekin, C.; Argadal, O.G.; Civan, M.N.; Kaya, I.S.; et al. Coexistence of TERT C228T mutation and MALAT1 dysregulation in primary glioblastoma: New prognostic and therapeutic targets. Neurol. Res. 2021, 43, 916–925. [Google Scholar] [CrossRef]
- Kurihara, S.; Hiyama, E.; Onitake, Y.; Yamaoka, E.; Hiyama, K. Clinical features of ATRX or DAXX mutated neuroblastoma. J. Pediatr. Surg. 2014, 49, 1835–1838. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Bi, W.L.; Coy, S.; Davis, C.; Gallia, G.L.; Santagata, S.; Rodriguez, F.J. Telomere length alterations and ATRX/DAXX loss in pituitary adenomas. Mod. Pathol. 2020, 33, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- de Tayrac, M.; Aubry, M.; Saïkali, S.; Etcheverry, A.; Surbled, C.; Guénot, F.; Galibert, M.-D.; Hamlat, A.; Lesimple, T.; Quillien, V.; et al. A 4-Gene Signature Associated with Clinical Outcome in High-Grade Gliomas. Clin. Cancer Res. 2011, 17, 317–327. [Google Scholar] [CrossRef]
- de Tayrac, M.; Saikali, S.; Aubry, M.; Bellaud, P.; Boniface, R.; Quillien, V.; Mosser, J. Prognostic Significance of EDN/RB, HJURP, p60/CAF-1 and PDLI4, Four New Markers in High-Grade Gliomas. PLoS ONE 2013, 8, e73332. [Google Scholar] [CrossRef] [PubMed]
- Valente, V.; Serafim, R.B.; de Oliveira, L.C.; Adorni, F.S.; Torrieri, R.; Tirapelli, D.P.d.C.; Espreafico, E.M.; Oba-Shinjo, S.M.; Marie, S.K.N.; Paçó-Larson, M.L.; et al. Modulation of HJURP (Holliday Junction-Recognizing Protein) Levels Is Correlated with Glioblastoma Cells Survival. PLoS ONE 2013, 8, e62200. [Google Scholar] [CrossRef]
- Lin, G.-J.; Huang, Y.-S.; Lin, C.-K.; Huang, S.-H.; Shih, H.-M.; Sytwu, H.-K.; Chen, Y.-W. Daxx and TCF4 interaction links to oral squamous cell carcinoma growth by promoting cell cycle progression via induction of cyclin D1 expression. Clin. Oral Investig. 2016, 20, 533–540. [Google Scholar] [CrossRef]
- Tsevegjav, B.; Takano, A.; Zhu, M.; Yoshitake, Y.; Shinohara, M.; Daigo, Y. Holliday junction recognition protein as a prognostic biomarker and therapeutic target for oral cancer. Int. J. Oncol. 2022, 60, 26. [Google Scholar] [CrossRef]
- Levidou, G.; Palamaris, K.; Sykaras, A.G.; Andreadakis, G.; Masaoutis, C.; Theochari, I.; Korkolopoulou, P.; Rontogianni, D.; Theocharis, S. Unraveling the Role of Histone Variant CENP-A and Chaperone HJURP Expression in Thymic Epithelial Neoplasms. Int. J. Mol. Sci. 2022, 23, 8339. [Google Scholar] [CrossRef]
- Ma, J.; Cai, X.; Kang, L.; Chen, S.; Liu, H. Identification of novel biomarkers and candidate small-molecule drugs in cutaneous melanoma by comprehensive gene microarrays analysis. J. Cancer 2021, 12, 1307–1317. [Google Scholar] [CrossRef]
- Buentzel, J.; Yao, S.; Elakad, O.; Lois, A.-M.; Brünies, J.; König, J.; Hinterthaner, M.; Danner, B.C.; Ströbel, P.; Emmert, A.; et al. Expression and prognostic impact of alpha thalassemia/mental retardation X-linked and death domain-associated protein in human lung cancer. Medicine 2019, 98, e16712. [Google Scholar] [CrossRef]
- Yin, Q.; Chen, W.; Zhang, C.; Wei, Z. A convolutional neural network model for survival prediction based on prognosis-related cascaded Wx feature selection. Lab. Investig. 2022, 102, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qu, J.; Liang, Y.; Zhao, D.; Rehman, F.U.; Qin, K.; Zhang, X. Identification and validation of key genes with prognostic value in non-small-cell lung cancer via integrated bioinformatics analysis. Thorac. Cancer 2020, 11, 851–866. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Ouyang, G.-L.; Yao, W.-X.; Zhu, Y.-J.; Li, X.; Huang, L.-X.; Yang, X.-W.; Jiang, W.-J. Knockdown of HJURP inhibits non-small cell lung cancer cell proliferation, migration, and invasion by repressing Wnt/beta-catenin signaling. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3847–3856. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zeng, C.; Yan, L.; Liao, W.; Zhen, C.; Yao, J. Prognostic value of Holliday junction-recognizing protein and its correlation with immune infiltrates in lung adenocarcinoma. Oncol. Lett. 2022, 24, 232. [Google Scholar] [CrossRef]
- Hussien, M.T.; Shaban, S.; Temerik, D.F.; Helal, S.R.; Mosad, E.; Elgammal, S.; Mostafa, A.; Hassan, E.; Ibrahim, A. Impact of DAXX and ATRX expression on telomere length and prognosis of breast cancer patients. J. Egypt. Natl. Cancer Inst. 2020, 32, 34. [Google Scholar] [CrossRef]
- Yanai, H.; Ishida, M.; Yoshikawa, K.; Tsuta, K.; Sekimoto, M.; Sugie, T. Immunohistochemical analyses of the expression profiles of INSM1, ATRX, DAXX and DLL3 in solid papillary carcinomas of the breast. Oncol. Lett. 2022, 23, 137. [Google Scholar] [CrossRef]
- Bravaccini, S.; Tumedei, M.M.; Scarpi, E.; Zoli, W.; Rengucci, C.; Serra, L.; Curcio, A.; Buggi, F.; Folli, S.; Rocca, A.; et al. New Biomarkers to Predict the Evolution of In Situ Breast Cancers. BioMed Res. Int. 2014, 2014, 159765. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Milioli, H.H.; Tishchenko, I.; Riveros, C.; Berretta, R.; Moscato, P. Basal-like breast cancer: Molecular profiles, clinical features and survival outcomes. BMC Med. Genom. 2017, 10, 19. [Google Scholar] [CrossRef]
- Hu, Z.; Huang, G.; Sadanandam, A.; Gu, S.; Lenburg, M.E.; Pai, M.; Bayani, N.; Blakely, E.A.; Gray, J.W.; Mao, J.-H. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010, 12, R18. [Google Scholar] [CrossRef]
- de Oca, R.M.; Gurard-Levin, Z.A.; Berger, F.; Rehman, H.; Martel, E.; Corpet, A.; de Koning, L.; Vassias, I.; Wilson, L.O.; Meseure, D.; et al. The histone chaperone HJURP is a new independent prognostic marker for luminal A breast carcinoma. Mol. Oncol. 2015, 9, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Panse, G.; Chrisinger, J.S.; Leung, C.H.; Ingram, D.R.; Khan, S.; Wani, K.; Lin, H.; Lazar, A.J.; Wang, W. Clinicopathological analysis of ATRX, DAXX and NOTCH receptor expression in angiosarcomas. Histopathology 2018, 72, 239–247. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Shi, X.; Chen, R.; Wu, Z.; Yang, Z.; Li, Z. Association of Mental Health-Related Proteins DAXX, DRD3, and DISC1 with the Progression and Prognosis of Chondrosarcoma. Front. Mol. Biosci. 2019, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Vargas, A.C.; Gray, L.-A.; White, C.L.; Maclean, F.M.; Grimison, P.; Ardakani, N.M.; Bonar, F.; Algar, E.M.; Cheah, A.L.; Russell, P.; et al. Genome wide methylation profiling of selected matched soft tissue sarcomas identifies methylation changes in metastatic and recurrent disease. Sci. Rep. 2021, 11, 667. [Google Scholar] [CrossRef]
- Liau, J.-Y.; Tsai, J.-H.; Jeng, Y.-M.; Lee, J.-C.; Hsu, H.-H.; Yang, C.-Y. Leiomyosarcoma with Alternative Lengthening of Telomeres Is Associated with Aggressive Histologic Features, Loss of ATRX Expression, and Poor Clinical Outcome. Am. J. Surg. Pathol. 2015, 39, 236–244. [Google Scholar] [CrossRef]
- Akaike, K.; Toda-Ishii, M.; Suehara, Y.; Mukaihara, K.; Kubota, D.; Mitani, K.; Takagi, T.; Kaneko, K.; Yao, T.; Saito, T. TERT promoter mutations are a rare event in gastrointestinal stromal tumors. SpringerPlus 2015, 4, 836. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, B.; Zhao, J.; Zhao, Y.; Ma, X.; Feng, H. Weighted Gene Co-Expression Network Analysis Identifies Five Hub Genes Associated with Metastasis in Synovial Sarcoma. Comb. Chem. High Throughput Screen. 2022, 25, 1767–1777. [Google Scholar] [CrossRef]
- Ko, T.Y.; Kim, J.I.; Park, E.S.; Mun, J.M.; Park, S.D. The Clinical Implications of Death Domain-Associated Protein (DAXX) Expression. Korean J. Thorac. Cardiovasc. Surg. 2018, 51, 187–194. [Google Scholar] [CrossRef]
- Yuen, H.-F.; Chan, Y.-P.; Law, S.; Srivastava, G.; El-Tanani, M.; Mak, T.-W.; Chan, K.-W. DJ-1 Could Predict Worse Prognosis in Esophageal Squamous Cell Carcinoma. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3593–3602. [Google Scholar] [CrossRef]
- Chen, C.; Sun, X.; Xie, W.; Chen, S.; Hu, Y.; Xing, D.; Xu, J.; Chen, X.; Zhao, Z.; Han, Z.; et al. Opposing biological functions of the cytoplasm and nucleus DAXX modified by SUMO-2/3 in gastric cancer. Cell Death Dis. 2020, 11, 514. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, Z.; Ye, L.; Zhuge, W.; Han, Z.; Zhang, T.; Ye, S.; Chen, W.; Zhu, S.; Shi, L.; et al. Prognostic significance of Daxx NCR (Nuclear/Cytoplasmic Ratio) in gastric cancer. Cancer Med. 2017, 6, 2063–2075. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Lee, T.-H.; Tzeng, S.-L. Reduced DAXX Expression Is Associated with Reduced CD24 Expression in Colorectal Cancer. Cells 2019, 8, 1242. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, F.; Zhu, X.; Guo, W.; Fu, T.; Wang, W. Death Domain-Associated Protein Promotes Colon Cancer Metastasis through Direct Interaction with ZEB1. J. Cancer 2020, 11, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, S.-L.; Cheng, Y.-W.; Li, C.-H.; Lin, Y.-S.; Hsu, H.-C.; Kang, J.-J. Physiological and Functional Interactions between Tcf4 and Daxx in Colon Cancer Cells. J. Biol. Chem. 2006, 281, 15405–15411. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-S.; Wu, C.-C.; Chang, C.-C.; Huang, S.-F.; Kuo, H.-Y.; Shih, H.-M. Reciprocal regulation of Daxx and PIK3CA promotes colorectal cancer cell growth. Cell. Mol. Life Sci. 2022, 79, 367. [Google Scholar] [CrossRef]
- Kang, D.H.; Woo, J.; Kim, H.; Kim, S.Y.; Ji, S.; Jaygal, G.; Ahn, T.S.; Kim, H.J.; Kwak, H.J.; Kim, C.-J.; et al. Prognostic Relevance of HJURP Expression in Patients with Surgically Resected Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 7928. [Google Scholar] [CrossRef]
- Li, C.; Ding, J.; Mei, J. Comprehensive Analysis of Epigenetic Associated Genes on Differential Gene Expression and Prognosis in Hepatocellular Carcinoma. J. Environ. Pathol. Toxicol. Oncol. 2022, 41, 27–43. [Google Scholar] [CrossRef]
- Li, Y.; Yi, Q.; Liao, X.; Han, C.; Zheng, L.; Li, H.; Yu, Q.; Yan, X.; Chen, X.; Zhu, H.; et al. Hypomethylation-driven overexpression of HJURP promotes progression of hepatocellular carcinoma and is associated with poor prognosis. Biochem. Biophys. Res. Commun. 2021, 566, 67–74. [Google Scholar] [CrossRef]
- Hu, B.; Wang, Q.; Wang, Y.; Chen, J.; Li, P.; Han, M. Holliday junction–recognizing protein promotes cell proliferation and correlates with unfavorable clinical outcome of hepatocellular carcinoma. OncoTargets Ther. 2017, 10, 2601–2607. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Yuan, J.; Liu, Z.; Cao, W.; Liu, P. The expression, clinical relevance, and prognostic significance of HJURP in cholangiocarcinoma. Front. Oncol. 2022, 12, 972550. [Google Scholar] [CrossRef]
- Jiao, Y.; Shi, C.; Edil, B.H.; de Wilde, R.F.; Klimstra, D.S.; Maitra, A.; Schulick, R.D.; Tang, L.H.; Wolfgang, C.L.; Choti, M.A.; et al. DAXX/ATRX, MEN1, and mTOR Pathway Genes Are Frequently Altered in Pancreatic Neuroendocrine Tumors. Science 2011, 331, 1199–1203. [Google Scholar] [CrossRef]
- Hackeng, W.M.; Brosens, L.A.; Kim, J.Y.; O’Sullivan, R.; Sung, Y.-N.; Liu, T.-C.; Cao, D.; Heayn, M.; Brosnan-Cashman, J.; An, S.; et al. Non-functional pancreatic neuroendocrine tumours: ATRX/DAXX and alternative lengthening of telomeres (ALT) are prognostically independent from ARX/PDX1 expression and tumour size. Gut 2022, 71, 961–973. [Google Scholar] [CrossRef]
- Ueda, H.; Akiyama, Y.; Shimada, S.; Mogushi, K.; Serizawa, M.; Matsumura, S.; Mitsunori, Y.; Aihara, A.; Ban, D.; Ochiai, T.; et al. Tumor suppressor functions of DAXX through histone H3.3/H3K9me3 pathway in pancreatic NETs. Endocr.-Relat. Cancer 2018, 25, 619–631. [Google Scholar] [CrossRef]
- Venugopal, A.; Michalczyk, A.; Khasraw, M.; Ackland, M.L. EMT Molecular Signatures of Pancreatic Neuroendocrine Neoplasms. Int. J. Mol. Sci. 2022, 23, 13645. [Google Scholar] [CrossRef]
- Ziv, E.; Rice, S.L.; Filtes, J.; Yarmohammadi, H.; Boas, F.E.; Erinjeri, J.P.; Petre, E.N.; Brody, L.A.; Brown, K.T.; Covey, A.M.; et al. Next generation sequencing of neuroendocrine tumors undergoing trans-arterial embolization reveals DAXX mutation status predicts shorter hepatic progression free survival and time to hepatic progression. J. Vasc. Interv. Radiol. 2018, 29, 1519–1526. [Google Scholar] [CrossRef]
- Park, J.K.; Paik, W.H.; Lee, K.; Ryu, J.K.; Lee, S.H.; Kim, Y.-T. DAXX/ATRX and MEN1 genes are strong prognostic markers in pancreatic neuroendocrine tumors. Oncotarget 2017, 8, 49796–49806. [Google Scholar] [CrossRef]
- Marinoni, I.; Kurrer, A.S.; Vassella, E.; Dettmer, M.; Rudolph, T.; Banz, V.; Hunger, F.; Pasquinelli, S.; Speel, E.; Perren, A. Loss of DAXX and ATRX Are Associated with Chromosome Instability and Reduced Survival of Patients with Pancreatic Neuroendocrine Tumors. Gastroenterology 2014, 146, 453–460.e5. [Google Scholar] [CrossRef]
- Pipinikas, C.P.; Dibra, H.; Karpathakis, A.; Feber, A.; Novelli, M.; Oukrif, D.; Fusai, G.; Valente, R.; Caplin, M.; Meyer, T.; et al. Epigenetic dysregulation and poorer prognosis in DAXX-deficient pancreatic neuroendocrine tumours. Endocr.-Relat. Cancer 2015, 22, L13–L18. [Google Scholar] [CrossRef]
- Yuan, F.; Shi, M.; Ji, J.; Shi, H.; Zhou, C.; Yu, Y.; Liu, B.; Zhu, Z.; Zhang, J. KRAS and DAXX/ATRX Gene Mutations Are Correlated with the Clinicopathological Features, Advanced Diseases, and Poor Prognosis in Chinese Patients with Pancreatic Neuroendocrine Tumors. Int. J. Biol. Sci. 2014, 10, 957–965. [Google Scholar] [CrossRef]
- Weisbrod, A.B.; Zhang, L.; Jain, M.; Barak, S.; Quezado, M.M.; Kebebew, E. Altered PTEN, ATRX, CHGA, CHGB, and TP53 Expression Are Associated with Aggressive VHL-Associated Pancreatic Neuroendocrine Tumors. Horm. Cancer 2013, 4, 165–175. [Google Scholar] [CrossRef]
- Sato, S.; Tsuchikawa, T.; Nakamura, T.; Sato, N.; Tamoto, E.; Okamura, K.; Shichinohe, T.; Hirano, S. Impact of the tumor microenvironment in predicting postoperative hepatic recurrence of pancreatic neuroendocrine tumors. Oncol. Rep. 2014, 32, 2753–2759. [Google Scholar] [CrossRef] [PubMed]
- Pea, A.; Yu, J.; Marchionni, L.; Noe, M.; Luchini, C.; Pulvirenti, A.; de Wilde, R.F.; Brosens, L.A.; Rezaee, N.; Javed, A.; et al. Genetic Analysis of Small Well-differentiated Pancreatic Neuroendocrine Tumors Identifies Subgroups with Differing Risks of Liver Metastases. Ann. Surg. 2020, 271, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Brosnan-Cashman, J.A.; An, S.; Kim, S.J.; Song, K.-B.; Kim, M.-S.; Kim, M.-J.; Hwang, D.W.; Meeker, A.K.; Yu, E.; et al. Alternative Lengthening of Telomeres in Primary Pancreatic Neuroendocrine Tumors Is Associated with Aggressive Clinical Behavior and Poor Survival. Clin. Cancer Res. 2017, 23, 1598–1606. [Google Scholar] [CrossRef]
- Uemura, J.; Okano, K.; Oshima, M.; Suto, H.; Ando, Y.; Kumamoto, K.; Kadota, K.; Ichihara, S.; Kokudo, Y.; Maeba, T.; et al. Immunohistochemically Detected Expression of ATRX, TSC2, and PTEN Predicts Clinical Outcomes in Patients with Grade 1 and 2 Pancreatic Neuroendocrine Tumors. Ann. Surg. 2021, 274, e949–e956. [Google Scholar] [CrossRef]
- Chou, A.; Itchins, M.; de Reuver, P.R.; Arena, J.; Clarkson, A.; Sheen, A.; Sioson, L.; Cheung, V.; Perren, A.; Nahm, C.; et al. ATRX loss is an independent predictor of poor survival in pancreatic neuroendocrine tumors. Hum. Pathol. 2018, 82, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Kakkar, A.; Sharma, A.; Malik, P.S.; Sharma, M.C. Study of clinicopathological features, hormone immunoexpression, and loss of ATRX and DAXX expression in pancreatic neuroendocrine tumors. Scand. J. Gastroenterol. 2016, 51, 994–999. [Google Scholar] [CrossRef]
- Chen, S.-F.; Kasajima, A.; Yazdani, S.; Chan, M.S.; Wang, L.; He, Y.-Y.; Gao, H.-W.; Sasano, H. Clinicopathologic significance of immunostaining of α-thalassemia/mental retardation syndrome X-linked protein and death domain–associated protein in neuroendocrine tumors. Hum. Pathol. 2013, 44, 2199–2203. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-J.; Li, X.; Shi, P.; Ding, H.-Y.; Liu, Y.-P.; Li, T.; Lin, P.-P.; Wang, Y.-S.; Zhang, G.-Q.; Cao, Y. Holliday junction recognition protein promotes pancreatic cancer growth and metastasis via modulation of the MDM2/p53 signaling. Cell Death Dis. 2020, 11, 386. [Google Scholar] [CrossRef]
- Pivovarcikova, K.; Agaimy, A.; Martinek, P.; Alaghehbandan, R.; Perez-Montiel, D.; Alvarado-Cabrero, I.; Rogala, J.; Kuroda, N.; Rychly, B.; Gasparov, S.; et al. Primary renal well-differentiated neuroendocrine tumour (carcinoid): Next-generation sequencing study of 11 cases. Histopathology 2019, 75, 104–117. [Google Scholar] [CrossRef]
- Wei, W.; Lv, Y.; Gan, Z.; Zhang, Y.; Han, X.; Xu, Z. Identification of key genes involved in the metastasis of clear cell renal cell carcinoma. Oncol. Lett. 2019, 17, 4321–4328. [Google Scholar] [CrossRef]
- Xu, T.; Ruan, H.; Gao, S.; Liu, J.; Liu, Y.; Song, Z.; Cao, Q.; Wang, K.; Bao, L.; Liu, D.; et al. ISG20 serves as a potential biomarker and drives tumor progression in clear cell renal cell carcinoma. Aging 2020, 12, 1808–1827. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, Z.; Wang, K.; Zhang, Z. Holliday junction-recognition protein modulates apoptosis, cell cycle arrest and reactive oxygen species stress in human renal cell carcinoma. Oncol. Rep. 2020, 44, 1246–1254. [Google Scholar] [CrossRef]
- Zhang, F.; Yuan, D.; Song, J.; Chen, W.; Wang, W.; Zhu, G.; Hu, B.; Chen, X.; Zhu, J. HJURP is a prognostic biomarker for clear cell renal cell carcinoma and is linked to immune infiltration. Int. Immunopharmacol. 2021, 99, 107899. [Google Scholar] [CrossRef] [PubMed]
- Zizzi, A.; Montironi, M.A.; Mazzucchelli, R.; Scarpelli, M.; Lopez-Beltran, A.; Cheng, L.; Paone, N.; Castellini, P.; Montironi, R. Immunohistochemical analysis of chromatin remodeler DAXX in high grade urothelial carcinoma. Diagn. Pathol. 2013, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Burgess, E.F.; Sanders, J.A.; Livasy, C.; Symanowski, J.; Gatalica, Z.; Steuerwald, N.M.; Arguello, D.; Brouwer, C.R.; Korn, W.M.; Grigg, C.M.; et al. Identification of potential biomarkers and novel therapeutic targets through genomic analysis of small cell bladder carcinoma and associated clinical outcomes. Urol. Oncol. Semin. Orig. Investig. 2022, 40, 383.e1–383.e10. [Google Scholar] [CrossRef] [PubMed]
- Kwan, P.S.; Lau, C.C.; Chiu, Y.T.; Man, C.; Liu, J.; Tang, K.D.; Wong, Y.C.; Ling, M.-T. Daxx regulates mitotic progression and prostate cancer predisposition. Carcinogenesis 2012, 34, 750–759. [Google Scholar] [CrossRef]
- Jamali, L.; Moradi, A.; Ganji, M.; Ayati, M.; Kazeminezhad, B.; Attar, Z.F.; Ghaedi, H.; Ghaderian, S.M.H.; Fallah-Karkan, M.; Ranjbar, A. Potential Prognostic Role for SPOP, DAXX, RARRES1, and LAMP2 as an Autophagy Related Genes in Prostate Cancer. Urol. J. 2020, 17, 156–163. [Google Scholar] [CrossRef]
- Tsourlakis, M.C.; Schoop, M.; Plass, C.; Huland, H.; Graefen, M.; Steuber, T.; Schlomm, T.; Simon, R.; Sauter, G.; Sirma, H.; et al. Overexpression of the chromatin remodeler death-domain–associated protein in prostate cancer is an independent predictor of early prostate-specific antigen recurrence. Hum. Pathol. 2013, 44, 1789–1796. [Google Scholar] [CrossRef]
- Puto, L.A.; Brognard, J.; Hunter, T. Transcriptional Repressor DAXX Promotes Prostate Cancer Tumorigenicity via Suppression of Autophagy. J. Biol. Chem. 2015, 290, 15406–15420. [Google Scholar] [CrossRef]
- Chen, Y.; Liang, Y.; Yang, J.; Yuan, D.; Li, J.; Zheng, S.; Wan, Y.; Wang, B.; Han, Z.; Zhong, W. Upregulation of Holliday junction recognition protein predicts poor prognosis and biochemical recurrence in patients with prostate cancer. Oncol. Lett. 2019, 18, 6697–6703. [Google Scholar] [CrossRef]
- Luo, C.; Chen, J.; Chen, L. Exploration of gene expression profiles and immune microenvironment between high and low tumor mutation burden groups in prostate cancer. Int. Immunopharmacol. 2020, 86, 106709. [Google Scholar] [CrossRef] [PubMed]
- Mete, O.; Gucer, H.; Kefeli, M.; Asa, S.L. Diagnostic and Prognostic Biomarkers of Adrenal Cortical Carcinoma. Am. J. Surg. Pathol. 2018, 42, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; McFadden, E.; Holth, A.; Brunetti, M.; Flørenes, V.A. Death domain-associated protein (DAXX) expression is associated with poor survival in metastatic high-grade serous carcinoma. Virchows Arch. 2020, 477, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Pontikakis, S.; Papadaki, C.; Tzardi, M.; Trypaki, M.; Sfakianaki, M.; Koinis, F.; Lagoudaki, E.; Giannikaki, L.; Kalykaki, A.; Kontopodis, E.; et al. Predictive value of ATP7b, BRCA1, BRCA2, PARP1, UIMC1 (RAP80), HOXA9, DAXX, TXN (TRX1), THBS1 (TSP1) and PRR13 (TXR1) genes in patients with epithelial ovarian cancer who received platinum-taxane first-line therapy. Pharmacogenom. J. 2017, 17, 506–514. [Google Scholar] [CrossRef]
- Dou, Z.; Qiu, C.; Zhang, X.; Yao, S.; Zhao, C.; Wang, Z.; Chu, R.; Chen, J.; Chen, Z.; Li, R.; et al. HJURP Promotes Malignant Progression and Mediates Sensitivity to Cisplatin and WEE1-inhibitor in Serous Ovarian Cancer. Int. J. Biol. Sci. 2022, 18, 1188–1210. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Meng, Q.; Khan, A.Q.; Chen, X. Increased Expression of Holliday Junction-Recognizing Protein (HJURP) as an Independent Prognostic Biomarker in Advanced-Stage Serous Ovarian Carcinoma. Experiment 2018, 24, 3050–3055. [Google Scholar] [CrossRef] [PubMed]
- DeLair, D.F.; Burke, K.A.; Selenica, P.; Lim, R.S.; Scott, S.N.; Middha, S.; Mohanty, A.S.; Cheng, D.T.; Berger, M.F.; Soslow, R.A.; et al. The genetic landscape of endometrial clear cell carcinomas. J. Pathol. 2017, 243, 230–241. [Google Scholar] [CrossRef]
- Slatter, T.L.; Hsia, H.; Samaranayaka, A.; Sykes, P.; Clow, W.; Devenish, C.J.; Sutton, T.; Royds, J.A.; Pc, P.; Cheung, A.N.; et al. Loss of ATRX and DAXX expression identifies poor prognosis for smooth muscle tumours of uncertain malignant potential and early stage uterine leiomyosarcoma. J. Pathol. Clin. Res. 2015, 1, 95–105. [Google Scholar] [CrossRef]
- Ahvenainen, T.V.; Mäkinen, N.; Von Nandelstadh, P.; Vahteristo, M.E.; Pasanen, A.M.; Bützow, R.C.; Vahteristo, P.M. Loss of ATRX/DAXX expression and alternative lengthening of telomeres in uterine leiomyomas. Cancer 2018, 124, 4650–4656. [Google Scholar] [CrossRef]
- Lv, S.; Xu, X.; Wu, Z. Identification of key candidate genes and pathways in endometrial cancer: Evidence from bioinformatics analysis. Oncol. Lett. 2019, 18, 6679–6689. [Google Scholar] [CrossRef]
- Tang, S.-Y.; Li, L.; Li, Y.-L.; Liu, A.-Y.; Yu, M.-J.; Wan, Y.-P. Distribution and location of Daxx in cervical epithelial cells with high risk human papillomavirus positive. Diagn. Pathol. 2014, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Jingqiao, F. Expression significance of Daxx protein in bone marrow cells of children with acute leukemia. Chin. Pediatr. Integr. Tradit. West. Med. 2016, 8, 161–163. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, L.-Q.; Hu, Q.; Lin, H.-H.; Liu, A.-G.; Tao, H.-F.; Song, Y.-Q.; Zhang, X.-L. Expression of Daxx in children with acute leukemia. Zhongguo dang dai er ke za zhi = Chin. J. Contemp. Pediatr. 2007, 9, 33–36. [Google Scholar]
- Han-hua, L.; Jing, L.; Liu-qing, Z. Expression of Daxx in pediatric acute leukemia and its relationship with expression of NF-κB. Chin. J. Pract. Pediatr. 2007, 22, 459–461. [Google Scholar]
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef]
- Attieh, Y.; Geng, Q.-R.; DiNardo, C.D.; Zheng, H.; Jia, Y.; Fang, Z.-H.; Gañán-Gómez, I.; Yang, H.; Wei, Y.; Kantarjian, H.; et al. Low frequency of H3.3 mutations and upregulated DAXX expression in MDS. Blood 2013, 121, 4009–4011. [Google Scholar] [CrossRef]
- Bojarska-Junak, A.; Sieklucka, M.; Hus, I.; Wąsik-Szczepanek, E.; Kusz, M.L.; Surdacka, A.; Chocholska, S.; Dmoszyńska, A.; Roliński, J. Assessment of the pathway of apoptosis involving PAR-4, DAXX and ZIPK proteins in CLL patients and its relationship with the principal prognostic factors. Folia Histochem. Cytobiol. 2011, 49, 98–103. [Google Scholar] [CrossRef]
- Horvilleur, E.; Sbarrato, T.; Hill, K.; Spriggs, R.V.; Screen, M.; Goodrem, P.J.; Sawicka, K.; Chaplin, L.C.; Touriol, C.; Packham, G.; et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia 2014, 28, 1092–1102. [Google Scholar] [CrossRef]
- Xiong, J.; Cui, B.-W.; Wang, N.; Dai, Y.-T.; Zhang, H.; Wang, C.-F.; Zhong, H.-J.; Cheng, S.; Ou-Yang, B.-S.; Hu, Y.; et al. Genomic and Transcriptomic Characterization of Natural Killer T Cell Lymphoma. Cancer Cell 2020, 37, 403–419.e6. [Google Scholar] [CrossRef]
- Jia, Y.; Zhou, J.; Tan, T.K.; Chung, T.-H.; Chen, Y.; Chooi, J.-Y.; Sanda, T.; Fullwood, M.J.; Xiong, S.; Toh, S.H.; et al. Super Enhancer-Mediated Upregulation of HJURP Promotes Growth and Survival of t(4;14)-Positive Multiple Myeloma. Cancer Res. 2022, 82, 406–418. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Kim, M.-Y. Cancer epigenetics: Past, present and future. Semin. Cancer Biol. 2022, 83, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Gupta, K.; Gupta, S. Cancer Epigenetics: An Introduction. In Methods in Molecular Biology; Verma, M., Ed.; Springer: New York, NY, USA, 2015; Volume 1238, pp. 3–25. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Psilopatis, I.; Garmpis, N.; Garmpi, A.; Vrettou, K.; Sarantis, P.; Koustas, E.; Antoniou, E.A.; Dimitroulis, D.; Kouraklis, G.; Karamouzis, M.V.; et al. The Emerging Role of Histone Deacetylase Inhibitors in Cervical Cancer Therapy. Cancers 2023, 15, 2222. [Google Scholar] [CrossRef]
- Psilopatis, I.; Pergaris, A.; Vrettou, K.; Theocharis, S.; Troungos, C. Thymic Epithelial Neoplasms: Focusing on the Epigenetic Alterations. Int. J. Mol. Sci. 2022, 23, 4045. [Google Scholar] [CrossRef]
- Luchini, C.; Scarpa, A. Neoplastic Progression in Neuroendocrine Neoplasms of the Pancreas. Arch. Pathol. Lab. Med. 2023. [Google Scholar] [CrossRef]
| Malignant Tissues | Benign Control Tissues | Methods | Results | Refs |
|---|---|---|---|---|
| BRAIN | ||||
| DAXX | ||||
| 55 intracranial meningioma samples | - | TMA IHC | Significantly higher DAXX expression (nuclear and cytoplasmic) in grade 2/grade 3 compared to grade 1 meningiomas | [10] |
| 74 glioblastoma multiforme (GBM) samples | - | NGS | Alterations in DAXX genes associated with
| [11] |
| 85 GBM samples | - | qRT-PCR | Loss of DAXX expression associated with short OS | [12] |
| 121 neuroblastomas (NBLs) in total (110 NBLs with normal or shortened telomeres and 11 NBLs with elongated telomeres by alternative lengthening of telomere (ALT)) | - | NGS |
| [13] |
| 106 pituitary adenomas | - | TMA IHC Telomere-specific FISH NGS |
| [14] |
| 32 recurrent pituitary adenomas from 22 patients |
| |||
| HJURP | ||||
| 267 malignant glioma specimens | - | qRT-PCR data collection from the GEO high-grade gliomas data sets | HJURP overexpression associated with decreased OS | [15] |
| 96 high-grade glioma samples (64 glioblastomas and 32 grade 3 gliomas) | 6 adult and 4 fetal normal brain samples | IHC |
| [16] |
| 40 astrocytoma samples (5 diffuse astrocytomas, 5 anaplastic astrocytomas and 30 GBMs) | 7 non-neoplastic white matter samples from patients undergoing temporal lobectomy for epilepsy treatment | qRT-PCR Western Blot |
| [17] |
| ORAL CANCER | ||||
| DAXX | ||||
| 25 oral squamous cell carcinoma (OSCC) samples | Matched normal tissues from the same patients | qRT-PCR Western Blot IHC | DAXX mRNA and protein expression elevated in OSCC samples in comparison to normal tissues | [18] |
| HJURP | ||||
| 152 oral cancer (OC) tissue specimens | Adjacent normal oral tissues | qPCR Western Blot TMA IHC |
| [19] |
| THYMUS | ||||
| HJURP | ||||
| 95 thymic epithelial tumor (TET) samples | - | TMA IHC |
| [20] |
| SKIN | ||||
| HJURP | ||||
| 458 melanoma patients | Adjacent normal tissues | Data from GEO database retrieval | HJURP expression in melanoma was
| [21] |
| Malignant Tissues | Benign Control Tissues | Methods | Results | Refs |
|---|---|---|---|---|
| LUNG | ||||
| DAXX | ||||
| 345 lung cancer samples in total (194 samples of squamous cell lung carcinoma (SQCLC), 111 samples of pulmonary adenocarcinoma (AC), 40 samples of small cell lung cancer (SCLC)) | - | IHC |
| [22] |
| HJURP | ||||
| 1090 lung cancer specimens in total (551 lung AC specimens 539 SQCLC) | - | Data collection from TCGA database | High HJURP expression is significantly associated with poor OS of lung AC patients | [23] |
| 154 non-SCLC (NSCLC) tissue samples in total | 92 paired nontumor samples in total | Data collection from the GEO database | HJURP upregulation associated with worse OS in NSCLC patients | [24] |
| 74 NSCLC samples | adjacent normal tissue specimens | RT-PCR Western blot |
| [25] |
| 519 lung AC specimens | 54 normal lung tissue specimens | Data collection from TCGA and the GEO databases | High HJURP expression was significantly associated with
| [26] |
| BREAST | ||||
| DAXX | ||||
| 220 breast cancer (BC) specimens | - | IHC |
| [27] |
| 39 solid papillary carcinoma (SPC) specimens (18 SPC in situ and 21 SPC invasive) | - | IHC | Loss of DAXX expression was significantly associated with lymphatic invasion | [28] |
| HJURP | ||||
| 44 in situ breast carcinoma specimens | - | IHC | A greater than sevenfold risk of relapse observed in patients highly expressing HJURP in stroma | [29] |
| 351 basal-like breast cancers (BLBCs) specimens | - | Genomic and transcriptomic profile data collection from METABRIC and ROCK data sets | High levels of HJURP expression in basal II basal-like subtype (BL2) [30]. BL1 and BL2 subtypes exhibited higher expression of cell cycle and DNA damage response genes, and representative cell lines preferentially responded to cisplatin. | [31] |
| 130 tumor specimens | - | Affymetrix Microarray | High HJURP mRNA levels were significantly associated with
| [32] |
| 71 BC samples | - | qRT-PCR IHC |
| [33] |
| SARCOMAS | ||||
| DAXX | ||||
| 106 angiosarcoma specimens | - | IHC | All specimens examined retained nuclear expression of DAXX | [34] |
| 80 chondrosarcoma specimens | 25 osteochondroma specimens | IHC |
| [35] |
| 32 soft tissue sarcoma specimens from 14 individuals (including leiomyosarcoma (LMS), myxofibrosarcoma (MFS), rhabdomyosarcoma (RMS) and synovial sarcoma (SS)) | - | Genome-wide methylation analysis | DAXX gene methylation detected in LMS specimens | [36] |
| 92 LMS (derived from the uterus, retroperitoneum/intra-abdomen, and various other sites) | - | IHC | None of the 92 cases lost DAXX expression | [37] |
| 92 gastrointestinal stromal tumor (GIST) specimens | - | IHC |
| [38] |
| HJURP | ||||
| 58 synovial sarcoma (SS) specimens | - | Data collection on SS gene expression profile from the GEO database | HJURP gene over-expressed in SS (along with four other genes, namely NCAPG, TPX2, CENPA, and NDC80) | [39] |
| Malignant Tissues | Benign Control Tissues | Methods | Results | Refs |
|---|---|---|---|---|
| ESOPHAGUS | ||||
| DAXX | ||||
| 60 esophageal squamous cell carcinoma (ESCC) specimens | Adjacent non-tumorous tissues | IHC |
| [40] |
| 81 ESCC and 19 paired ESCC LN metastases specimens | 31 paired non-neoplastic esophageal epithelia | TMA IHC |
| [41] |
| STOMACH | ||||
| DAXX | ||||
| 323 gastric cancer (GC) specimens | 20 paired adjacent normal tissues | TMA IHC |
| [42] |
| 70 GC specimens | 70 paired adjacent normal tissue specimens | IHC |
| [43] |
| 522 GC specimens | - | TMA IHC | ||
| COLON | ||||
| DAXX | ||||
| 106 colorectal carcinoma (CRC) samples | Matched nontumor-surrounding tissues | Western blot |
| [44] |
| 8 pairs of matched primary CRC tissue and liver metastatic CRC tissue specimens | - | Real-time RT-PCR IHC | DAXX expression lower in liver metastases than in primary CRC tissues | [45] |
| 8 CRC specimens | Matched adjacent tissue specimens | Western blot | DAXX expression reduced in CRC tissues compared to normal colon tissues | [46] |
| 28 CRC specimens | Adjacent normal tissues | Data collection from TCGA data sets | DAXX expression increased in 19 of the 28 CRC specimens, compared with the adjacent normal tissues | [47] |
| 15 CRC specimens | Adjacent normal tissues | qRT-PCR IHC |
| |
| HJURP | ||||
| 162 CRC specimens | - | IHC |
| [48] |
| LIVER | ||||
| HJURP | ||||
| 621 hepatocellular carcinoma (HCC) specimens in total | 292 non-tumor specimens | Data collection from TCGA and the GEO datasets | HJURP mRNA expression was significantly associated with
| [49] |
| 176 HCC specimens | 21 adjacent normal tissue specimens | Real-time PCR IHC Data collection from the GEO, TCGA and the ICGC databases |
| [50] |
| 164 HCC specimens | - | IHC real-time PCR | High HJURP expression was significantly associated with
| [51] |
| BILIARY TRACT | ||||
| HJURP | ||||
| 127 Cholangiocarcinoma (CCA) specimens in total, including 32 intrahepatic (iCCA), 71 perihilar (pCCA) and 24 distal (dCCA) CCA specimens | - | TMA IHC | HJURP overexpression was associated with lower OS of iCCA and pCCA patients but not in dCCA patients | [52] |
| - | - | Data collection from TCGA database | HJURP expression upregulated in CCAs compared with adjacent non-tumorous tissues | |
| PANCREAS | ||||
| DAXX | ||||
| 68 sporadic pancreatic neuroendocrine tumors (PanNETs) specimens (that were not part of a familial syndrome associated with PanNETs) | - | Whole-exome sequencing | Mutations of DAXX genes were detected in 12 out of 68 cases and associated with better OS | [53] |
| 1322 NETs in total 561 primary nonfunctional pancreatic neuroendocrine tumours (NF-PanNETs), 107 NF-PanNET metastases and 654 primary, non-pancreatic non-functional NETs and NET metastases | - | IHC |
| [54] |
| 44 PanNET specimens | - | IHC | Low DAXX expression was correlated with
| [55] |
| 13 pancreatic neuroendocrine neoplasm (NEN) specimens (4 G1, 5 G2 and 4 G3 tumor samples) | Matched non-tumor tissues | qRT-PCR IHC |
| [56] |
| 51 neuroendocrine liver metastases specimens | - | NGS |
| [57] |
| 76 PanNET specimens | - | TMA IHC | Loss of DAXX protein expression was associated with
| [58] |
| 243 well-differentiated (G1 and G2) primary PanNET specimens | - | TMA IHC | The loss of DAXX was associated with
| [59] |
| 53 PanNET specimens (46 primaries and 7 liver metastases from 39 cases) | - | IHC | The loss of DAXX expression was associated with poorer 5-year DFS | [60] |
| 37 PanNET specimens | - | Sanger sequencing | DAXX/ATRX mutations were
| [61] |
| PanNET specimens from patients with Von Hippel-Lindau (VHL) syndrome | - | IHC | No statistically significant difference in tumor DAXX protein expression via pathologic diagnosis, clinical data, VHL exon mutation, and functional imaging results was identified. | [62] |
| 16 PanNET specimens | - | IHC | DAXX loss was associated with
| [63] |
| 87 small primary PanNET (<3 cm) specimens | - | IHC NGS | The presence of DAXX mutation was associated with an increased risk of metastasis. | [64] |
| 269 primary PanNET specimens | 19 sporadic microadenomas | TMA IHC |
| [65] |
| 100 Grade 1 and 2 primary PanNET specimens | - | IHC | The loss of DAXX was detected in 64 out of 100 cases. | [66] |
| 105 PanNET specimens | - | IHC |
| [67] |
| 46 PanNET specimens | - | IHC | The loss of ATRX/DAXX expression was
| [68] |
| 164 tumor specimens in total (10 gastric, 15 duodenal, 20 rectal, 70 pancreatic, and 22 pulmonary NETs and 27 pancreatic adenocarcinomas (PDACs)) | 15 nonneoplastic pancreas specimens | IHC |
| [69] |
| HJURP | ||||
| 177 PDAC specimens | Adjacent normal tissues | Data collection from the GEO dataset | HJURP expression was
| [70] |
| 219 PDAC specimens | Adjacent normal tissues | IHC qRT-PCR Western blot | ||
| Malignant Tissues | Benign Control Tissues | Methods | Results | Refs |
|---|---|---|---|---|
| KIDNEY | ||||
| DAXX | ||||
| 11 primary renal well-differentiated neuroendocrine tumors (NET) specimens | - | NGS | No DAXX mutations were detected in any of the cases | [71] |
| HJURP | ||||
| 416 clear cell renal cell carcinoma (ccRCC) specimens | - | Data collection from TCGA dataset | HJURP overexpression was associated with
| [72] |
| 52 ccRCC specimens | Adjacent normal tissues | Data collection from TCGA dataset | No association between HJURP expression and clinicopathological parameters observed | [73] |
| 15 ccRCC specimens | Adjacent paracancerous renal tissue samples | qRT-PCR Western blot | HJURP expression lower in ccRCC tissues compared with that in the adjacent paracancerous ones | [74] |
| 539 ccRCC specimens | 72 paracancerous normal tissue samples | Data collection from TCGA dataset |
| [75] |
| BLADDER | ||||
| DAXX | ||||
| 5 pT1 bladder urothelial carcinoma (UC) specimens | 5 normal bladder urothelium samples | IHC |
| [76] |
| 31 small cell bladder carcinoma specimens | - | NGS | loss of DAXX expression observed in 5 (16.1%) cases, including 4 patients who presented with limited stage disease. All 4 patients who presented with limited-stage disease underwent chemotherapy and remained alive during follow-up, implying that loss of DAXX may sensitize tumor cells to chemotherapy. | [77] |
| PROSTATE | ||||
| DAXX | ||||
| 115 prostate cancer specimens, 8 prostatic intraepithelial neoplasia specimens | 37 benign prostatic hyperplasia samples | TMA IHC |
| [78] |
| 50 prostate cancer tissue specimens | 50 normal adjacent tissue samples from the same cases with prostate cancer 50 benign prostatic hyperplasia (BPH) tissue samples | qRT-PCR | DAXX expression significantly increased compared to both control groups (normal adjacent and BPH tissues) | [79] |
| 5718 prostate cancer specimens | Normal prostate tissues | TMA IHC |
| [80] |
| Prostate cancer tissues | Normal prostate tissues | Data collection on DAXX mRNA expression levels from the Oncomine database |
| [81] |
| HJURP | ||||
| 99 prostate cancer specimens | 81 benign prostate tissue samples | TMA IHC |
| [82] |
| 22 prostate cancer specimens | 10 benign prostate tissue samples | qRT-PCR | ||
| 484 prostate cancer specimens | - | Data collection from TCGA dataset | High HJURP expression (along with high expression of UBE2C, PLK1, CDC20, BUB1 and CDK1 genes) was associated with worse biochemical recurrence-free survival | [83] |
| ADRENAL GLANDS | ||||
| DAXX | ||||
| 43 adrenal cortical carcinoma specimens | 50 cortical adenoma samples | TMA IHC | Global loss of DAXX expression was associated with longer DFS | [84] |
| Malignant Tissues | Benign Control Tissues | Methods | Results | Refs |
|---|---|---|---|---|
| OVARY | ||||
| DAXX | ||||
| 59 tumor specimens (36 serous cystadenoma, 13 endometrioid carcinoma, 3 clear cell carcinoma, 4 mucinous cystadenoma, and 3 granular cell tumor samples) | 3 normal ovary tissue samples | IHC |
| [8] |
| 400 high-grade serous carcinoma serous effusion specimens | - | IHC |
| [85] |
| 81 of the 400 high-grade serous carcinoma effusion specimens | - | Western blot | ||
| 187 epithelial ovarian cancer specimens | - | qRT-PCR | No statistically significant association between DAXX mRNA expression and clinicopathological parameters was observed | [86] |
| HJURP | ||||
| 156 ovarian cancer specimens | Fallopian tube tissue samples from patients undergoing salpingectomy for benign diseases | TMA IHC |
| [87] |
| 98 advanced-stage (stage IIIB–IV) serous ovarian carcinoma specimens | - | IHC |
| [88] |
| UTERUS | ||||
| DAXX | ||||
| Endometrial clear cell carcinomas specimens | - | NGS | DAXX homozygous deletions were detected on 11% of cases | [89] |
| 18 Uterine smooth muscle tumours of uncertain malignant potential (STUMP) and 43 leiomyosarcoma specimens | - | IHC |
| [90] |
| 142 various uterine leiomyoma (UL) subtype specimens (consisting of 35 cellular ULs, 45 highly cellular ULs, 31 mitotically active ULs (including 15 tumors also displaying cellularity (n = 7) or high cellularity (n = 8) and 31 ULs with bizarre nuclei) and 64 conventional UL specimens | - | IHC | The loss of DAXX expression was observed only in 1 case of cellular UL | [91] |
| HJURP | ||||
| 552 endometrial carcinoma specimens | 23 normal endometrium samples | Data collection from the GEO dataset | High HJURP mRNA expression was associated with
| [92] |
| CERVIX | ||||
| DAXX | ||||
| Cervical intraepithelial neoplasia grade 1 (CIN1), CIN2, CIN3, and cervical cancer specimens | Normal cervical epithelial tissue samples | IHC | DAXX expression was located
| [93] |
| Malignant Tissues | Control Tissues | Methods | Results | Refs |
|---|---|---|---|---|
| LEUKEMIA | ||||
| DAXX | ||||
| 100 BM specimens of pediatric acute leukemia (AL) (49 acute lymphoblastic leukemia (ALL), 21 acute non-lymhocytic leukemia (ANLL), 30 acute myeloid leukemia (AML)) | 30 BM specimens of healthy children | IHC | Increased rate of DAXX protein expression compared to controls
| [94] |
| 50 BM specimens of pediatric AL (34 ALL, 16 ANLL) | 20 BM specimens of healthy children | IHC | Increased rate of DAXX protein expression
| [95] |
| 50 BM specimens of pediatric AL | 20 BM specimens of healthy children | IHC | Increased rate of DAXX protein expression
| [96] |
| BM specimens of 8 AML cases at diagnosis and relapse | - | WGS | Recurrent somatic DAXX mutations in relapsed AML cases | [97] |
| MYELODYSPLASTIC NEOPLASMS | ||||
| DAXX | ||||
| 77 BM aspiration specimens | Control CD34+ BM cells | Real-time PCR | Increased DAXX mRNA expression in myelodysplastic neoplasms (MDS) CD34+ cells | [98] |
| CHRONIC LYMPHOCYTIC LEUKEMIA | ||||
| DAXX | ||||
| PB and BM aspiration specimens from 62 untreated patients | - | Flow cytometry | Positive correlation between Par-4 and DAXX protein expression | [99] |
| LYMPHOMA | ||||
| DAXX | ||||
| Purified B cells derived from cell suspensions of lymph nodes from 5 diffuse large B cell lymphoma (DLBCL) patients | Purified B cells derived from cell suspensions of tonsils from 2 controls | Western blot | Ιncreased DAXX protein expression in DLBCL B cells compared to tonsil B cells | [100] |
| 196 DLBCL tissues | 76 follicular lymphoma (FL) tissues, 52 chronic lymphocytic leukemia (CLL) tissues | Tissue microarray analysis |
| |
| 93 biopsy samples of newly diagnosed natural killer T cell lymphoma (NKTCL) | - | IHC |
| [101] |
| MULTIPLE MYELOMA | ||||
| HJURP | ||||
| - | Gene Expression Omnibus datasets: Expression profiling by array |
| [102] |
| MM patient datasets (CoMMpass, HOVON, and UAMS) | - | WGS | High HJURP value was associated with worse OS and PFS | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pergaris, A.; Genaris, I.; Stergiou, I.E.; Klijanienko, J.; Papadakos, S.P.; Theocharis, S. The Clinical Impact of Death Domain-Associated Protein and Holliday Junction Recognition Protein Expression in Cancer: Unmasking the Driving Forces of Neoplasia. Cancers 2023, 15, 5165. https://doi.org/10.3390/cancers15215165
Pergaris A, Genaris I, Stergiou IE, Klijanienko J, Papadakos SP, Theocharis S. The Clinical Impact of Death Domain-Associated Protein and Holliday Junction Recognition Protein Expression in Cancer: Unmasking the Driving Forces of Neoplasia. Cancers. 2023; 15(21):5165. https://doi.org/10.3390/cancers15215165
Chicago/Turabian StylePergaris, Alexandros, Ioannis Genaris, Ioanna E. Stergiou, Jerzy Klijanienko, Stavros P. Papadakos, and Stamatios Theocharis. 2023. "The Clinical Impact of Death Domain-Associated Protein and Holliday Junction Recognition Protein Expression in Cancer: Unmasking the Driving Forces of Neoplasia" Cancers 15, no. 21: 5165. https://doi.org/10.3390/cancers15215165
APA StylePergaris, A., Genaris, I., Stergiou, I. E., Klijanienko, J., Papadakos, S. P., & Theocharis, S. (2023). The Clinical Impact of Death Domain-Associated Protein and Holliday Junction Recognition Protein Expression in Cancer: Unmasking the Driving Forces of Neoplasia. Cancers, 15(21), 5165. https://doi.org/10.3390/cancers15215165








