Prediction of Radiation Treatment Response for Locally Advanced Rectal Cancer via a Longitudinal Trend Analysis Framework on Cone-Beam CT
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview of Longitudinal Radiomic Trend (LRT) Framework
2.1.1. Automatic Registration and Evaluation
2.1.2. Feature Extraction and Normalization
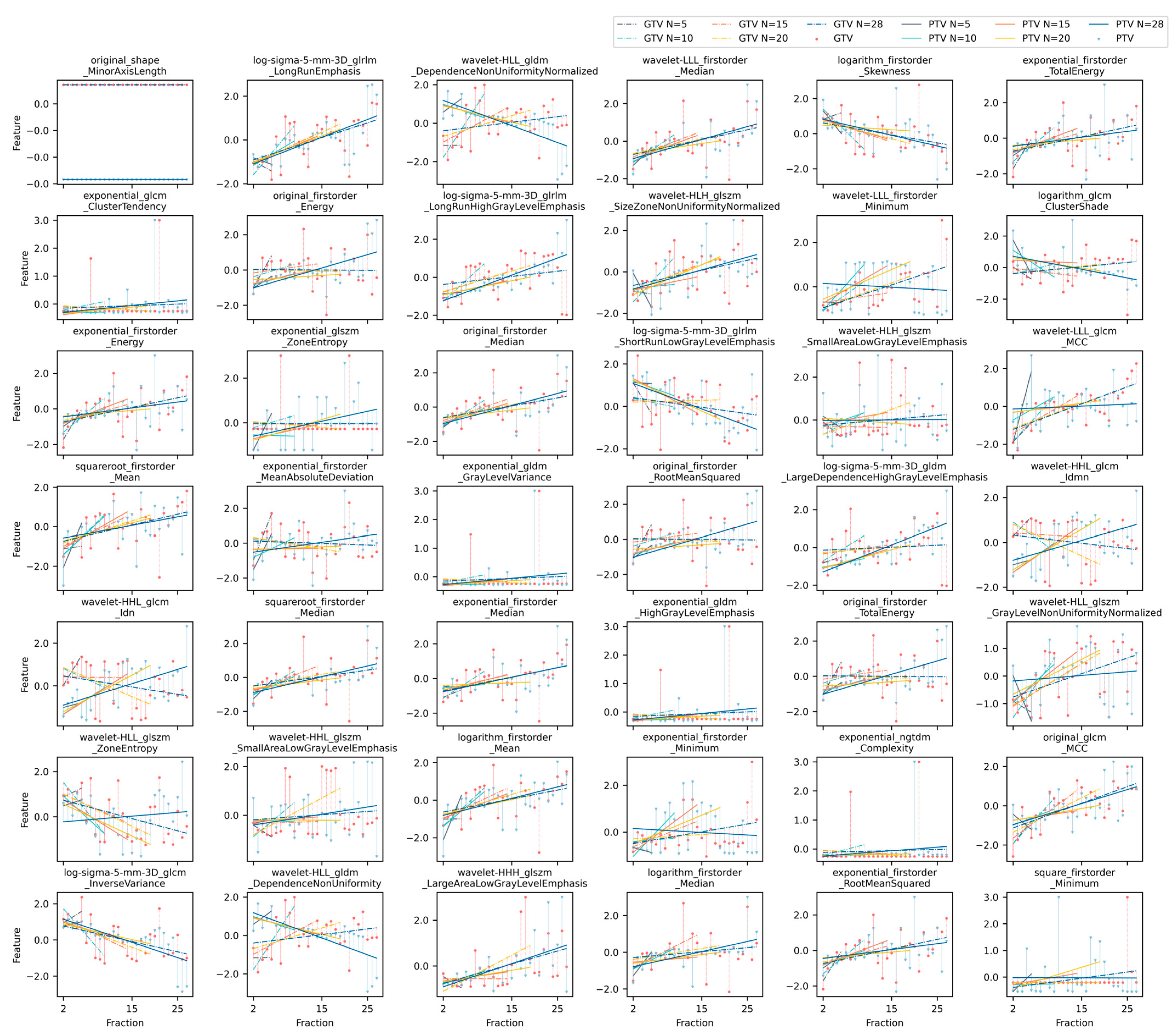
2.1.3. Longitudinal Radiomic Feature Trending
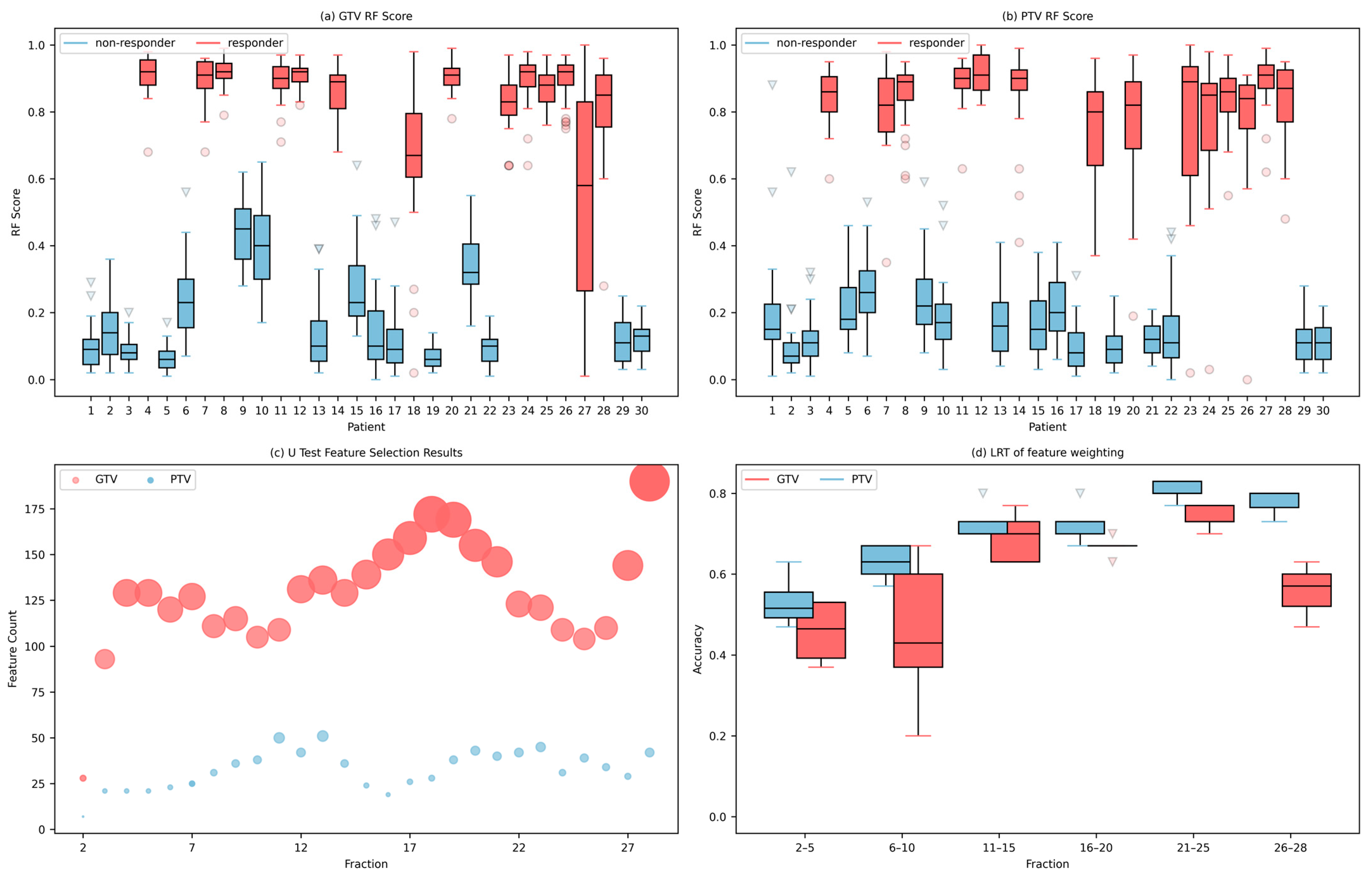
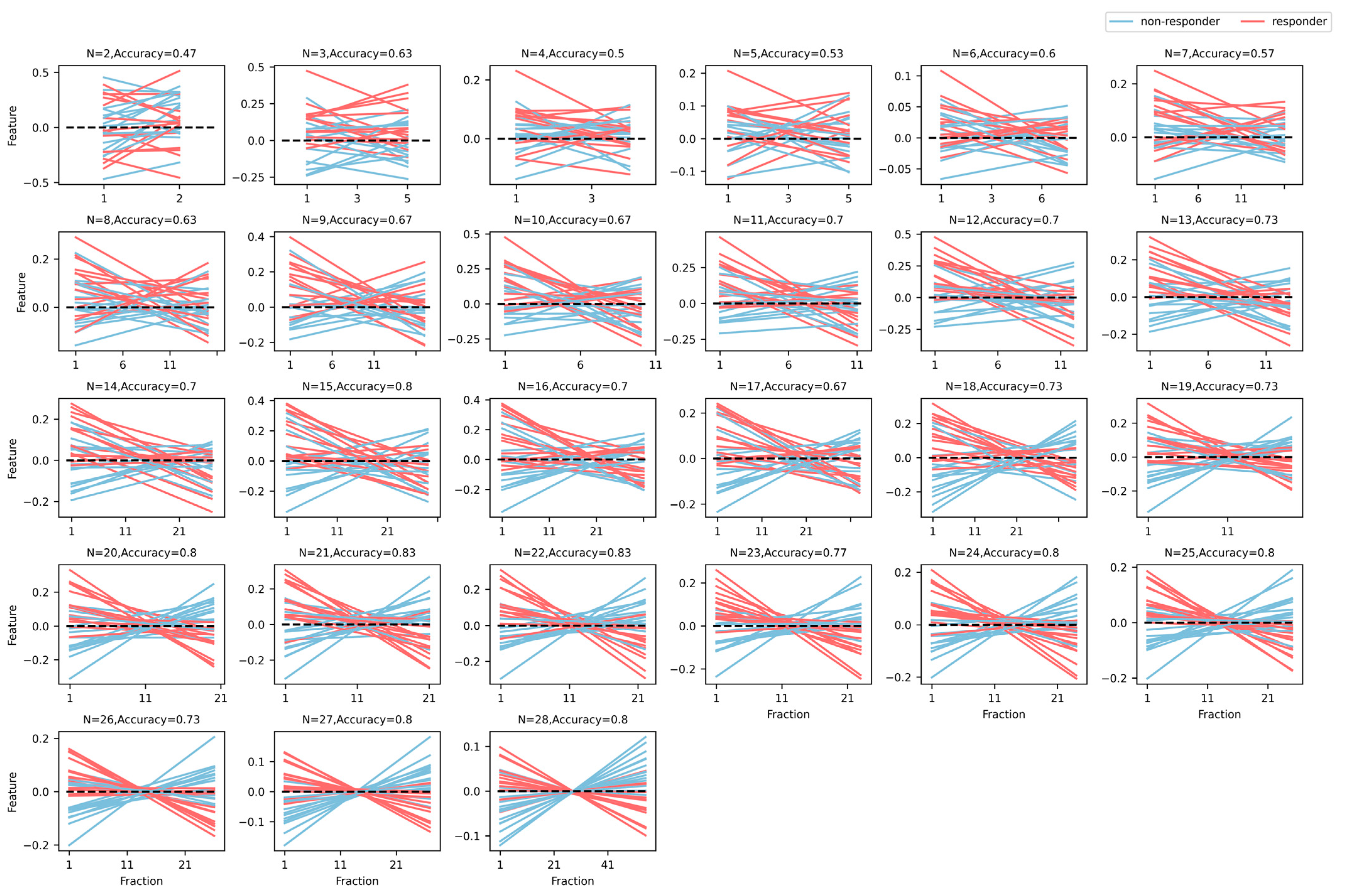
2.1.4. Feature Reduction and Modeling
2.2. Validation
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Breugom, A.J.; Swets, M.; Bosset, J.-F.; Collette, L.; Sainato, A.; Cionini, L.; Glynne-Jones, R.; Counsell, N.; Bastiaannet, E.; Broek, C.B.M.v.D.; et al. Adjuvant chemotherapy after preoperative (chemo) radiotherapy and surgery for patients with rectal cancer: A systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015, 16, 200–207. [Google Scholar] [CrossRef]
- Ferrari, L.; Fichera, A. Neoadjuvant chemoradiation therapy and pathological complete response in rectal cancer. Gastroenterol. Rep. 2015, 3, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Janjan, N.A.; Khoo, V.S.; Abbruzzese, J.; Pazdur, R.; Dubrow, R.; Cleary, K.R.; Allen, P.K.; Lynch, P.M.; Glober, G.; Wolff, R.; et al. Tumor downstaging and sphincter preservation with preoperative chemoradiation in locally advanced rectal cancer: The MD anderson cancer center experience. Int. J. Radiat. Oncol. Biol. Phys. 1999, 44, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Maas, M.; Nelemans, P.J.; Valentini, V.; Das, P.; Rödel, C.; Kuo, L.-J.; Calvo, F.; García-Aguilar, J.; Glynne-Jones, R.; Haustermans, K.; et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol. 2010, 11, 835–844. [Google Scholar] [CrossRef]
- Rombouts, A.J.M.; Al-Najami, I.; Abbott, N.L.; Appelt, A.; Baatrup, G.; Bach, S.; Bhangu, A.; Garm Spindler, K.L.; Gray, R.; Handley, K.; et al. Can we save the rectum by watchful waiting or TransAnal microsurgery following (chemo) radiotherapy versus total mesorectal excision for early REctal cancer (STAR-TREC study)?: Protocol for a multicentre, randomised feasibility study. BMJ Open 2017, 7, e019474. [Google Scholar] [CrossRef]
- Park, S.H.; Cho, S.H.; Choi, S.H.; Jang, J.K.; Kim, M.J.; Kim, S.H.; Lim, J.S.; Moon, S.K.; Park, J.H.; Seo, N.; et al. MRI assessment of complete response to preoperative chemoradiation therapy for rectal cancer: 2020 guide for practice from the korean society of abdominal radiology. Korean J. Radiol. 2020, 21, 812. [Google Scholar] [CrossRef]
- Gambacorta, M.A.; Masciocchi, C.; Chiloiro, G.; Meldolesi, E.; Macchia, G.; van Soest, J.; Peters, F.; Collette, L.; Gérard, J.P.; Ngan, S.; et al. Timing to achieve the highest rate of pCR after preoperative radiochemotherapy in rectal cancer: A pooled analysis of 3085 patients from 7 randomized trials. Radiother. Oncol. 2021, 154, 154–160. [Google Scholar] [CrossRef]
- López-Campos, F.; Martín-Martín, M.; Fornell-Pérez, R.; García-Pérez, J.C.; Die-Trill, J.; Fuentes-Mateos, R.; López-Durán, S.; Domínguez-Rullán, J.; Ferreiro, R.; Riquelme-Oliveira, A.; et al. Watch and wait approach in rectal cancer: Current controversies and future directions. World J. Gastroenterol. 2020, 26, 4218. [Google Scholar] [CrossRef]
- Lin, Z.; Cai, W.; Hou, W.; Chen, Y.; Gao, B.; Mao, R.; Wang, L.; Li, Z. CT-guided survival prediction of esophageal cancer. IEEE J. Biomed. Health Inform. 2021, 26, 2660–2669. [Google Scholar] [CrossRef]
- Cui, S.; Ten Haken, R.K.; El Naqa, I. Integrating multiomics information in deep learning architectures for joint actuarial outcome prediction in non-small cell lung cancer patients after radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Lou, B.; Doken, S.; Zhuang, T.; Wingerter, D.; Gidwani, M.; Mistry, N.; Ladic, L.; Kamen, A.; Abazeed, M.E. An image-based deep learning framework for individualising radiotherapy dose: A retrospective analysis of outcome prediction. Lancet Digit. Health 2019, 1, e136–e147. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, Y.; Dong, D.; Li, C.; Liang, X.; Zhang, C.; Wan, L.; Zhao, X.; Xu, K.; Zhou, C.; et al. Novel radiomic signature as a prognostic biomarker for locally advanced rectal cancer. J. Magn. Reson. Imaging 2018, 48, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Cusumano, D.; Dinapoli, N.; Boldrini, L.; Chiloiro, G.; Gatta, R.; Masciocchi, C.; Lenkowicz, J.; Casà, C.; Damiani, A.; Azario, L.; et al. Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. La Radiol. Med. 2018, 123, 286–295. [Google Scholar] [CrossRef]
- Horvat, N.; Veeraraghavan, H.; Khan, M.; Blazic, I.; Zheng, J.; Capanu, M.; Sala, E.; Garcia-Aguilar, J.; Gollub, M.J.; Petkovska, I. MR imaging of rectal cancer: Radiomics analysis to assess treatment response after neoadjuvant therapy. Radiology 2018, 287, 833–843. [Google Scholar] [CrossRef]
- Bonomo, P.; Socarras Fernandez, J.; Thorwarth, D.; Casati, M.; Livi, L.; Zips, D.; Gani, C. Simulation CT-based radiomics for prediction of response after neoadjuvant chemo-radiotherapy in patients with locally advanced rectal cancer. Radiat. Oncol. 2022, 17, 84. [Google Scholar] [CrossRef]
- Rosen, B.S.; Hawkins, P.G.; Polan, D.F.; Balter, J.M.; Brock, K.K.; Kamp, J.D.; Lockhart, C.M.; Eisbruch, A.; Mierzwa, M.L.; Haken, R.K.T.; et al. Early changes in serial CBCT-measured parotid gland biomarkers predict chronic xerostomia after head and neck radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1319–1329. [Google Scholar] [CrossRef]
- Du, F.; Tang, N.; Cui, Y.; Wang, W.; Zhang, Y.; Li, Z.; Li, J. A novel nomogram model based on cone-beam CT radiomics analysis technology for predicting radiation pneumonitis in esophageal cancer patients undergoing radiotherapy. Front. Oncol. 2020, 10, 596013. [Google Scholar] [CrossRef]
- Posiewnik, M.; Piotrowski, T. A review of cone-beam CT applications for adaptive radiotherapy of prostate cancer. Phys. Med. 2019, 59, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Le, W.T.; Bang, C.; Cordelle, P.; Markel, D.; Nguyen-Tan, P.F.; Bahig, H.; Kadoury, S. Comparing 3D deformations between longitudinal daily CBCT acquisitions using CNN for head and neck radiotherapy toxicity prediction. arXiv 2023, arXiv:230303965. [Google Scholar]
- Nardone, V.; Reginelli, A.; Grassi, R.; Boldrini, L.; Vacca, G.; D’ippolito, E.; Annunziata, S.; Farchione, A.; Belfiore, M.P.; Desideri, I.; et al. Delta radiomics: A systematic review. La Radiol. Med. 2021, 126, 1571–1583. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Yang, P.-F.; Chen, S.; Shao, Y.-Y.; Xu, L.-M.; Wu, Y.; Teng, W.; Zhou, X.-Z.; Li, B.-H.; Luo, C.; et al. A delta-radiomics model for preoperative evaluation of neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imaging 2020, 20, 7. [Google Scholar] [CrossRef]
- Crombé, A.; Périer, C.; Kind, M.; De Senneville, B.D.; Le Loarer, F.; Italiano, A.; Buy, X.; Saut, O. T2-based MRI delta-radiomics improve response prediction in soft-tissue sarcomas treated by neoadjuvant chemotherapy. J. Magn. Reson. Imaging 2019, 50, 497–510. [Google Scholar] [CrossRef]
- Shayesteh, S.; Nazari, M.; Salahshour, A.; Sandoughdaran, S.; Hajianfar, G.; Khateri, M.; Yaghobi Joybari, A.; Jozian, F.; Fatehi Feyzabad, S.H.; Arabi, H.; et al. Treatment response prediction using MRI-based pre-, post-, and delta-radiomic features and machine learning algorithms in colorectal cancer. Med. Phys. 2021, 48, 3691–3701. [Google Scholar] [CrossRef]
- Alahmari, S.S.; Cherezov, D.; Goldgof, D.B.; Hall, L.O.; Gillies, R.J.; Schabath, M.B. Delta radiomics improves pulmonary nodule malignancy prediction in lung cancer screening. IEEE Access 2018, 6, 77796–77806. [Google Scholar] [CrossRef]
- Plautz, T.E.; Zheng, C.; Noid, G.; Li, X.A. Time stability of delta-radiomics features and the impact on patient analysis in longitudinal CT images. Med. Phys. 2019, 46, 1663–1676. [Google Scholar] [CrossRef]
- Nasief, H.; Zheng, C.; Schott, D.; Hall, W.; Tsai, S.; Erickson, B.; Li, X.A. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. NPJ Precis. Oncol. 2019, 3, 25. [Google Scholar] [CrossRef]
- Boldrini, L.; Cusumano, D.; Chiloiro, G.; Casà, C.; Masciocchi, C.; Lenkowicz, J.; Cellini, F.; Dinapoli, N.; Azario, L.; Teodoli, S.; et al. Delta radiomics for rectal cancer response prediction with hybrid 0.35 t magnetic resonance-guided radiotherapy (MRgRT): A hypothesis-generating study for an innovative personalized medicine approach. La Radiol. Med. 2019, 124, 145–153. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and reproducibility of radiomic features: A systematic review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Reginelli, A.; Guida, C.; Belfiore, M.P.; Biondi, M.; Mormile, M.; Buonamici, F.B.; Di Giorgio, E.; Spadafora, M.; Tini, P.; et al. Delta-radiomics increases multicentre reproducibility: A phantom study. Med. Oncol. 2020, 37, 38. [Google Scholar] [CrossRef] [PubMed]
- Delgadillo, R.; Spieler, B.O.; Ford, J.C.; Kwon, D.; Yang, F.; Studenski, M.; Padgett, K.R.; Abramowitz, M.C.; Pra, A.D.; Stoyanova, R.; et al. Repeatability of CBCT radiomic features and their correlation with CT radiomic features for prostate cancer. Med. Phys. 2021, 48, 2386–2399. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.S.; Manoj, K.; Arumugam, S. Labeling methods for identifying outliers. Int. J. Stat. Syst. 2015, 10, 231–238. [Google Scholar]
- York, D. Least-squares fitting of a straight line. Can. J. Phys. 1966, 44, 1079–1086. [Google Scholar] [CrossRef]
- Johnson, M.L. Why, when, and how biochemists should use least squares. Anal. Biochem. 1992, 206, 215–225. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Pan, X.; Feng, T.; Liu, C.; Savjani, R.R.; Chin, R.K.; Sharon Qi, X. A survival prediction model via interpretable machine learning for patients with oropharyngeal cancer following radiotherapy. J. Cancer Res. Clin. Oncol. 2023, 149, 6813–6825. [Google Scholar] [CrossRef]
- Fu, J.; Zhong, X.; Li, N.; Van Dams, R.; Lewis, J.; Sung, K.; Raldow, A.C.; Jin, J.; Qi, X.S. Deep Learning-based Radiomic Features for Improving Neoadjuvant Chemoradiation Response Prediction in Locally Advanced Rectal Cancer. Phys. Med. Biol. 2020, 65, 075001. [Google Scholar] [CrossRef]
- Yamashita, H.; Takenaka, R.; Sakumi, A.; Haga, A.; Otomo, K.; Nakagawa, K. Analysis of motion of the rectum during preoperative intensity modulated radiation therapy for rectal cancer using cone-beam computed tomography. Radiat. Oncol. 2015, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Alickikus, Z.A.; Kuru, A.; Aydin, B.; Akcay, D.; Gorken, I.B. The importance of mesorectum motion in determining PTV margins in rectal cancer patients treated with neoadjuvant radiotherapy. J. Radiat. Res. 2020, 61, 335–342. [Google Scholar] [CrossRef] [PubMed]


| Clinical Characteristics | Good Responder (GR) | Poor Responder (PR) |
|---|---|---|
| # of cases | 14 | 16 |
| Gender, n (%) | ||
| Male | 9 (64.3%) | 12 (75%) |
| Female | 5 (35.7%) | 4 (25%) |
| Average age (standard deviation) (yr) | 52.3 ± 11.7 | 59.2 ± 9.3 |
| Stage, n (%) | ||
| II | 3 (21.4%) | 4 (25%) |
| III | 8 (57.2%) | 9 (56.3%) |
| IV | 3 (21.4%) | 1 (6.3%) |
| Unknown | 2 (12.4%) | |
| Response scores, n (%) | ||
| 0 | 7 (50%) | |
| 1 | 7 (50%) | |
| 2 | 11 (68.8%) | |
| 3 | 5 (31.2%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Raldow, A.C.; Weidhaas, J.B.; Zhou, Q.; Qi, X.S. Prediction of Radiation Treatment Response for Locally Advanced Rectal Cancer via a Longitudinal Trend Analysis Framework on Cone-Beam CT. Cancers 2023, 15, 5142. https://doi.org/10.3390/cancers15215142
Li Z, Raldow AC, Weidhaas JB, Zhou Q, Qi XS. Prediction of Radiation Treatment Response for Locally Advanced Rectal Cancer via a Longitudinal Trend Analysis Framework on Cone-Beam CT. Cancers. 2023; 15(21):5142. https://doi.org/10.3390/cancers15215142
Chicago/Turabian StyleLi, Zirong, Ann C. Raldow, Joanne B. Weidhaas, Qichao Zhou, and X. Sharon Qi. 2023. "Prediction of Radiation Treatment Response for Locally Advanced Rectal Cancer via a Longitudinal Trend Analysis Framework on Cone-Beam CT" Cancers 15, no. 21: 5142. https://doi.org/10.3390/cancers15215142
APA StyleLi, Z., Raldow, A. C., Weidhaas, J. B., Zhou, Q., & Qi, X. S. (2023). Prediction of Radiation Treatment Response for Locally Advanced Rectal Cancer via a Longitudinal Trend Analysis Framework on Cone-Beam CT. Cancers, 15(21), 5142. https://doi.org/10.3390/cancers15215142








