Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Search Strategy
2.3. Eligibility Criteria
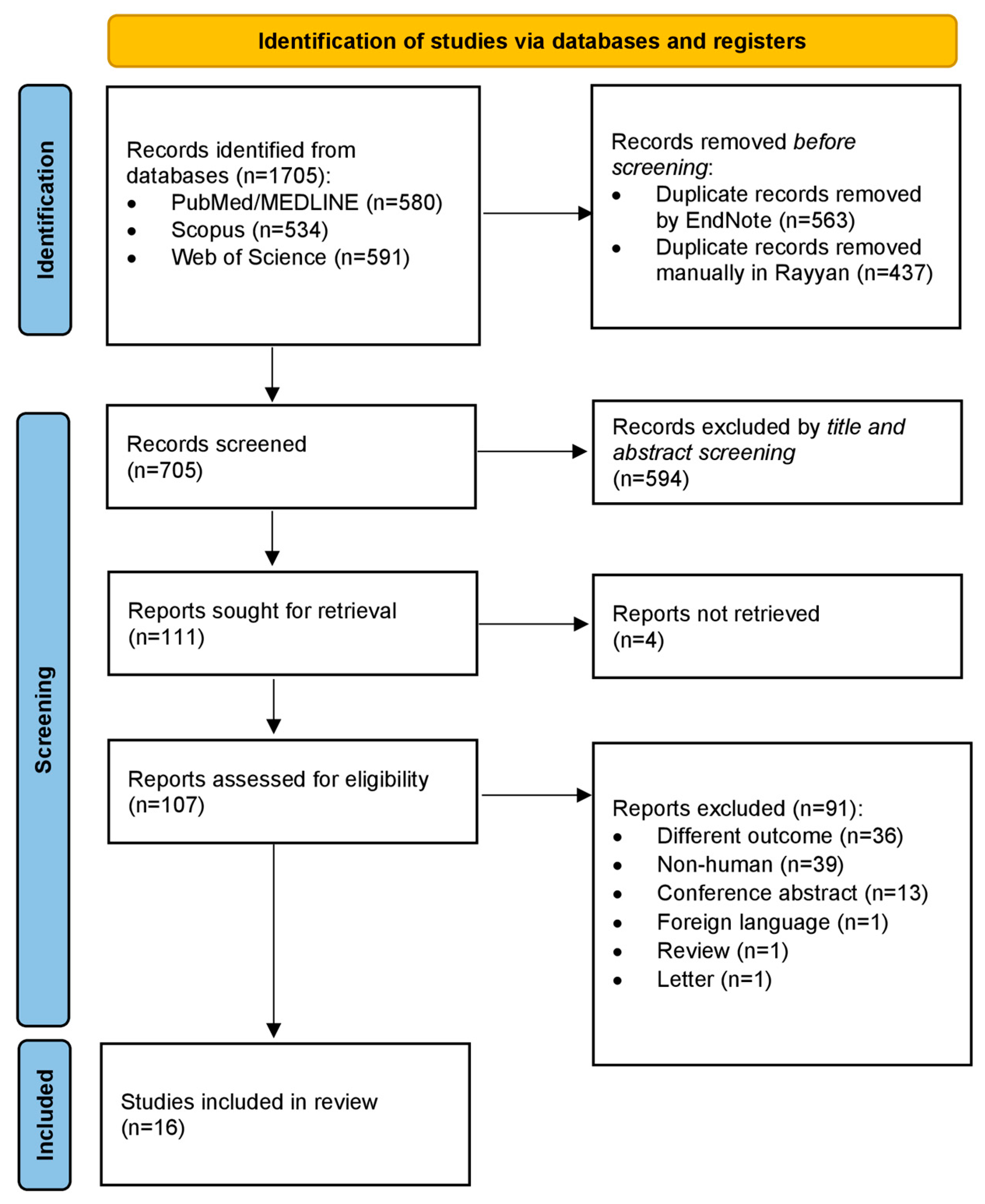
2.4. Study Selection and Screening
2.5. Data Extraction
2.6. Quality of Studies
2.7. Outcomes
3. Results
3.1. Study Selection
3.2. Study Characteristics and Data Collection
3.3. Quality Assessment
3.4. Proteomic Findings in SCLC Compared with Controls
3.5. Differential Proteomic Findings between SCLC and LCNEC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaspar, L.E.; McNamara, E.J.; Gay, E.G.; Putnam, J.B.; Crawford, J.; Herbst, R.S.; Bonner, J.A. Small-cell lung cancer: Prognostic factors and changing treatment over 15 years. Clin. Lung Cancer 2012, 13, 115–122. [Google Scholar] [CrossRef]
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers 2021, 7, 3. [Google Scholar] [CrossRef]
- Who, E.B. WHO Classification Thoracic Tumours; IARC Press: Lyon, France, 2021. [Google Scholar]
- Szeitz, B.; Megyesfalvi, Z.; Woldmar, N.; Valko, Z.; Schwendenwein, A.; Barany, N.; Paku, S.; Laszlo, V.; Kiss, H.; Bugyik, E.; et al. In-depth proteomic analysis reveals unique subtype-specific signatures in human small-cell lung cancer. Clin. Transl. Med. 2022, 12, e1060. [Google Scholar] [CrossRef]
- Derks, J.L.; Leblay, N.; Lantuejoul, S.; Dingemans, A.C.; Speel, E.M.; Fernandez-Cuesta, L. New Insights into the Molecular Characteristics of Pulmonary Carcinoids and Large Cell Neuroendocrine Carcinomas, and the Impact on Their Clinical Management. J. Thorac. Oncol. 2018, 13, 752–766. [Google Scholar] [CrossRef]
- Bobos, M.; Hytiroglou, P.; Kostopoulos, I.; Karkavelas, G.; Papadimitriou, C.S. Immunohistochemical distinction between merkel cell carcinoma and small cell carcinoma of the lung. Am. J. Dermatopathol. 2006, 28, 99–104. [Google Scholar] [CrossRef]
- Hiroshima, K.; Iyoda, A.; Shida, T.; Shibuya, K.; Iizasa, T.; Kishi, H.; Tanizawa, T.; Fujisawa, T.; Nakatani, Y. Distinction of pulmonary large cell neuroendocrine carcinoma from small cell lung carcinoma: A morphological, immunohistochemical, and molecular analysis. Mod. Pathol. 2006, 19, 1358–1368. [Google Scholar] [CrossRef]
- Kontogianni, K.; Nicholson, A.G.; Butcher, D.; Sheppard, M.N. CD56: A useful tool for the diagnosis of small cell lung carcinomas on biopsies with extensive crush artefact. J. Clin. Pathol. 2005, 58, 978–980. [Google Scholar] [CrossRef]
- Krpina, K.; Vranic, S.; Tomic, K.; Samarzija, M.; Baticic, L. Small Cell Lung Carcinoma: Current Diagnosis, Biomarkers, and Treatment Options with Future Perspectives. Biomedicines 2023, 11, 1982. [Google Scholar] [CrossRef]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- George, J.; Walter, V.; Peifer, M.; Alexandrov, L.B.; Seidel, D.; Leenders, F.; Maas, L.; Müller, C.; Dahmen, I.; Delhomme, T.M.; et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 2018, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.L.; Thornton, R.H. Personalized medicine for non-small-cell lung cancer: Implications of recent advances in tissue acquisition for molecular and histologic testing. Clin. Lung Cancer 2012, 13, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Castiglia, M.; Hong, D.; Alessandro, R.; Mertens, I.; Baggerman, G.; Zwaenepoel, K.; Gil-Bazo, I.; Passiglia, F.; Carreca, A.P.; et al. Liquid biopsies in lung cancer: The new ambrosia of researchers. Biochim. Biophys. Acta 2014, 1846, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-Manzo, R.; Rios-Castro, E.; Hernandez-Hernandez, J.M.; Oza, G.; Medina, M.A.; Tapia-Ramirez, J. Identification of Transferrin Receptor 1 (TfR1) Overexpressed in Lung Cancer Cells, and Internalization of Magnetic Au-CoFe2O4 Core-Shell Nanoparticles Functionalized with Its Ligand in a Cellular Model of Small Cell Lung Cancer (SCLC). Pharmaceutics 2022, 14, 1715. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.M.; Sanders, S.; Carter, M.; Honeyman, D.; Cleo, G.; Auld, Y.; Booth, D.; Condron, P.; Dalais, C.; Bateup, S.; et al. Improving the translation of search strategies using the Polyglot Search Translator: A randomized controlled trial. J. Med. Libr. Assoc. 2020, 108, 195–207. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Lumbreras, B.; Porta, M.; Marquez, S.; Pollan, M.; Parker, L.A.; Hernandez-Aguado, I. QUADOMICS: An adaptation of the Quality Assessment of Diagnostic Accuracy Assessment (QUADAS) for the evaluation of the methodological quality of studies on the diagnostic accuracy of ‘-omics’-based technologies. Clin. Biochem. 2008, 41, 1316–1325. [Google Scholar] [CrossRef]
- Whiting, P.; Rutjes, A.W.; Reitsma, J.B.; Bossuyt, P.M.; Kleijnen, J. The development of QUADAS: A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med. Res. Methodol. 2003, 3, 25. [Google Scholar] [CrossRef]
- Ahn, J.M.; Sung, H.J.; Yoon, Y.H.; Kim, B.G.; Yang, W.S.; Lee, C.; Park, H.M.; Kim, B.J.; Lee, S.Y.; An, H.J.; et al. Integrated Glycoproteomics Demonstrates Fucosylated Serum Paraoxonase 1 Alterations in Small Cell Lung Cancer. Mol. Cell. Proteom. 2014, 13, 30–48. [Google Scholar] [CrossRef]
- Bharti, A.; Ma, P.C.; Maulik, G.; Singh, R.; Khan, E.; Skarin, A.T.; Salgia, R. Haptoglobin alpha-subunit and hepatocyte growth factor can potentially serve as serum tumor biomarkers in small cell lung cancer. Anticancer Res. 2004, 24, 1031–1038. [Google Scholar]
- Fahrmann, J.F.; Katayama, H.; Irajizad, E.; Chakraborty, A.; Kato, T.; Mao, X.; Park, S.; Murage, E.; Rusling, L.; Yu, C.Y.; et al. Plasma Based Protein Signatures Associated with Small Cell Lung Cancer. Cancers 2021, 13, 3972. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Dai, J.; Zhang, Y.; Lin, Q.; Jiang, M.; Xu, X.; Liu, Q.; Jia, J. Support vector machines coupled with proteomics approaches for detecting biomarkers predicting chemotherapy resistance in small cell lung cancer. Oncol. Rep. 2012, 28, 2233–2238. [Google Scholar] [CrossRef] [PubMed]
- Hye-Cheol, J.; Gwang-Il, K.; Sang-Ho, C.; Kwang-Hyung, L.; Jung-Jae, K.; Jeong-Hee, K.; Kwang-Hoe, C. Proteomic analysis of human small cell lung cancer tissues: Up-regulation of coactosin-like protein-1. J. Proteome Res. 2011, 10, 269–276. [Google Scholar] [CrossRef]
- Kang, S.M.; Sung, H.J.; Ahn, J.M.; Park, J.Y.; Lee, S.Y.; Park, C.S.; Cho, J.Y. The Haptoglobin β chain as a supportive biomarker for human lung cancers. Mol. Biosyst. 2011, 7, 1167–1175. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, J.W.; Chertov, O.; Colantonio, S.; Simpson, J.T.; Fivash, M.J.; Yoo, C.W.; Lee, G.K.; Zo, J.I.; Kim, H.T.; et al. Matrix-assisted laser desorption/ionization mass spectrometry reveals decreased calcylcin expression in small cell lung cancer. Pathol. Int. 2012, 62, 28–35. [Google Scholar] [CrossRef]
- Lv, P.; Liu, Z.; Xu, B.; Tang, C.; Li, X.; Qin, H.; Yang, S.; Gao, H.; He, K.; Liu, X. Exploratory study on application of MALDI-TOF-MS to detect serum and urine peptides related to small cell lung carcinoma. Mol. Med. Rep. 2020, 21, 51–60. [Google Scholar] [CrossRef]
- Pedersen, S.; Jensen, K.P.; Honore, B.; Kristensen, S.R.; Pedersen, C.H.; Szejniuk, W.M.; Maltesen, R.G.; Falkmer, U. Circulating microvesicles and exosomes in small cell lung cancer by quantitative proteomics. Clin. Proteom. 2022, 19, 2. [Google Scholar] [CrossRef]
- Shah, A.; Singh, H.; Sachdev, V.; Lee, J.; Yotsukura, S.; Salgia, R.; Bharti, A. Differential Serum Level of Specific Haptoglobin Isoforms in Small Cell Lung Cancer. Curr. Proteom. 2010, 7, 49–65. [Google Scholar] [CrossRef]
- Sugar, S.; Bugyi, F.; Toth, G.; Papay, J.; Kovalszky, I.; Tornoczky, T.; Drahos, L.; Turiak, L. Proteomic Analysis of Lung Cancer Types-A Pilot Study. Cancers 2022, 14, 2629. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Cheng, X.Y.; Jiang, H.L.; Gu, J.Y.; Yin, Y.F.; Shen, Z.J.; Xu, C.G.; Pu, Z.J.; Li, J.B.; Xu, G.Q. Quantitative proteomic analysis of glycosylated proteins enriched from urine samples with magnetic ConA nanoparticles identifies potential biomarkers for small cell lung cancer. J. Pharm. Biomed. Anal. 2021, 206, 114352. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Yang, W.M.; Ao, M.H.; Hoti, N.; Gabrielson, E.; Chan, D.W.; Zhang, H.; Li, Q.K. Proteomic Analysis of the Air-Way Fluid in Lung Cancer. Detection of Periostin in Bronchoalveolar Lavage (BAL). Front. Oncol. 2020, 10, 1072. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Nomura, M.; Kato, Y.; Tojo, H.; Fujii, K.; Nagao, T.; Bando, Y.; Fehniger, T.E.; Marko-Varga, G.; Nakamura, H.; et al. A selected reaction monitoring mass spectrometric assessment of biomarker candidates diagnosing large-cell neuroendocrine lung carcinoma by the scaling method using endogenous references. PLoS ONE 2017, 12, e0176219. [Google Scholar] [CrossRef]
- Nakamura, H.; Fujii, K.; Gupta, V.; Hata, H.; Koizumu, H.; Hoshikawa, M.; Naruki, S.; Miyata, Y.; Takahashi, I.; Miyazawa, T.; et al. Identification of key modules and hub genes for small-cell lung carcinoma and large-cell neuroendocrine lung carcinoma by weighted gene co-expression network analysis of clinical tissue-proteomes. PLoS ONE 2019, 14, e0217105. [Google Scholar] [CrossRef]
- Nomura, M.; Fukuda, T.; Fujii, K.; Kawamura, T.; Tojo, H.; Kihara, M.; Bando, Y.; Gazdar, A.F.; Tsuboi, M.; Oshiro, H.; et al. Preferential expression of potential markers for cancer stem cells in large cell neuroendocrine carcinoma of the lung. An FFPE proteomic study. J. Clin. Bioinform. 2011, 1, 23. [Google Scholar] [CrossRef] [PubMed]
- Turner, G.A. Haptoglobin. A potential reporter molecule for glycosylation changes in disease. Adv. Exp. Med. Biol. 1995, 376, 231–238. [Google Scholar]
- Thompson, S.; Turner, G.A. Elevated levels of abnormally-fucosylated haptoglobins in cancer sera. Br. J. Cancer 1987, 56, 605–610. [Google Scholar] [CrossRef]
- Ahmed, N.; Barker, G.; Oliva, K.T.; Hoffmann, P.; Riley, C.; Reeve, S.; Smith, A.I.; Kemp, B.E.; Quinn, M.A.; Rice, G.E. Proteomic-based identification of haptoglobin-1 precursor as a novel circulating biomarker of ovarian cancer. Br. J. Cancer 2004, 91, 129–140. [Google Scholar] [CrossRef]
- Kossowska, B.; Ferens-Sieczkowska, M.; Gancarz, R.; Passowicz-Muszynska, E.; Jankowska, R. Fucosylation of serum glycoproteins in lung cancer patients. Clin. Chem. Lab. Med. 2005, 43, 361–369. [Google Scholar] [CrossRef]
- Chen, J.; Cheuk, I.W.; Siu, M.T.; Yang, W.; Cheng, A.S.; Shin, V.Y.; Kwong, A. Human haptoglobin contributes to breast cancer oncogenesis through glycolytic activity modulation. Am. J. Cancer Res. 2020, 10, 2865–2877. [Google Scholar]
- Provost, P.; Doucet, J.; Stock, A.; Gerisch, G.; Samuelsson, B.; Radmark, O. Coactosin-like protein, a human F-actin-binding protein: Critical role of lysine-75. Biochem. J. 2001, 359, 255–263. [Google Scholar] [CrossRef]
- Rakonjac, M.; Fischer, L.; Provost, P.; Werz, O.; Steinhilber, D.; Samuelsson, B.; Radmark, O. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc. Natl. Acad. Sci. USA 2006, 103, 13150–13155. [Google Scholar] [CrossRef] [PubMed]
- Pidgeon, G.P.; Lysaght, J.; Krishnamoorthy, S.; Reynolds, J.V.; O’Byrne, K.; Nie, D.; Honn, K.V. Lipoxygenase metabolism: Roles in tumor progression and survival. Cancer Metastasis Rev. 2007, 26, 503–524. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Xiao, X.; Liu, W.L.; Song, Y.; Liu, T.J.J.; Li, Y.J.; Zacksenhaus, E.; Hao, X.J.; Ben-David, Y. Coactosin-like protein CLP/Cotl1 suppresses breast cancer growth through activation of IL-24/PERP and inhibition of non-canonical TGFβ signaling. Oncogene 2018, 37, 323–331. [Google Scholar] [CrossRef]
- Song, Q.B.; Hu, W.G.; Wang, P.; Yao, Y.; Zeng, H.Z. Identification of serum biomarkers for lung cancer using magnetic bead-based SELDI-TOF-MS. Acta Pharmacol. Sin. 2011, 32, 1537–1542. [Google Scholar] [CrossRef][Green Version]
- Park, J.H.; Kim, Y.S.; Lee, H.L.; Shim, J.Y.; Lee, K.S.; Oh, Y.J.; Shin, S.S.; Choi, Y.H.; Park, K.J.; Park, R.W.; et al. Expression of peroxiredoxin and thioredoxin in human lung cancer and paired normal lung. Respirology 2006, 11, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, S.; Tang, X.; Liu, W.; Chen, H.; Gao, H. Prognostic value of baseline hemoglobin-to-red blood cell distribution width ratio in small cell lung cancer: A retrospective analysis. Thorac. Cancer 2020, 11, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, Y.; Cui, T.; Knosel, T.; Zhang, Q.; Albring, K.F.; Huber, O.; Petersen, I. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/beta-catenin signaling pathway in human lung cancer. Carcinogenesis 2012, 33, 1863–1870. [Google Scholar] [CrossRef]
- Lin, C.C.; Huang, Y.K.; Cho, C.F.; Lin, Y.S.; Lo, C.C.; Kuo, T.T.; Tseng, G.C.; Cheng, W.C.; Chang, W.C.; Hsiao, T.H.; et al. Targeting positive feedback between BASP1 and EGFR as a therapeutic strategy for lung cancer progression. Theranostics 2020, 10, 10925–10939. [Google Scholar] [CrossRef]
- Li, L.; Meng, Q.; Li, G.; Zhao, L. BASP1 Suppresses Cell Growth and Metastasis through Inhibiting Wnt/β-Catenin Pathway in Gastric Cancer. BioMed Res. Int. 2020, 2020, 8628695. [Google Scholar] [CrossRef]
- Liao, X.; Li, Z.; Zheng, H.; Qian, W.; Zhang, S.; Chen, S.; Li, X.; Tang, M.; Xu, Y.; Yu, R.; et al. Downregulation of BASP1 Promotes Temozolomide Resistance in Gliomas via Epigenetic Activation of the FBXO32/NF-κB/MGMT Axis. Mol. Cancer Res. 2023, 21, 648–663. [Google Scholar] [CrossRef]
- Asad, M.; Wajid, S.; Katare, D.P.; Mani, R.J.; Jain, S.K. Differential Expression of TOM34, AL1A1, PADI2 and KLRBA in NNK Induced Lung Cancer in Wistar Rats and their Implications. Curr. Cancer Drug Targets 2019, 19, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Guo, D.; Zhang, X.; Zhu, Y.; Wang, Z.; Jin, Y.; Guo, W.; Zhang, S. ENO3 Inhibits Growth and Metastasis of Hepatocellular Carcinoma via Wnt/β-Catenin Signaling Pathway. Front. Cell Dev. Biol. 2021, 9, 797102. [Google Scholar]
- Zhou, M.; Wang, Z.; Yao, Y.; Zhou, H.; Liu, M.; Sun, J. Neuron-specific enolase and response to initial therapy are important prognostic factors in patients with small cell lung cancer. Clin. Transl. Oncol. 2017, 19, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Baykara, Y.; Xiao, Y.; Yang, D.; Yakirevich, E.; Maleki, S.; Garcia-Moliner, M.; Wang, L.J.; Huang, C.K.; Lu, S. Utility of secretagogin as a marker for the diagnosis of lung neuroendocrine carcinoma. Virchows Arch. 2022, 481, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Sauerbrei, W.; McShane, L.M. REMARK guidelines for tumour biomarker study reporting: A remarkable history. Br. J. Cancer 2023, 128, 443–445. [Google Scholar] [CrossRef]
- Harris, A.L. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. Cancer 2005, 93, 385–386. [Google Scholar] [CrossRef][Green Version]
- Hayes, D.F. Defining Clinical Utility of Tumor Biomarker Tests: A Clinician’s Viewpoint. J. Clin. Oncol. 2021, 39, 238–248. [Google Scholar] [CrossRef]
- Freidlin, B.; McShane, L.M.; Korn, E.L. Randomized clinical trials with biomarkers: Design issues. J. Natl. Cancer Inst. 2010, 102, 152–160. [Google Scholar] [CrossRef]
- Sargent, D.J.; Conley, B.A.; Allegra, C.; Collette, L. Clinical trial designs for predictive marker validation in cancer treatment trials. J. Clin. Oncol. 2005, 23, 2020–2027. [Google Scholar] [CrossRef]
- Simon, R.; Altman, D.G. Statistical aspects of prognostic factor studies in oncology. Br. J. Cancer 1994, 69, 979–985. [Google Scholar] [CrossRef]
| Author Year | Sample Size | Country | Type of Sample | Proteomic Method Utilized | Type of Analysis | Proteins Identified |
|---|---|---|---|---|---|---|
| Ahn et al., 2014 [20] | SCLC = 50 Controls = 29 | Republic of Korea | Serum | LC/MRM-MS | Expressional | Upregulated in SCLC: SAMP, Fuscosylated-SAMP, CO9, Fucosylated-CO9, PON1, Fucosylated-PON1, Fusosylated- KAIN |
| Bharti et al., 2004 [21] | SCLC = 17 Controls = 5 | USA | Serum | MALDI-TOF-MS | Expressional | Upregulated in SCLC: HPA, HGF |
| Fahrmann et al., 2021 [22] | SCLC = 15 Controls = 15 | USA | Plasma | LC MS/MS | Expressional | Upregulated in SCLC: ENOG, CTRO, DIAP3, FBX11, TSP1, 1433Z, ACTB, ATCBL2, ACTC, COTL1, URP2, PROF1, TPM3, TPM4, VINC |
| Han et al., 2012 [23] | SCLC = 60 Controls = 48 | China | Serum | SELDI-TOF MS | Expressional | Upregulated in SCLC: S10A9 |
| Hye-Cheol et al., 2011 [24] | SCLC = 6 Controls = 6 | Republic of Korea | Tumor tissue (FFPE) | MALDI-TOF MS | Expressional | Upregulated in SCLC: ACTG, TUBA1B, LAMB1, COTL1, UCH1, UBE2K, CAH11 |
| Kang et al., 2010 [25] | SCLC = 40 Controls = 201 | Republic of Korea | Blood serum | Untargeted LC-ESI-MS/MS | Expressional | Upregulated in SCLC: HPB |
| Lee et al., 2012 [26] | SCLC = 7 Controls = 13 | Republic of Korea | Tumor tissue (FFPE) | MALDI-TOF MS | Expressional | Upregulated in SCLC: H4 Downregulated in SCLC: S10A6 |
| Lv et al., 2020 [27] | SCLC = 72 Controls = 72 | China | serum and urine samples | MALDI-TOF MS | Expressional | Upregulated in SCLC: FIBA, G6PI, CDK1 |
| Pedersen et al., 2022 [28] | SCLC = 24 Controls = 24 | Denmark | Plasma-derived microvesicles and exosomes | Nano LC-MS/MS | Expressional | Micovesicular proteins Upregulated in SCLC: SAA1, CRP, TFR1, AMPN, LG3BP Downregulated in SCLC: PGRP2, HBD, HBB, GELS, BGH3 Exosomal proteins Upregulated in SCLC: SAA1, SAA2, AMPN, HPT, FHR4 Downregulated in SCLC: KV401, FCN2, FA11, F13A, HBA |
| Shah et al., 2010 [29] | SCLC = 8 Controls = 8 | USA | Serum | MALDI-TOF-MS, ES-MS-MS | Expressional | Upregulated in SCLC: HPT |
| Sugár et al., 2022 [30] | SCLC = 10 Controls = 9 | Hungary | FFPE human tissue sections | nanoUHPLC-MS | Expressional Functional | Upregulated in SCLC: DESP, PSPC1, SSRP1, ACL6A, GORS2, NP1L1 Downregulated in SCLC: VWF, UTRN, EHD4, FKBP2, SUMF2, CO6A1, C4BPA, TPM2, S10A4, EIF1, CO6A2, GILT, GAPR1, ANK1, CO1A2, CATA, MEAK7, CAVN2, PDLI2, HBA, FHL1, NID1, LAMC1, HBB, CAH1, ANXA3, LYSC, AOC3, CAV1, ADH1B, CATZ, CAVN1, DESM, TENX |
| Zhang et al., 2021 [31] | SCLC = 9 Controls = 9 | China | Urine | LC-MS/MS | Expressional | Upregulated glycosylated proteins in SCLC: CATC, MA2B2, GNS, CATD, IGHG1, PCP, IGHV3, HEXA, KV133, PLBL2 Downregulated glycosylated proteins in SCLC: DSC1, QPCT, TGC, ANXA2, DESP |
| Zhou et al., 2020 [32] | SCLC = 4 Controls = 3 | USA | Bronchoalveolar lavage | LC-MS/MS | Expressional | Upregulated in SCLC: GNPTG, PI16, PERM, DIAC, POSTN, ITAL, PLXB2 |
| Author Year | Sample Size | Country | Type of Sample | Proteomic Method Utilized | Type of Analysis | Proteins Identified |
|---|---|---|---|---|---|---|
| Fukuda et al., 2017 [33] | LCNEC = 10 SCLC = 10 | Japan | FFPE tumor tissue | LC-MS/MS | Expressional | Upregulated in SCLC: BASP1, ENOG Downregulated in SCLC: 4F2, AL1A1, APOA1, ENOB, KCRB, LG3BP, PEBP1 |
| Nakamura et al., 2019 [34] | SCLC = 6 LCNEC = 6 | Japan | FFPE tumor tissue | LC-MS/MS | Expressional | Upregulated in SCLC: PARP9, DTX3L, HLTF, E2AK2, CNOT1, CDN2C, KAPCA, TOP1, NICA, RABP2 Downregulated in SCLC: KCD12, KCRU, SHLB2, EMAL2, CMGA, PRKRA, ERF1, HXK1, SEGN, MK01, MP2K1, RING1 |
| Nomura et al., 2011 [35] | SCLC = 5 LCNEC = 5 | Republic of Korea | FFPE tumor tissue | LC-MS/MS | Expressional | Upregulated in SCLC: BASP1, SEGN, FSCN1, NCAM1 Downregulated in SCLC: AL1A1, AK1C1, AK1C3, CD44, FABP7, ENOB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshoeibi, A.M.; Elsayed, B.; Kaleem, M.Z.; Elhadary, M.R.; Abu-Haweeleh, M.N.; Haithm, Y.; Krzyslak, H.; Vranic, S.; Pedersen, S. Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review. Cancers 2023, 15, 5005. https://doi.org/10.3390/cancers15205005
Elshoeibi AM, Elsayed B, Kaleem MZ, Elhadary MR, Abu-Haweeleh MN, Haithm Y, Krzyslak H, Vranic S, Pedersen S. Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review. Cancers. 2023; 15(20):5005. https://doi.org/10.3390/cancers15205005
Chicago/Turabian StyleElshoeibi, Amgad Mohamed, Basel Elsayed, Muhammad Zain Kaleem, Mohamed Ragab Elhadary, Mohannad Natheef Abu-Haweeleh, Yunes Haithm, Hubert Krzyslak, Semir Vranic, and Shona Pedersen. 2023. "Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review" Cancers 15, no. 20: 5005. https://doi.org/10.3390/cancers15205005
APA StyleElshoeibi, A. M., Elsayed, B., Kaleem, M. Z., Elhadary, M. R., Abu-Haweeleh, M. N., Haithm, Y., Krzyslak, H., Vranic, S., & Pedersen, S. (2023). Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review. Cancers, 15(20), 5005. https://doi.org/10.3390/cancers15205005







