Towards the Definition of Radiomic Features and Clinical Indices to Enhance the Diagnosis of Clinically Significant Cancers in PI-RADS 4 and 5 Lesions
Abstract
:Simple Summary
Abstract: Objectives
1. Introduction
2. Materials and Methods
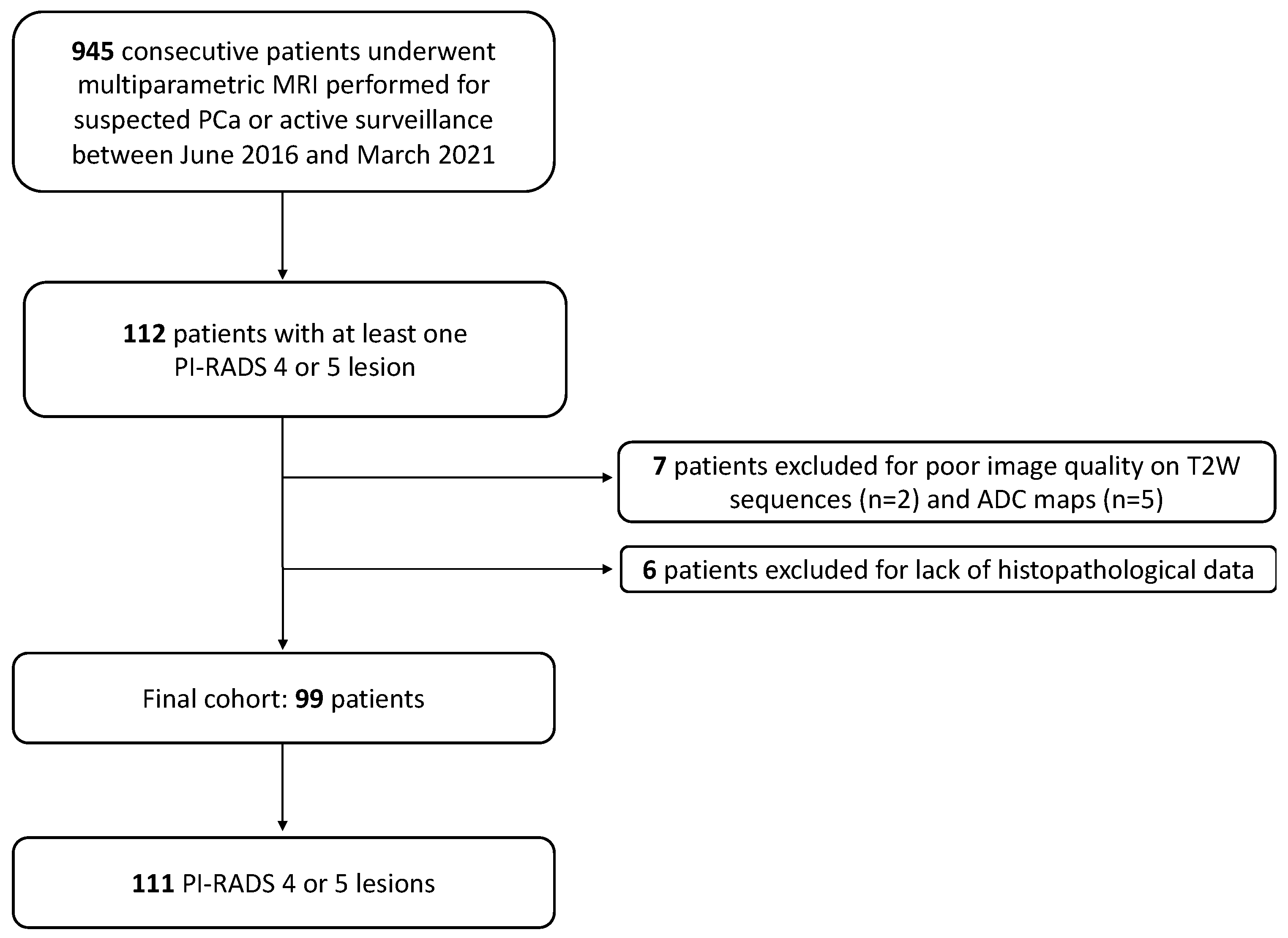
2.1. Patient Sample
2.2. Definition of the Reference Standard
2.3. MRI Acquisition
2.4. Image Analysis and Lesion Segmentation
2.5. Radiomic Feature Extraction, Selection, and Analysis
2.6. Statistical Analysis
3. Results
3.1. Patient and Lesion Characteristics
3.2. Selected Features and Univariate Model Performance
3.3. Clinical and Radiomic Multivariate Model Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AS | active surveillance |
| ADC | apparent diffusion coefficient |
| BPH | benign prostate hyperplasia |
| csPC | clinically significant prostate cancer |
| DICOM | Digital Imaging and Communication in Medicine |
| DWI | diffusion-weighted imaging |
| ISUP | International Society of Urological Pathology |
| GS | Gleason score |
| mpMRI | multiparametric magnetic resonance imaging |
| PACS | Picture Archiving and Communication System |
| PI-RADS | Prostate Imaging–Reporting and Data System |
| PC | prostate cancer |
| PPV | positive predictive value |
| PSA | prostate-specific antigen |
| PZ | peripheral zone |
| ROI | region of interest |
| T1W | T1-weighted |
| T2W | T2-weighted |
| TZ | transition zone |
| VOI | volume of interest |
References
- Pernar, C.H.; Ebot, E.M.; Wilson, K.M.; Mucci, L.A. The epidemiology of prostate cancer. Cold Spring Harb. Perspect Med. 2018, 8, a030361. [Google Scholar] [CrossRef]
- Kimura, T.; Egawa, S. Epidemiology of prostate cancer in Asian countries. Int. J. Urol. 2018, 25, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.; Castro, E.; Fizazi, K.; Heidenreich, A.; Ost, P.; Procopio, G.; Tombal, B.; Gillessen, S. Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- O’connor, L.P.; Lebastchi, A.H.; Horuz, R.; Rastinehad, A.R.; Siddiqui, M.M.; Grummet, J.; Kastner, C.; Ahmed, H.U.; Pinto, P.A.; Turkbey, B. Role of multiparametric prostate MRI in the management of prostate cancer. World J. Urol. 2021, 39, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Richenberg, J.; Løgager, V.; Panebianco, V.; Rouviere, O.; Villeirs, G.; Schoots, I.G. The primacy of multiparametric MRI in men with suspected prostate cancer. Eur. Radiol. 2019, 29, 6940–6952. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- Matoso, A.; I Epstein, J. Defining clinically significant prostate cancer on the basis of pathological findings. Histopathology 2019, 74, 135–145. [Google Scholar] [CrossRef]
- Kasivisvanathan, V.; Stabile, A.; Neves, J.B.; Giganti, F.; Valerio, M.; Shanmugabavan, Y.; Clement, K.D.; Sarkar, D.; Philippou, Y.; Thurtle, D.; et al. Magnetic Resonance Imaging-targeted Biopsy Versus Systematic Biopsy in the Detection of Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. 2019, 76, 284–303. [Google Scholar] [CrossRef]
- Oerther, B.; Engel, H.; Bamberg, F.; Sigle, A.; Gratzke, C.; Benndorf, M. Cancer detection rates of the PI-RADSv2.1 assessment cate-gories: Systematic review and meta-analysis on lesion level and patient level. Prostate Cancer Prostatic Dis. 2022, 25, 256–263. [Google Scholar] [CrossRef]
- Mazzone, E.; Stabile, A.; Pellegrino, F.; Basile, G.; Cignoli, D.; Cirulli, G.O.; Sorce, G.; Barletta, F.; Scuderi, S.; Bravi, C.A.; et al. Positive Predictive Value of Prostate Imaging Reporting and Data System Version 2 for the Detection of Clinically Significant Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. Oncol. 2021, 4, 697–713. [Google Scholar] [CrossRef]
- Syed, J.S.; Nguyen, K.A.; Nawaf, C.B.; Bhagat, A.M.; Huber, S.; Levi, A.; Humphrey, P.; Weinreb, J.C.; Schulam, P.G.; Sprenkle, P.C. Prostate zonal anatomy correlates with the detection of prostate cancer on multiparametric magnetic resonance imaging/ultrasound fusion–targeted biopsy in patients with a solitary PI-RADS v2–scored lesion. Urol. Oncol. Semin. Orig. Investig. 2017, 35, 542.e19–542.e24. [Google Scholar] [CrossRef] [PubMed]
- Gaudiano, C.; Rustici, A.; Corcioni, B.; Ciccarese, F.; Bianchi, L.; Schiavina, R.; Giunchi, F.; Fiorentino, M.; Brunocilla, E.; Golfieri, R. PI-RADS version 2.1 for the evaluation of transition zone lesions: A practical guide for radiologists. Br. J. Radiol. 2022, 95, 20210916. [Google Scholar] [CrossRef] [PubMed]
- Hupe, M.C.; Offermann, A.; Tharun, L.; Fürschke, A.; Frydrychowicz, A.; Garstka, N.; Shariat, S.F.; Barkhausen, J.; Merseburger, A.S.; Kramer, M.W.; et al. Histomorphological analysis of false pos-itive PI-RADS 4 and 5 lesions. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 636.e7–636.e12. [Google Scholar]
- Padhani, A.R.; Barentsz, J.; Villeirs, G.; Rosenkrantz, A.B.; Margolis, D.J.; Turkbey, B.; Thoeny, H.C.; Cornud, F.; Haider, M.A.; Macura, K.J.; et al. PI-RADS Steering Committee: The PI-RADS Multiparametric MRI and MRI-directed Biopsy Pathway. Radiology 2019, 292, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Midiri, F.; Vernuccio, F.; Purpura, P.; Alongi, P.; Bartolotta, T.V. Multiparametric MRI and Radiomics in Prostate Cancer: A Review of the Current Literature. Diagnostics 2021, 11, 1829. [Google Scholar] [CrossRef]
- Epstein, J.I.; Allsbrook, W.C., Jr.; Amin, M.B.; Egevad, L.L.; ISUP Grading Committee. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2005, 29, 1228–1242. [Google Scholar] [CrossRef]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- Sarradin, M.; Lepiney, C.; Celhay, O.; Delpech, P.O.; Charles, T.; Pillot, P.; Bernardeau, S.; Tasu, J.P.; Irani, J. Évaluation prospective du délai minimum de réalisation de l’IRM prostatique après une biopsie de prostate : Facteurs prédictifs cliniques et anatomopathologiques de remaniements hémorragiques. Progrès En Urol. 2018, 28, 85–93. [Google Scholar] [CrossRef]
- Scalco, E.; Belfatto, A.; Mastropietro, A.; Rancati, T.; Avuzzi, B.; Messina, A.; Valdagni, R.; Rizzo, G. T2w-MRI signal normalization affects radiomics features reproducibility. Med. Phys. 2020, 47, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; de Cobelli, O.; Musi, G.; del Giudice, F.; Carrieri, G.; Busetto, G.M.; Falagario, U.G.; Sciarra, A.; Maggi, M.; Crocetto, F.; et al. Radiomics in prostate cancer: An up-to-date review. Ther. Adv. Urol. 2022, 14, 175628722211090. [Google Scholar] [CrossRef] [PubMed]
- Fehr, D.; Veeraraghavan, H.; Wibmer, A.; Gondo, T.; Matsumoto, K.; Vargas, H.A.; Sala, E.; Hricak, H.; Deasy, J.O. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc. Natl. Acad. Sci. USA 2015, 112, E6265–E6273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, W.; Yue, X.; Shen, J.; Gao, C.; Pang, P.; Cui, F.; Xu, M. Development of a Novel, Multi-Parametric, MRI-Based Radiomic Nomogram for Differentiating Between Clinically Significant and Insignificant Prostate Cancer. Front. Oncol. 2020, 10, 888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.S.; Schelb, P.; Kohl, S.; Radtke, J.P.; Wiesenfarth, M.; Schimmöller, L.; Kuder, T.A.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.-P.; et al. Improvement of PI-RADS-dependent prostate cancer classification by quantitative image assessment using radiomics or mean ADC. Magn. Reson. Imaging 2021, 82, 9–17. [Google Scholar] [CrossRef]
- Giambelluca, D.; Cannella, R.; Vernuccio, F.; Comelli, A.; Pavone, A.; Salvaggio, L.; Galia, M.; Midiri, M.; Lagalla, R.; Salvaggio, G. PI-RADS 3 Lesions: Role of Prostate MRI Texture Analysis in the Identification of Prostate Cancer. Curr. Probl. Diagn. Radiol. 2021, 50, 175–185. [Google Scholar] [CrossRef]
- Hou, Y.; Bao, M.-L.; Wu, C.-J.; Zhang, J.; Zhang, Y.-D.; Shi, H.-B. A radiomics machine learning-based redefining score robustly identifies clinically significant prostate cancer in equivocal PI-RADS score 3 lesions. Abdom. Radiol. 2020, 45, 4223–4234. [Google Scholar] [CrossRef]
- Corsi, A.; De Bernardi, E.; Bonaffini, P.A.; Franco, P.N.; Nicoletta, D.; Simonini, R.; Ippolito, D.; Perugini, G.; Occhipinti, M.; Da Pozzo, L.F.; et al. Radiomics in PI-RADS 3 Multiparametric MRI for Prostate Cancer Identification: Literature Models Re-Implementation and Proposal of a Clinical–Radiological Model. J. Clin. Med. 2022, 11, 6304. [Google Scholar] [CrossRef]
- Ma, L.; Zhou, Q.; Yin, H.; Ang, X.; Li, Y.; Xie, G.; Li, G. Texture analysis based on PI-RADS 4/5-scored magnetic resonance images combined with machine learning to distinguish benign lesions from prostate cancer. Transl. Cancer Res. TCR 2022, 11, 1146–1161. [Google Scholar] [CrossRef]
- Laschkar, S.; Montagne, S.; De Kerviler, E.; Roupret, M.; Lucidarme, O.; Cussenot, O.; Penna, R.R. Zonal anatomy of the prostate using magnetic resonance imaging, morphometrics, and radiomic features: Impact of age-related changes. Br. J. Radiol. 2022, 95, 20210156. [Google Scholar] [CrossRef]
- Ginsburg, S.B.; Algohary, A.; Pahwa, S.; Gulani, V.; Ponsky, L.; Aronen, H.J.; Boström, P.J.; Böhm, M.; Haynes, A.; Brenner, P.; et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study. J. Magn. Reson. Imaging 2016, 46, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Krishna, S.; Thornhill, R.E.; Flood, T.A.; McInnes, M.D.F.; Schieda, N. Transition zone prostate cancer: Logistic regression and machine-learning models of quantitative ADC, shape and texture features are highly accurate for diagnosis: Machine-Learning Diagnosis of PZ PCa. J. Magn. Reson. Imaging 2019, 50, 940–950. [Google Scholar] [CrossRef] [PubMed]
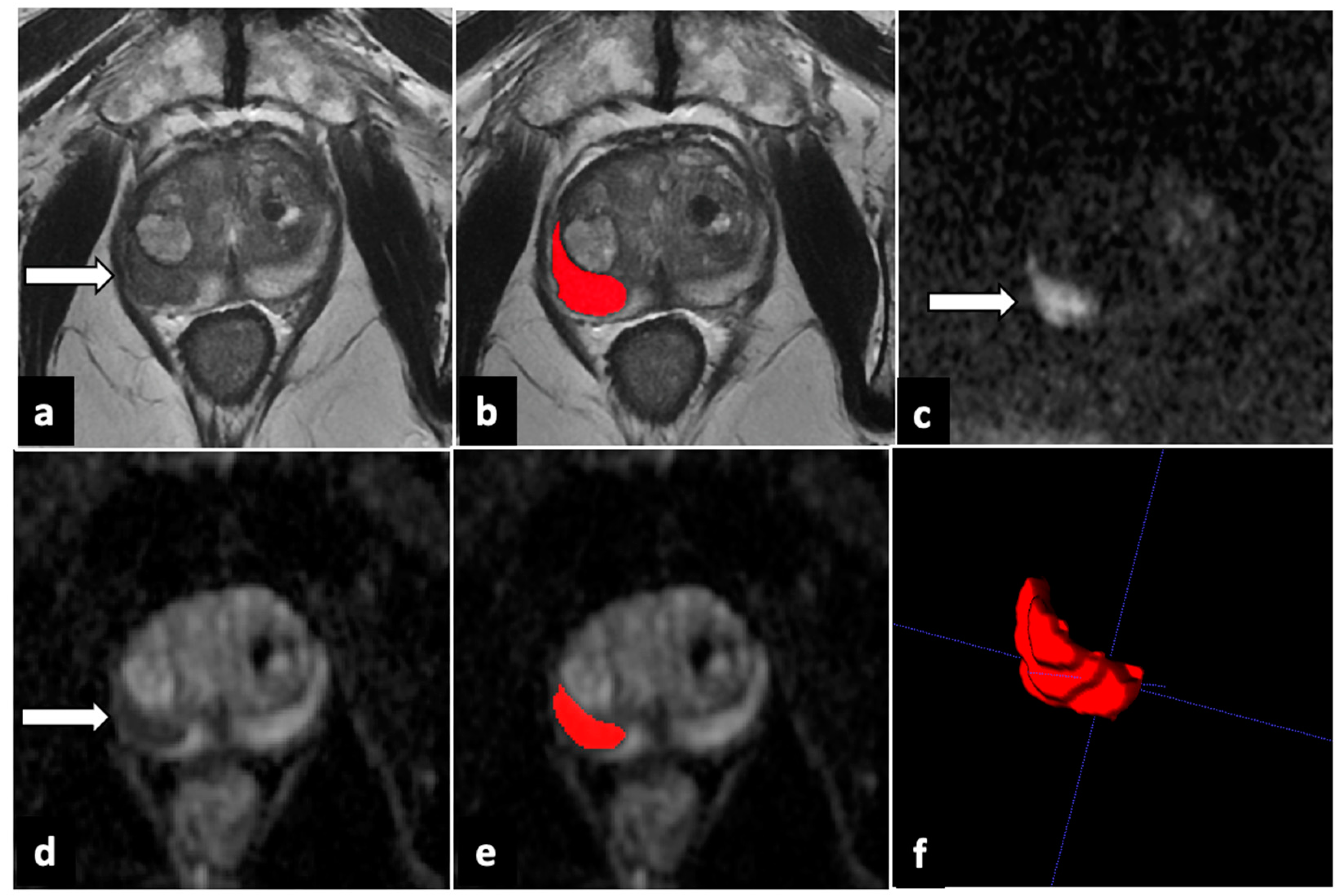
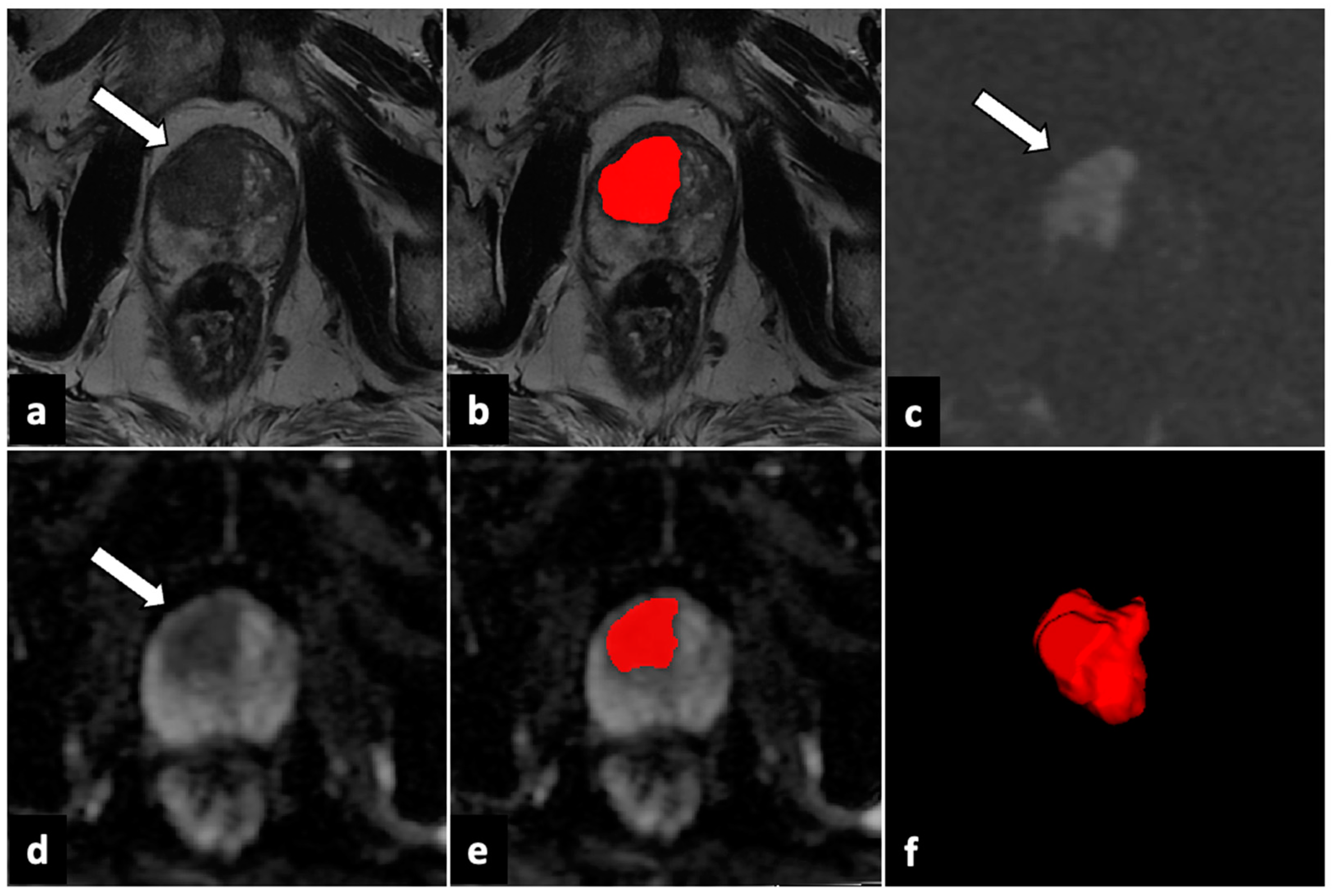
| Sequence | Plane | TR/TE (ms) | FOV (mm) | Slice Thickness (mm) | Gap (mm) |
|---|---|---|---|---|---|
| T2W SSFSE | axial | 3290/90 | 320 × 240 | 4 | 0.4 |
| T2W FRFSE | axial | 7480/150 | 220 × 220 | 3 | 0 |
| T2W FRFSE | sagittal | 7861/150 | 220 × 220 | 3 | 0 |
| T2W FRFSE | coronal | 6583/150 | 220 × 220 | 3 | 0 |
| T1W SSFSE | axial | 620/10 | 512 × 256 | 4 | 0.4 |
| T1W 3D FSPGR (DCE) Fat-saturated | axial | 5.3/2.2 | 380 × 350 | 3 | 0 |
| DWI (b-values: 100, 1000, 2000 s/mm2) | axial | 7775/91 | 120 × 120 | 3 | 0 |
| Patients (n = 99) | |||
|---|---|---|---|
| Age (y, IQR) | 69 (59–79) | ||
| PSA (ng/mL) mean ± SD | 7.9 ± 4.8 | ||
| PSA density (ng/mL2) mean ± SD | 0.18 ± 0.15 | ||
| Prostate volume (mL) mean ± SD | 51.6 ± 27.3 | ||
| Lesions (n = 111) | |||
| ADC value (10−6 mm2/s) mean ± SD | 653 ± 223 | ||
| Positive at biopsy (n, %) | Negative at biopsy (n, %) | ||
| Overall (n, %) | 111 (100) | 79 (71.2) | 32 (28.8) |
| PI-RADS 4 (n, %) | 74 (66.7) | 52 (70.3) | 22 (29.7) |
| Peripheral zone (n, %) | 68 (91.9) | 49 (72) | 19 (28) |
| Transition zone (n, %) | 6 (8.1) | 3 (50) | 3 (50) |
| PI-RADS 5 (n, %) | 37 (33) | 27 (73) | 10 (37) |
| Peripheral zone (n, %) | 18 (48.7) | 15 (83) | 3 (17) |
| Transition zone (n, %) | 19 (51.3) | 12 (63) | 7 (37) |
| ISUP prostate cancer grade group (n, %) | |||
| 1 (GS ≤ 6) | 32 (28.8) | ||
| 2 (GS = 3 + 4) | 46 (41.5) | ||
| 3 (GS = 4 + 3) | 20 (18.0) | ||
| 4 (GS = 8) | 11 (9.9) | ||
| 5 (GS ≥ 9) | 2 (1.8) | ||
| Features | % Choices |
|---|---|
| Peripheral and transitional zone | |
| PSA density | 56% |
| T2-wavelet-HHH_glszm_GrayLevelVariance | 51% |
| T2-wavelet-LLL_glszm_GrayLevelVariance | 21% |
| ADC-original_glcm_ClusterShade | 24% |
| ADC-wavelet-HHH_firstorder_Minimum | 31% |
| ADC-wavelet-LLL_glrlm_LongRunHighGrayLevelEmphasis | 26% |
| Peripheral zone | |
| PSA density | 21% |
| T2-wavelet-HHH_glszm_GrayLevelVariance | 41% |
| T2-wavelet-HHH_glszm_LowGrayLevelZoneEmphasis | 36% |
| T2-wavelet-LLL_glszm_GrayLevelVariance | 80% |
| ADC-original_glrlm_ShortRunHighGrayLevelEmphasis | 27% |
| ADC-wavelet-LLL_gldm_HighGrayLevelEmphasis | 23% |
| Transition zone | |
| ADC-wavelet-LLL_glcm_ClusterShade | 10% |
| Features | Sensitivity (% ± SD) | Specificity (% ± SD) | PPV (% ± SD) | NPV (% ± SD) | Accuracy (%) |
|---|---|---|---|---|---|
| Peripheral and transitional zone | |||||
| PSA density | 49 ± 12 | 91 ± 11 | 93 ± 8 | 43 ± 7 | 61 |
| T2-wavelet-HHH_glszm_GrayLevelVariance | 74 ± 14 | 52 ± 20 | 80 ± 7 | 47 ± 17 | 68 |
| T2-wavelet-LLL_glszm_GrayLevelVariance | 64 ± 11 | 63 ± 19 | 82 ± 8 | 42 ± 10 | 64 |
| ADC-original_glcm_ClusterShade | 81 ± 10 | 62 ± 18 | 84 ± 7 | 58 ± 16 | 76 |
| ADC-wavelet-HHH_firstorder_Minimum | 31 ± 30 | 71 ± 22 | 44 ± 37 | 31 ± 10 | 42 |
| ADC-wavelet-LLL_glrlm_LongRunHighGrayLevelEmphasis | 49 ± 23 | 57 ± 30 | 77 ± 13 | 31 ± 11 | 51 |
| Peripheral zone | |||||
| PSA density | 46 ± 14 | 89 ± 18 | 94 ± 10 | 36 ± 9 | 57 |
| T2-wavelet-HHH_glszm_GrayLevelVariance | 70 ± 15 | 57 ± 25 | 83 ± 9 | 41 ± 17 | 67 |
| T2-wavelet-HHH_glszm_LowGrayLevelZoneEmphasis | 64 ± 17 | 61 ± 25 | 84 ± 9 | 38 ± 15 | 63 |
| T2-wavelet-LLL_glszm_GrayLevelVariance | 67 ± 12 | 81 ± 18 | 92 ± 8 | 47 ± 12 | 70 |
| ADC-original_glrlm_ShortRunHighGrayLevelEmphasis | 72 ± 12 | 68 ± 24 | 87 ± 8 | 46 ± 15 | 71 |
| ADC-wavelet-LLL_gldm_HighGrayLevelEmphasis | 82 ± 14 | 51 ± 23 | 83 ± 7 | 54 ± 24 | 74 |
| Transition zone | |||||
| ADC-wavelet-LLL_glcm_ClusterShade | 80 ± 23 | 70 ± 30 | 82 ± 19 | 74 ± 29 | 76 |
| Features | Sensitivity (% ± SD) | Specificity (% ± SD) | PPV (% ± SD) | NPV (% ± SD) | Accuracy (%) |
|---|---|---|---|---|---|
| Multivariate model for peripheral and transitional zone | |||||
| PSA density | 79 ± 10 | 80 ± 15 | 91 ± 6 | 63 ± 13 | 79 |
| T2-wavelet-HHH_glszm_GrayLevelVariance | |||||
| T2-wavelet-LLL_glszm_GrayLevelVariance | |||||
| ADC-wavelet-LLL_glrlm_LongRunHighGrayLevelEmphasis | |||||
| Multivariate model for peripheral zone lesions | |||||
| T2-wavelet-HHH_glszm_GrayLevelVariance T2-wavelet-LLL_glszm_GrayLevelVariance ADC-wavelet-LLL_gldm_HighGrayLevelEmphasis | 86 ± 12 | 80 ± 19 | 93 ± 6 | 70 ± 19 | 84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonaffini, P.A.; De Bernardi, E.; Corsi, A.; Franco, P.N.; Nicoletta, D.; Muglia, R.; Perugini, G.; Roscigno, M.; Occhipinti, M.; Da Pozzo, L.F.; et al. Towards the Definition of Radiomic Features and Clinical Indices to Enhance the Diagnosis of Clinically Significant Cancers in PI-RADS 4 and 5 Lesions. Cancers 2023, 15, 4963. https://doi.org/10.3390/cancers15204963
Bonaffini PA, De Bernardi E, Corsi A, Franco PN, Nicoletta D, Muglia R, Perugini G, Roscigno M, Occhipinti M, Da Pozzo LF, et al. Towards the Definition of Radiomic Features and Clinical Indices to Enhance the Diagnosis of Clinically Significant Cancers in PI-RADS 4 and 5 Lesions. Cancers. 2023; 15(20):4963. https://doi.org/10.3390/cancers15204963
Chicago/Turabian StyleBonaffini, Pietro Andrea, Elisabetta De Bernardi, Andrea Corsi, Paolo Niccolò Franco, Dario Nicoletta, Riccardo Muglia, Giovanna Perugini, Marco Roscigno, Mariaelena Occhipinti, Luigi Filippo Da Pozzo, and et al. 2023. "Towards the Definition of Radiomic Features and Clinical Indices to Enhance the Diagnosis of Clinically Significant Cancers in PI-RADS 4 and 5 Lesions" Cancers 15, no. 20: 4963. https://doi.org/10.3390/cancers15204963
APA StyleBonaffini, P. A., De Bernardi, E., Corsi, A., Franco, P. N., Nicoletta, D., Muglia, R., Perugini, G., Roscigno, M., Occhipinti, M., Da Pozzo, L. F., & Sironi, S. (2023). Towards the Definition of Radiomic Features and Clinical Indices to Enhance the Diagnosis of Clinically Significant Cancers in PI-RADS 4 and 5 Lesions. Cancers, 15(20), 4963. https://doi.org/10.3390/cancers15204963






