Simple Summary
YAP and TAZ are transcriptional activators, which play critical roles in the regulation of cell/organ growth and regeneration. Accordingly, they are important mediators and modulators of cancer formation and spread, impacting a wide range of processes from cancer stem cell differentiation through the control of proliferation, phenotype determination, and migration, till the induction of antitumor drug resistance. These widespread activities are due to their profound effect on gene expression. Importantly, to exert these physiological and pathological functions, they must enter the nucleus, while for the termination of their transcriptional effects, they must exit. However, the critical structural requirements (nuclear localization signal and nuclear export signal) as well as their passageway(s) and mechanism(s) for crossing the nuclear pore are only beginning to emerge. This review summarizes the current knowledge about the nuclear traffic of YAP and TAZ, a fundamental, novel, and potentially exploitable aspect of their biology.
Abstract
Yes-associated Protein (YAP) and its paralog Transcriptional Coactivator with PDZ-binding Motif (TAZ) are major regulators of gene transcription/expression, primarily controlled by the Hippo pathway and the cytoskeleton. Integrating an array of chemical and mechanical signals, they impact growth, differentiation, and regeneration. Accordingly, they also play key roles in tumorigenesis and metastasis formation. Their activity is primarily regulated by their localization, that is, Hippo pathway- and/or cytoskeleton-controlled cytosolic or nuclear sequestration. While many details of such prevailing retention models have been elucidated, much less is known about their actual nuclear traffic: import and export. Although their size is not far from the cutoff for passive diffusion through the nuclear pore complex (NPC), and they do not contain any classic nuclear localization (NLS) or nuclear export signal (NES), evidence has been accumulating that their shuttling involves mediated and thus regulatable/targetable processes. The aim of this review is to summarize emerging information/concepts about their nucleocytoplasmic shuttling, encompassing the relevant structural requirements (NLS, NES), nuclear transport receptors (NTRs, karyophererins), and NPC components, along with the potential transport mechanisms and their regulation. While dissecting retention vs. transport is often challenging, the emerging picture suggests that YAP/TAZ shuttles across the NPC via multiple, non-exclusive, mediated mechanisms, constituting a novel and intriguing facet of YAP/TAZ biology.
1. Introduction: A Central Enigma in the Biology of YAP and TAZ
YAP and TAZ are paralogous transcriptional coactivators that are primarily regulated by the Hippo pathway [1,2,3] and the cytoskeleton [4,5,6]. They play key roles in a plethora of physiological processes—such as tissue growth, regeneration, differentiation, and matrix deposition [1,7,8,9,10,11], and, accordingly, in related pathologies, including organ fibrosis [12,13,14,15,16] and cancer [17,18,19,20].
1.1. Types of YAP/TAZ Contribution to Cancer
The types (and root causes) of YAP/TAZ dysregulation in cancer fall in the following three categories.
(1) Wild type YAP and/or TAZ are often activated and overexpressed in cancer cells or cancer stem cells, as observed in human malignancies, including breast, liver, colon, prostate, pancreatic, ovarian, uterine, and renal carcinomas, as well as melanoma, sarcomas, and neuronal tumors [17,18,21,22,23,24,25,26,27,28,29,30,31]. In a small subset of tumors (<10%), overexpression or activations of wild type YAP/TAZ is due to genetic alterations [18,32]. These include the amplification of YAP/TAZ genes themselves [32,33], or mutations in their upstream suppressors, primarily components of the Hippo pathway [18,32]. Accordingly, certain tumors harbor loss-of-function mutations in the core Hippo kinases (MST1/2, LATS1/2), their adaptors (SAV1), or their upstream regulators. The most prevalent of the latter are germline or somatic mutations of the Neurofibromatosis-2 (NF2) gene, encoding Merlin [34], a cytoskeletal protein and potent activator of the Hippo pathway [32]. In addition to neurofibromatosis, a hereditary tumor syndrome [34], somatic NF2 mutations are found in 30–40% of malignant mesotheliomas, and less frequently in other tumors (renal cancer and cholangiocarcinoma) [32,35,36,37,38]. Nevertheless, in the vast majority of cases, overactivation of wild type YAP/TAZ is not due to Hippo-related mutations but due to dysregulation of a large variety of signaling events affecting their transcription, stability, and transcriptional activity. Moreover, these phenomena occur not only in the tumor cells themselves, but also in the surrounding cell types, including cancer-associated fibroblasts (CAFs) and the endothelium [22,39,40,41]. These are critical for shaping the tumor microenvironment (TME) [40,42], which, in turn, promotes tumor growth and spread through mechanical (extracellular matrix) and paracrine (cytokines) cues [43,44]. In many cancers, dysregulated YAP/TAZ signaling shows strong correlation with poor survival [19,32]. Finally, it is worth noting that in addition to human malignancies, a large variety of experimental murine cancer models also revealed increased nuclear accumulation of wild type YAP/TAZ by immunostaining. These include breast cancer [45], non-small cell lung cancer [46], colorectal cancer [47], gastric cancer [48], hepatocellular carcinoma [49], pancreatic adenocarcinoma [50], cutaneous squamous cell and basal cell carcinoma [51], melanoma [52], and soft-tissue sarcoma [53].
(2) Cancer-related point mutation(s) in the coding region of YAP/TAZ proteins per se seem exceedingly rare [32]; so far, the YAP1 R331W missense mutant was reported to predispose for lung adenocarcinoma [54] and activating somatic mutations of regulatory phosphorylation sites were detected in YAP in a melanoma patient [55].
(3) The presence of various YAP or TAZ fusion proteins is the direct cause for tumor formation in a set of rare malignances, exemplified by epithelioid hemangioendothelioma [56,57,58]. The resulting oncoproteins are composed of an N-terminal portion of YAP or TAZ fused to a C-terminal sequence of another protein (e.g., TAZ-CAMTA1, YAP-TFE3); they are resistant to upstream regulatory signals and constantly nuclear, as dictated by the fusion partner.
1.2. Key Importance of Nucleocytoplasmic Shutting
In all the above scenarios, YAP/TAZ initiate transcriptional reprograming (YAP/TAZ signature) [29,32,59,60], primarily by activating the TEAD family of transcription factors [61,62,63,64]. This in turn impacts a large cohort of tumor-promoting processes, including stemness, epithelial-mesenchymal transition, proliferation, metabolism, invasion/metastasis, angiogenesis, and drug resistance [19,32,33,65]. In aggregate, all of the studies (Section 1.2) prompt two main conclusions, relevant to the topic of this review. The first is that in most cases, the tumor-promoting role of YAP/TAZ is associated with the dysregulation of their wild type versions; the second is that the nuclear presence of YAP/TAZ is indispensable for these effects in all tumor categories. Thus, understanding the nucleocytoplasmic traffic of YAP/TAZ, and the contributions of its various determinants, that is, cytosolic and nuclear retention and nuclear import and export, is of paramount importance.
1.3. Retention Models
There are two prerequisites for the nuclear accumulation of YAP/TAZ: they must be “free” (sufficiently mobile), and they must enter the nucleus (Figure 1).
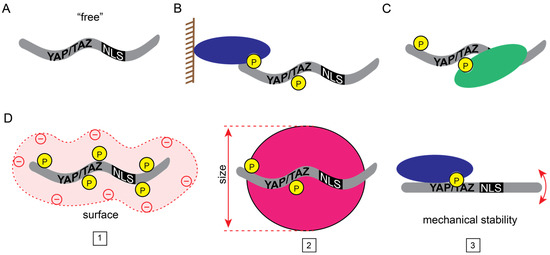
Figure 1.
Transport states of YAP/TAZ. (A) “Free”, transport-competent YAP/TAZ. (B–D) Posttranslational modifications and interactions with other proteins could (B) tether YAP/TAZ to macromolecular complexes (e.g., plasma membrane, chromatin), (C) mask their transport signals (i.e., NLS or NES), or (D) change their physicochemical properties (1: surface charge, 2: size, 3: mechanical stability) to diminish nucleocytoplasmic shuttling.
The overwhelming majority of studies so far have focused on the first requirement and, accordingly, their nuclear accumulation was interpreted in the context of retention models. Indeed, the Hippo pathway is a major regulator of their cytosolic retention. When this cascade is active, MST1/2 kinase-mediated activation of LATS1/2 kinase induces phosphorylation of YAP/TAZ [1,2,3]. This results in their interaction with “cytosolic retention factors”, such as members of the 14-3-3 family, thereby preventing their nuclear accumulation [66,67]. YAP/TAZ sequestration is also regulated by large protein complexes associated with the plasma membrane and the underlying cytoskeleton. In polarized cells, these include members of the Crumbs complex, the Par complex, and tight junctions (e.g., angiomotins, Kibra, Merlin, ZO2) in the apical compartment, and the Scribble complex and adherens junctions (e.g., Scrib, α-cadherin, PTPN14, Merlin, Kibra) in the basolateral compartment. These components were shown to act as activators or scaffolds of the Hippo pathway, orchestrating the inactivation of YAP/TAZ, and/or as direct binders of YAP/TAZ (for reviews see [68,69]). The cytoskeleton was also shown to regulate the free pool, independently of the Hippo pathway: F-actin and YAP compete for the cytosolic retention factor angiomotin; thus, an increase in F-actin can liberate YAP for nuclear uptake [5,70].
Since a multitude of conditions, including the presence of extracellular ligands, the integrity of intercellular contacts, metabolic state, matrix stiffness, and other mechanical cues, as well as genetic defects can affect Hippo pathway activity and/or cytoskeletal organization [71,72,73,74,75,76], all these inputs can impact the mobile pool of YAP/TAZ. Conversely, nuclear retention is dependent on YAP/TAZ binding to intranuclear partners, such as TEAD proteins, which in turn can result in net nuclear accumulation [77,78,79,80].
Finally, it is worth noting that in addition to the classic Hippo kinase-mediated Ser phosphorylation, YAP/TAZ are subject to a variety of other posttranslational modifications [81,82,83], including Ser/Thr phosphorylation by AMPK [84], JNK [85], and MST4 [45], tyrosine phosphorylation (e.g., by Yes, Src, and Abl kinases [86,87,88,89]), acetylation [90], methylation [91], and sumoylation [92]. Many of these modifications affect localization, e.g., AMPK and the methyl transferase Set7 are negative, whereas MST4 and Abl are positive regulators of nuclear accumulation. However, in most cases (except for MST4, see Section 3.3.2) it remains to be clarified whether these modifications act on cytoplasmic/nuclear retention or alter nuclear transport as well.
1.4. The Fate of “Free” YAP/TAZ: Nuclear Flux
But once “free”, how do YAP and TAZ enter (and leave) the nucleus? Although their molecular mass is not far from the reported cutoff limit for passive diffusion through the nuclear pore complex (NPC), evidence is accumulating that such a mechanism (alone) is unlikely to account for their nucleocytoplasmic shuttling (see below). However, YAP/TAZ do not contain a conventional nuclear localization signal (NLS) or a prototypic nuclear export signal (NES). Thus, the possibility of mediated transport (import and export) posed intriguing questions about the identity of these sequences in YAP/TAZ. Further, recent observations suggest that in addition to the availability of YAP/TAZ for transport, mechanical forces may regulate both the shape of these molecules and their passageway, the NPC [93]. While this field is in a primordial state yet, it is becoming increasingly clear that the import of YAP/TAZ may involve multiple mechanisms. Accordingly, the purpose of this review is to summarize and organize the emerging knowledge on YAP/TAZ nuclear shuttling. First, we will provide a brief overview of the “classic” and other karyopherin-dependent nuclear transport pathways, and then discuss the different transport modalities whereby YAP/TAZ is thought to shuttle across the nuclear membrane.
2. Nuclear Import and Export of Proteins, a General Overview
Proteins access the nucleus through NPCs which are embedded in the nuclear envelope, a double lipid bilayer separating cytoplasm and nucleoplasm. NPCs are 60–120 MDa structures assembled from multiple copies of 30 different nucleoporins, totaling approximately 1000 proteins [94,95,96,97]. The central pore of NPCs is lined with intrinsically disordered phenylalanine-glycine (FG) repeat domains that establish a sieve-like permeability barrier with a soft exclusion size threshold [98,99,100,101,102]. In consequence, passive diffusion of proteins beyond 30–60 kD is dramatically suppressed. To overcome this size restriction and allow for nucleocytoplasmic distributions of proteins beyond simple diffusional equilibration, specialized nuclear transport receptors (NTR) exist that shuttle protein cargoes across the nuclear envelope [103,104,105,106,107,108,109]. NTR recognize nuclear import cargoes via nuclear import signals (NLS) and export cargoes through nuclear export sequences (NES) (Figure 2).
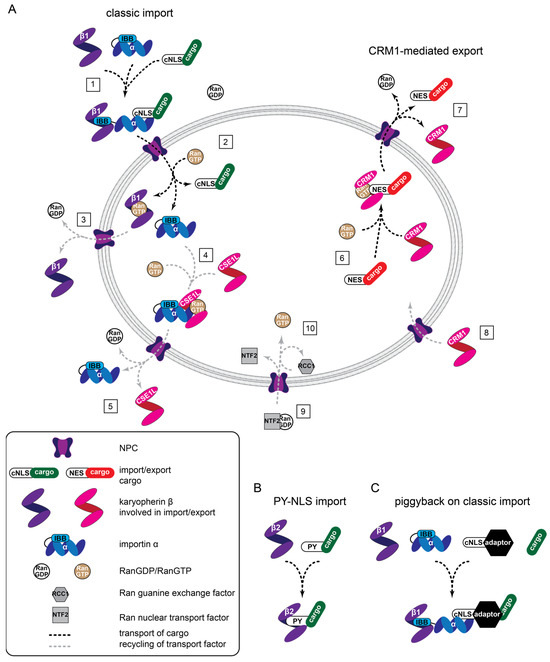
Figure 2.
Nuclear transport machines and different transport modalities. (A) General overview of classic protein import (1, 2), CRM1-mediated export (6, 7), and the recycling of factors supporting transport (3–5, 8–10). (1) Assembly of a heterotrimeric import complex, NPC passage, and (2) RanGTP-driven disassembly. Importin β1-RanGTP complexes cycle back to the cytoplasm where (3) hydrolysis of GTP to GDP triggers Ran dissociation. (4) CSE1L associates with RanGTP and importin α, passes the NPC, and (5) releases importin α into the cytoplasm. (6) Similarly, CRM1 associates with RanGTP and an NES-containing cargo for (7) nuclear export, followed by (8) CRM1 reentering the nucleus. (9) RanGDP is transported into the nucleus by NTF2 and RCC1 catalysis is the exchange of GTP for GDP (10) to maintain the RanGDP/RanGTP gradient across the nuclear envelope. (B) Transportin-mediated import involves direct binding between karyopherin-β2 (transportin) and cargo. (C) Classic, piggyback import involves an NLS containing an adaptor that bridges importin α and the cargo. IBB: importin β binding.
Direct interactions of the NTR with FG repeat domains allows the NTR-cargo complexes to translocate through the NPC by facilitated diffusion [102]. A sharp concentration gradient of the small Ran GTPase bound to GDP (RanGDP) vs. GTP (RanGTP) exists across the nuclear envelope, and this is the driving force for vectorial protein transport [110,111]. Mechanistically, RanGTP, which dominates in the nucleus, triggers the disassembly of incoming NTR-cargo complexes [112] (Figure 1, step 2) and the assembly of complexes designated for nuclear export [113] (Figure 1, step 4, 6). In the cytoplasm where RanGDP is prevailing, exported NTR-Cargo-RanGTP complexes dissociate, driven by the hydrolysis of bound GTP to GDP (Figure 1 step 5, 7). Low levels of RanGTP in the cytoplasm also permit the formation of NTR-cargo complexes destined for import (Figure 1 step 1). The asymmetric distribution of Ran GTPase activating proteins in the cytoplasm, boosting Ran GTPase activity, and the chromatin-bound guanine-nucleotide exchange factor RCC1, which facilitates GTP loading of Ran, maintains the steep RanGDP/RanGTP gradient. The RanGDP-specific transport receptor NTF2 replenishes Ran levels in the nucleus [114] (Figure 1, step 9, 10).
2.1. Nuclear Import Pathways
2.1.1. Classic Protein Import
The most extensively used protein import mechanism in cells is known as the classic nuclear import pathway. It depends on importin β1, a member of the karyopherin-β type NTR family, which recognizes cargoes with classic NLS indirectly. It binds protein adaptors belonging to the importin α NTR family (Figure 2), the direct receptors for classic NLS (Table 1). In the nucleus, the importin β-importin α-cargo complexes are disassembled through RanGTP (see Section 2), and the importin α-specific (karyopherin-β type) NTR CAS/CSE1L shuttles importin α back to the cytoplasm. Classic NLS come in two major flavors: monopartite comprising one stretch of positively charged (basic) residues (Lys, Arg), or bipartite containing two stretches, separated by a linker (Table 1) [104,105,115].

Table 1.
List of karyopherin-β NTR (modified from [106]).
Cargo specificity in the classic nuclear import pathway is attained by the seven importin α subtypes (in humans), which show differences in their cargo recognition and their expression in distinct cell types, tissue, and developmental stages. As a variation to this theme, importin β1 can bind directly to certain cargo proteins [107,115].
2.1.2. Protein Import by Other Karyopherin-β Type NTR
The full complexity of nuclear import systems becomes more tangible when looking at the entire family of karyopherin-β NTR, which encompasses twenty members in humans [106] (Table 1). Besides importin β1, nine others are also involved in protein import. Of these, transportin and transportin 2 are the best characterized and recognize a non-classic, so called PY-type NLS (stretches of 20–30 amino acids harboring a C-terminal R/K/H(X)2-5PY sequence [105]). Another member, importin 7 (IPO7) is of particular interest since it has been recently implicated in mechanosensitive import of YAP [116]. Importin 7 can function as a genuine karyopherin-β type NTR or it acts as an adaptor protein for importin β1-mediated import. Three additional karyopherin-β members are called biportins, due to their capacity to mediate both nuclear import and export. Since a full characterization of the cargo specificities for all known NTR is still missing [106], it remains an open question whether the increasing number of “unconventional” NLS [80,117,118] are indicators for novel, undiscovered import pathways or can be assigned to one of the less well-characterized karyopherin-β NTR.
2.2. Nuclear Export Pathways
Four members of the karyopherin-β NTR family are involved in protein export, in addition to the bidirectional biportins. Amongst them, CRM1 appears to mediate most protein export from the nucleus [106]. CRM1 recognizes well-characterized, hydrophobic NES in cargo proteins (Table 1). However, recent experiments suggest that a large portion of CRM1-export cargoes lack a typical NES [106,119]. Conveniently, the specific CRM1 inhibitor Leptomycin B (LMB) facilitates the designation of CRM1 dependency. CAS/CSE1L is a karyopherin-β type NTR that is mainly involved in the export of importin α subtypes, as mentioned above. As for the import pathways, further research is warranted to better characterize the specificities of these export pathways. For example, it awaits elucidation whether RanBP6 and RanBP17, the last two members of the karyopherin-β NTR family, are involved at all in protein export and/or import.
2.3. Additional Complexities of Import and Export and the Relevance for YAP and TAZ
Several NPC components themselves have been found to directly bind cargo proteins and promote their import, independent from NTRs [120,121,122,123]. These findings blur the distinction between NPC components and NTR. Moreover, importin α subtypes were found to function in an importin β1- and Ran-independent manner in some instances [124]. Lastly, proteins can be transported through the central pore of NPCs as part of larger complexes. This process is referred to as piggyback transport and alleviates the requirement of specific localization signals for individual components of the complex [125]. All these complexities make it difficult to predict the protein transport systems responsible for cargoes that lack defined NLS and NES. Accordingly, for YAP and TAZ, notoriously lacking recognizable NLS and NES, and with molecular weights around 60 kD, the relevant nuclear import and export processes remained unknown for a long time. This has changed in recent years.
3. Nuclear Import of Yorkie, YAP, and TAZ
3.1. Import of Yorkie—A Special Case of Classic Import
As with many Hippo signaling pathway components, the first downstream effector of the pathway was discovered in Drosophila: Yorkie. Wang et al. [126] found that the N-terminal 55 amino acids of Yorkie (isoform H, which contains 23 additional amino acids at the N-terminus compared to isoform F) are essential for the protein to localize to the nucleus, when Scalloped (Sd), the Drosophila TEAD homolog was coexpressed. The N-terminus was also required to drive a Sd-reporter luciferase construct in cells and to induce an overgrowth phenotype in vivo, when expressed as a transgene in the Drosophila eye. Loss of the overgrowth phenotype upon N-terminal deletion could be rescued by fusing a classic NLS to the Yorkie construct. Using the N-terminus for pull-down experiments, they identified by mass spectrometry Drosophila importin α1 and importin β as binding partners and showed that importin α1 overexpression led to the nuclear enrichment of both the isolated N-terminus and full-length Yorkie in cells. Furthermore, they convincingly showed that importin α1 was required for the overgrowth phenotypes of the Drosophila eye in vivo, driven by Yorkie expression or hpo (MST1/2 homolog) ablation. Detailed mutational analysis revealed that Arg 15 in the N-terminus is part of the NLS in Yorkie and critical for importin α1 binding, nuclear localization, and the overgrowth phenotype. Concerning the regulation of nuclear localization, they found that the expression of hpo not only resulted in Yorkie cytoplasmic sequestration, but also reduced importin α1 binding. Similarly, expression of the constitutively GTP-loaded Ran mutant Q69L, but not the GDP-loaded T24N mutant, decreased Yorkie-importin α1 association. Therefore, this work identified the key components of the classic nuclear import pathway, importin α1, importin β, and Ran, as critical factors for Yorkie nuclear localization, albeit the discovered NLS differs significantly from the classic one. However, this import mechanism is likely not conserved in mammals, as the homologs YAP and TAZ lack this N-terminal region [127], and co-expression of KPNA6, the human homolog to drosophila importin α1, failed to affect YAP localization [126].
3.2. Piggyback Mechanisms
The absence of a conventional NLS in YAP/TAZ suggested that their uptake is mediated via indirect or alternative mechanisms. Of these, the most plausible is that their import still occurs in an importin α/β-dependent manner but utilizes the classic NLS of a YAP/TAZ binding partner. In other words, it is brought about by a piggyback mechanism (see Section 2.3, Figure 2C). Results of some studies compatible with such mechanisms are described below.
3.2.1. MAML1/2
Members of the Mastermind-like family (MAML1-3 in mammals, homologs of Drosophila MAM) [128,129,130], were identified as key interactors and effectors of the Notch signaling pathway. Later, these nuclear proteins were found to facilitate the transcription of Wnt, Hedgehog, and NFκB pathway target genes as well [131,132,133,134], indicating that they are pivotal regulators of a wide spectrum of developmental programs. MAML proteins have a significant role in tumorigenesis, too [130,135,136]. Screening for proteins that promote the nuclear localization YAP/TAZ, Kim et al. [137] identified MAML1 and 2 whose ectopic expression induced strong nuclear accumulation of both YAP and TAZ and promoted the expression of their target genes, ANKRD1, CTGF, and CYR61. Conversely, knockdown of MAML1/2 shifted YAP/TAZ from the nucleus to cytosol. Importantly, the PPxY motif in MAML1 directly bound to the WW domain of YAP/TAZ. In addition, the deletion of the classic NLS from MAML1, as well as deletion or inactivation of the WW domain in YAP, abolished the MAML1-induced rise in nuclear YAP accumulation. MAML1 was binding equally well wild type YAP and YAP 5SA (mimicking the active, dephosphorylated form), and was also able to induce nuclear accumulation of the YAP 5SA, but not its WW mutant. This observation indicated that the effect of MAML1 on YAP is Hippo pathway-independent. Taken together, these results were consistent with a model wherein cytosolic MAML1 or 2 binds YAP or TAZ and carries the partner into the nucleus using its own classic NLS. However, the results are also consistent with MAML1/2-induced nuclear retention of YAP/TAZ imported by separate mechanisms, and this is the preferred interpretation by the authors themselves. This view is based on their observations that the kinetics of MAML1 and YAP nuclear entry are different, the former being faster than the latter. Further support for this view comes from the fact that MAML1 forms a trimeric complex with YAP/TAZ and TEAD. The authors also suppose that if import were the critical mechanism, then YAP phosphomimic mutants, which show cytosolic retention would have lower affinity for MAML1; however, this is not the case. Finally, their finding that downregulation of MAML1 does not prevent nuclear YAP accumulation in Hippo pathway-inactivated (LATS KO) cells, although it inhibits the transcriptional output of YAP, is also in line with uncoupled uptake mechanisms. Nonetheless, it cannot be excluded that a small pool of YAP/TAZ co-translocated with MAML1/2, but there is no current evidence to support this. In any case, nuclear enrichment, as well as the oncogenic properties of YAP/TAZ (proliferation, colony formation, invasion) is highly potentiated by overexpression and mitigated by the absence of MAML1. Whether this effect involves any import modulation—in addition to nuclear retention and augmented transcriptional activity—remains to be determined. The authors of this work elegantly showcase the difficulties to distinguish between retention and transport control, albeit clearly observable effects on nuclear localization and biological functions.
3.2.2. Mask
Another example of possible piggyback regards the Mask family of ankyrin-repeat- and KH-domain-containing proteins, the Drosophila Mask, and it its mammalian homologs, Mask1 (ANKHD1) and Mask2 (ANKRD17) [138,139]. Mask was originally identified as a component of receptor tyrosine kinase signaling in Drosophila eye development [138], providing the initial example for the widespread roles of the family. Ankyrin repeat proteins are important scaffolds for a variety of protein–protein interactions, while KH domains exhibit RNA/ssDNA-binding [140,141,142]. Accordingly, these proteins have been implicated in multiple functions, including proliferation, differentiation, cytoskeletal organization, and transcriptional control [141,143,144,145]. Moreover, their dysregulation and role are increasingly recognized in the context of tumorigenesis/metastasis in various malignancies [139,143,144,146,147,148]. Parallel discoveries in two laboratories have revealed that Mask binds Yorkie and acts as an important cofactor strongly potentiating the transcriptional activity of Yorkie and YAP [78,149]. Subsequent studies by Sidor et al. [150] have shown that the knockdown of Mask prevents or strongly mitigates nuclear accumulation of Yorkie during the developmental stretch (flattening) of Drosophila ovarian follicle cells, and that the classic NLS in Mask is necessary for its capacity to induce the nuclear enrichment of Yorkie. The addition of the classic NLS to Yorkie bypasses the requirement of Mask expression for proper Yorkie localization. Analogous observations were made for overexpressed GFP-Yorkie in the columnar cells of the wing. Furthermore, Mask1/2 downregulation in mammalian cells (siRNA) or in organoids (from conditional Mask1/2 KO mice) reduced both the nuclear localization and the total expression of YAP. The authors interpreted these findings to mean that Mask proteins, via their classic NLS, mediate the nuclear import of Yorkie/YAP/TAZ. This is certainly plausible; however, the results are also consistent with Mask-induced nuclear retention and accumulation of Yorkie/YAP/TAZ. Indeed, overexpression of Mask1/2 results in the formation of granular aggregates of YAP both in the cytosol and in the nucleus. The authors suggest that Mask promotes clustering/polymerization and consequent colloidal phase separation, as well as the stabilization of YAP. These effects are fully compatible with (and suggestive of) a retention model. This study did not follow the kinetics of Mask vs. YAP/TAZ translocation, which could potentially differentiate piggyback transport from retention. However, Sensores-Garcia et al. [78] reported that hpo inactivation or Sd (TEAD homolog) overexpression translocated Yorkie but not Mask into the nucleus, indicating that these factors can move (accumulate) separately. Finally, the finding that an artificial classic NLS fused to Yorkie is necessary to overcome the effect of Mask depletion is curious, given that the N-terminus of Yorkie contains an NLS [126]. Arguably, retention-augmented nuclear accumulation can be mimicked by robust, energy-dependent nuclear import, driven by a strong NLS. Taken together, future studies are necessary to verify the exact nature of the effects of Mask on import, the YAP/TAZ region necessary for Mask binding, and its relationship to sequences affecting import.
3.2.3. Selective Piggyback?
In principle, selective binding partners for YAP and TAZ could support selective import of these molecules. This has been proposed for the p300 acetyltransferase I (p300), which binds TAZ but not YAP [151]. Accordingly, p300 was suggested to shuttle TAZ (and its other binding partner Smad3) but not YAP into the nucleus via a piggyback mechanism, requiring the classic NLS within p300. It must be mentioned, however, that nuclear retention by the predominantly nuclear p300 could also explain these findings. In either case, p300 potentiated the fibrogenic capacity of TAZ and Smad3 in TGFβ-stimulated hepatic stellate cells which are the main mediators of liver fibrosis.
3.2.4. MRTF: A Caution
It is worth mentioning that the binding of YAP/TAZ to a classic NLS-containing protein by no means guarantees piggyback-type import or even nuclear retention. In this regard, we have shown that TAZ binds to myocardin-related transcription (MRTF) partly via the interaction of the TAZ WW domain with the PPXY sequence in MRTF [13]. This was an exciting observation because it raised the possibility of uncovering a key mechanism whereby the cytoskeleton regulates the nuclear localization of YAP/TAZ. MRTF is a monomer actin-binding protein, whose classic NLS is masked by G-actin. Upon actin polymerization, G-actin dissociates from MRTF, thereby liberating its NLS and inducing nuclear accumulation [152,153,154]. Since similar cytoskeleton-remodeling stimuli promote the nuclear uptake of MRTF and YAP/TAZ, it was a plausible hypothesis that MRTF drags along YAP/TAZ to the nucleus. However, overexpression of constitutively nuclear (G-actin-binding deficient) MRTF did not redistribute TAZ to the nucleus, and MRTF downregulation did not inhibit, but facilitated nuclear TAZ accumulation. Thus, MRTF acted as a cytosolic TAZ retention factor and vice versa. These proteins antagonize each other at the level of nuclear accumulation, while once in the nucleus, they can functionally synergize, often driving the same genes via adjacent cis elements [13,155,156]. This fine-tuned regulation may establish a stimulatory threshold, above which efficient amplification occurs. From a transport standpoint, the take-home message is that co-import vs. compartmental retention (cytosolic or nuclear) is dependent on the binding partner, the stimulus, and the cellular context. For example, the association of the partners may reduce import by elevating the molecular weight or by masking the corresponding NLS (see Figure 1).
3.3. YAP/TAZ-Import Involving Importin α
3.3.1. CSE1L/Importin α
The possibility of a direct involvement of classic import components in YAP/TAZ uptake was raised by pharmacological studies. Nagashima et al. [157] found that a TAZ inhibitor, named TI-4, potently abrogated the nuclear accumulation of TAZ without affecting its phosphorylation. Searching for the underlying mechanism of this Hippo-independent effect, they identified CSE1L (exportin-2) as the major target of TI-4. In keeping with this, CSE1L silencing also suppressed TAZ accumulation and eliminated any (additional) TI-4 effect. Both CSE1 knockdown and TI-4 reduced the rate of recovery of nuclear TAZ levels after photobleaching (FRAP), suggesting slower import. Since CSE1L is known to mediate the nuclear export of importin α5 (human KPNA1), the authors tested the potential involvement of this NTR. Both CSE1L and TAZ were found to bind importin α5, and TI-4 strengthened this interaction with the former, while it was weakened with the latter. TI-4 redistributed importin α5 from the nucleus to the cytosol, presumably by blocking its release from CSE1L. Finally, ivermectin, an inhibitor of classic (importin α/β1-mediated) import, slowed TAZ entry as measured by FRAP. CSE1L was shown to regulate YAP1 localization in a similar manner. Based on these findings, the authors proposed the following model: CSE1L exports importin α5 from the nucleus, which in turn binds TAZ in the cytosol and facilitates its import. TI-4 inhibits the dissociation of importin α5 from CSE1L, and therefore its association with TAZ; CSE1L downregulation abrogates the cytosolic availability of importin α5. This is certainly an intriguing model that warrants further investigation. Unfortunately, the impact of importin α5 downregulation was not tested in this study. A recent publication using a different cell type (RPE-1) found a mild (15%) but significant reduction in the nucleocytoplasmic ratio of YAP upon importin α5 silencing, while importin β1 silencing had no effect [116]. Thus, it remains unclear whether this pathway requires importin β1 or may represent an example for the importin α-dependent/β1-independent uptake, as mentioned in Section 2.3 (also described for casein kinase IV [124] or the Ran-independent import of importin α per se [158]).
3.3.2. Importin α5/α8
Further findings supporting the role of importin α5 in YAP import, and its potential regulation, were obtained by An et al. [45]. These authors showed that MST4, a member of the mammalian sterile-like kinase family, also affects YAP localization, similarly to its classic Hippo kinase relatives MST1/2 but via a different mechanism. Namely, MST4 induces direct phosphorylation of YAP on Thr 83 (distinct form the classic Hippo cascade targets, e.g., Ser 127) and this weakens the association between YAP and importin α5 as well as importin α8 (KPNA7). The authors proposed that this results in suppressed nuclear entry and thus enhanced cytosolic accumulation. Consistent with this view, phosphomimetic (T83D) and non-phosphorylatable (T83A) YAP mutants accumulate in the cytosol and the nucleus, respectively. They also show that dephosphorylation of Thr 83 promotes YAP/TAZ target gene activation and tumorigenesis in vitro and in vivo in a gastric cancer context. Taken together, MST4-mediated Thr 83 phosphorylation of YAP negatively correlates with YAP and importin α5 association and YAP nuclear localization. A confounding factor is the location of Thr 83 within the TEAD binding domain of YAP. Phosphorylation of this site reduces TEAD binding, and it remains to be seen whether this is rather the consequence of reduced import or the cause of diminished nuclear retention.
Overall, future studies should address the exact nature, mechanism, and structural basis (YAP/TAZ sequence) of importin α5-mediated YAP import, as well as its relation to the other import mechanisms and its pathophysiological significance.
3.4. YAP/Yorkie Nuclear Import by a Karyopherin-β Type NTR
Very recently, a nuclear import pathway for YAP has been identified that involves importin 7 [116]. The authors of this elegant paper started off with an unbiased approach, involving isobaric labeling of cyto- and nucleoplasmic fractions together with quantitative mass spectrometry to identify proteins with mechanoresponsive (i.e., cell density-sensitive) nucleocytoplasmic shuttling. The NTR importin 7 was one of the proteins with increased nuclear presence at low confluence. Other mechanical cues such as growth on large spreading areas or on stiff substrates and mechanical stretching also increased the nuclear accumulation of importin 7, whereas pharmacological treatments that diminished actomyosin-controlled tension or mechanical uncoupling of cyto- and nucleoskeleton, via a mutation in the connecting complex (LINC complex), had the opposite effect. The researchers recognized the similarity in mechanoresponsive nuclear localization of importin 7 and YAP, and these proteins had been shown to interact in a previous publication [159]. Immunoprecipitation experiments, proximity ligation assays, interaction studies with purified proteins, and localization studies in cells confirmed that the interaction between importin 7 and YAP occurred within cells, was direct, and depended on three regions in YAP which are also critical for nuclear localization: unphosphorylated Ser 127 (the major Hippo-regulated phosphorylation site), amino acids 314–320 (resembling a proposed NLS for importin 7 [160]), and the C-terminal PDZ-binding motif (PBM), which was reported to play a role in supporting the nuclear localization of YAP and TAZ [80,161,162]. Concerning the regulation of the importin 7-YAP interaction, they found mechanical cues to operate upstream of the Hippo kinase MST1/2: high cell density or low actomyosin-controlled tension inhibited the interaction, which could be rescued by the MST1/2 inhibitor XMU-MP-1. To determine whether importin 7 was a bona fide NTR for YAP, as opposed to sequestration-based nuclear accumulation, Garcia et al. tested the capacity of importin 7 to help YAP partitioning into a hydrogel formed by recombinant FG domains and to mediate nuclear import in digitonized cells. These experiments indicated that YAP transport is Ran sensitive and depends on importin 7 and energy. Correspondingly, depletion of importin 7 using siRNAs strongly diminished nuclear accumulation of YAP/TAZ and reduced their transcriptional activity, as measured by a TEAD luciferase reporter construct and the expression of YAP/TAZ target genes ANKRD1, CTGF, and CYR61. In comparison, when individually depleting the other karyopherin-β type NTRs involved in import, including two of the three biportins, or the full set of importin α subtypes, only a slight reduction of YAP and TAZ nuclear localization was observable upon elimination of importin αs KPNA1, KPNA2, and KPNA7. This is consistent with earlier reports [45,157] implicating these importin α subtypes in YAP nuclear localization based on observed interactions (Section 3.2). Intriguingly, YAP, in return, controlled localization and function of importin 7: depletion of YAP or inhibition of the importin 7-YAP interaction by mechanical cues decreased importin 7 nuclear localization, whereas the MST1/2 inhibitor XMU-MP-1 had the opposite effect. However, unlike YAP, depletion of other importin 7 cargoes—Smad3 and Erk2—did not alter importin 7 localization. YAP also reduced the association between importin 7 and Smad3 or Erk2 and the nuclear presence of these cargoes. These intriguing findings add another mechanism to the complex functional interplay between YAP and other transcription factors such as Smad3; thus, in addition to sequestration and expression control, altered transport can also play a role. Lastly, the researchers confirmed that the interactions and functions of importin 7 were conserved in Drosophila. The importin 7 homolog Msk bound Yorkie which required unphosphorylated Ser 168 (the equivalent to YAP Ser 127) and the C-terminal residues of Yorkie. Moreover, Msk was important for the nuclear localization of Yorkie and a Yorkie-driven overgrowth phenotype in the dorsal compartment of the wing imaginal disc.
3.5. YAP/TAZ Nuclear Accumulation Involving Unconventional Import Routes
Our approach to investigate the process of TAZ nucleocytoplasmic shuttling [80,163] involved a molecular toolkit of fluorescent tags and sequestration systems we developed to study the mediated nuclear import and export of TAZ in cells, unbiased by contributions from passive diffusion-driven flux and retention-based compartmentalization. The tags consisted of multiple mCitrine fluorophores, as described elsewhere [100], to increase the molecular weight of TAZ beyond the exclusion size limit of the NPC and, thereby, minimize the passive passage through the central pore. Since the barrier function of the NPC displays a soft, gradual exclusion size threshold with residual entry of even large fluorophore-multimers beyond 150 kD, rather than a sharp cutoff at 60 kD [98,99,100], we utilized the mCitrine homopentamers as tag (5C) and carefully determined the ratio of fluorescence found in the nucleus and the cytoplasm (N/C) of transfected cells. With these precautions, the 5C-tag diminished passive influx sufficiently to detect even weak and highly specific import signals, however, at the cost of reduced import dynamics. These limitations were overcome by our RIS’N (Rapamycin-Induced Sequestration in the Nucleus) system. It is based on the Rapamycin-induced strong interactions between an FRB domain, fused to mCitrine and a NES (1C-NES-FRB), and a FKBP domain of a nuclear anchor construct (H2B-2xFKBP-mCherry). When the 1C-NES-FRB is fused to a fragment with NLS function, it shuttles between nucleus and cytoplasm, but the potent NES maintains a steady-state cytoplasmic localization. Rapamycin-triggered nuclear sequestration blocks export, and the dynamics of nuclear import can be unmasked and monitored by the increase in nuclear fluorescence. Equipped with these tools, and using the TAZ 4SA mutant, in which all LATS phosphorylation sites were mutated to alanine to eliminate cytoplasmic retention, we identified a 15-amino acid long region (327–341) in the TAZ C-terminus as NLS. This segment works as a bone fide import signal, in that it is both necessary and sufficient for nuclear entry. Moreover, two features of this NLS proved to be indispensable for import: a central methionine residue flanked by negative charges ([80] and submitted manuscript). These unique features distinguish the TAZ NLS (which we term M-motif because of the essential methionine) from any other identified NLS. Importantly, mutation of the critical residues abrogated import in the context of full-length (otherwise intact) TAZ as well. In addition, the three hydrophobic motifs FLxxΦ present in the NLS, region 360–365 and the C-terminal residues, cumulatively support import, but this motif is dispensable for the function of the isolated identified NLS ([80] and submitted manuscript). The mediated import of TAZ, as measured by the steady-state distribution of 5C-fusion proteins, was insensitive to Ran depletion or the expression of constitutively active Ran mutant G19V, in contrast to a 5C construct with classic NLS. Consistent with Ran independence, the identified unusual NLS does not engender a large net nuclear accumulation of TAZ over the cytosolic level in the absence of nuclear retention. The NLS is evolutionarily conserved both in YAP and TAZ. In addition, our recent studies have uncovered the presence of similar and transport-component sequences (M-motifs) in various cellular and viral proteins as well, suggesting that the TAZ NLS may be a prototypic member of a new NLS family (manuscript submitted). Taken together, our studies define an independent and functionally important import motif in YAP/TAZ, which is necessary for basal, mediated diffusion across the nuclear pore. Future studies should test its functional relationship (or the lack thereof) with other transport-affecting sequences in YAP/TAZ and the associated mechanisms.
3.6. YAP/TAZ Interactions with the NPC
3.6.1. Nup37
A growing list of articles report interactions between YAP/TAZ and components of the NPC and show functional consequences in that the downregulation of these affect the nuclear localization and transcriptional activity of the two transcriptional coactivators [120,121,159]. One group studied the role of Nup37, a component of the NPC scaffold [164], in hepatocellular carcinomas (HCC). They found elevated expression of Nup37 in this carcinoma which promoted growth, migration, and invasion [120]. Nup37 downregulation inhibited intrahepatic metastasis in a mouse model. Given the importance of YAP-signaling in HCC, they further examined the interplay between Nup37 and YAP. The authors showed that Nup37 is critical for YAP transcriptional activity, as demonstrated by the expression of CTGF, CYR61, and Cyclin E. Nup37 expression increased nuclear YAP, and the migration-promoting effects of Nup37 expression could be suppressed by YAP silencing. Moreover, immunoprecipitations detected binding between Nup37 and YAP, which depended on the YAP WW domain. These results identified Nup37 as a significant component for YAP nuclear localization and function, promoting the authors’ conclusion that it enhances either transport or retention.
3.6.2. Nup205
In a very recent publication [121], researchers concentrated on the role of YAP and TAZ in podocytes, a cell type critical for the maintenance of the glomerular filter. They identified by quantitative label-free mass spectrometry several karyopherins (IPO5, 7, 8, 9, 11, Xpo2, 5, Xpot) as well as Nup107, Nup133, and Nup205 in immunoprecipitates of both YAP and TAZ. Interestingly, Nup205, the most abundant Nup amongst the YAP/TAZ interactors, was found to be mutated in families suffering from steroid-resistant nephrotic syndrome [165]. Downregulation of Nup205 decreased YAP/TAZ nuclear localization, YAP-TEAD interaction, and the expression of YAP/TAZ target genes ANKRD1, DIAPH3, CTGF, and CYR61. Moreover, depletion of either Nup205 or YAP reduced cell proliferation and increased oxidative stress-induced cell death. On the other hand, depletion of TAZ diminished Nup205 expression and nuclear localization. Correspondingly, loss of TAZ negatively impacted YAP nuclear localization via its effects on Nup205, revealing an interesting TAZ-dependent regulation of YAP activity. The authors assigned Nup205 a direct role in YAP/TAZ shuttling, consistent with a publication that found Nup205 to enter the nucleus in a NTR- and Ran-independent manner and that identified extensive structural similarities between Nup205 and NTRs [123]. This work adds a further mechanism for YAP/TAZ import, but additional studies are warranted to better distinguish effects on transport from changes in nuclear (Nup205-mediated) retention or consequences on NPC conformations upon Nup205 depletion.
3.6.3. Involvement of Other Nups?
Nup43, Nup98, Nup153, and Nup155, in addition to importin 7, were identified as YAP interactors in an article researching mechanotransduction through a Caveolin-1/YAP signaling axis [159]. The study was conducted in the context of pancreatitis, since YAP is required for pancreatitis-induced acinar-to-ductal metaplasia, and upregulation of Caveolin-1 expression decreases survival of pancreatic cancer. Nup98 and Nup155 were also amongst the top ten genes, whose siRNA mediated knockdown blunted YAP nuclear translocation. While this report did not focus on transport aspects of YAP regulation, it served as a resource for a later study [116] involving importin 7 in YAP transport (see Section 3.4).
3.7. Regulation of YAP/TAZ Nuclear Import
3.7.1. Import Regulation by Posttranslational Modifications
Retention mechanisms are critical determinants of YAP/TAZ localization as they control the pool of “free”, transport-competent species (see Section 1.3 and Section 1.4, and Figure 1). Inherently, regulation of retention and import are tightly interlinked: phosphorylation of site Ser 127 in YAP, for example, is suggested to negatively affect importin 7 binding [116] and, at the same time, it establishes the association with 14-3-3 proteins. Similarly, phosphorylation of Thr 83 diminishes importin α5/α8 binding but might also affect interactions with TEAD [45]. However, further studies are needed to understand how the ever-growing list of YAP/TAZ posttranslational modifications (Section 1.3) affect localization and what the underlying mechanisms are.
3.7.2. Import Regulation by Mechanical Forces
Transduction of signals emanating from mechanical forces emerged as an essential part of many aspects of cell (and tissue) fate and behavior in as much that it developed into the distinct field of mechanobiology [166]. In general, mechanical cues generated outside or within the cell can be sensed via force-sensitive structures in the cell periphery (cilia, focal adhesions, cell–cell contacts, individual (transmembrane) proteins) or through direct deformation of internal organelles, primarily the nucleus. The latter can result from migration through confined space or osmotic stress [167,168].
Concerning the regulation of YAP/TAZ import by mechanical forces, these cues can be transduced through biochemical signals (posttranslational modifications) which is addressed in the previous Section 3.7.1. Here, we focus on mechanical cues (stiffness, shear stress, tensile, contractile, and osmotic forces) that affect cell and nuclear shapes via transmission through the cytoskeleton-LINC complex–nuclear lamina axis or through direct nuclear deformation (in general [169,170,171,172,173,174], specific for YAP/TAZ [175,176,177]).
Novel insights into the mechanoresponsive regulation of YAP import came from the groundbreaking work of the Roca-Cusachs lab [93,109]. The group showed that mechanical cues are transmitted to the nucleus via the cytoskeleton-LINC complex axis, leading to deformation of the nuclear envelope and, thereby, widening the pore diameter of the embedded NPCs (Figure 3).
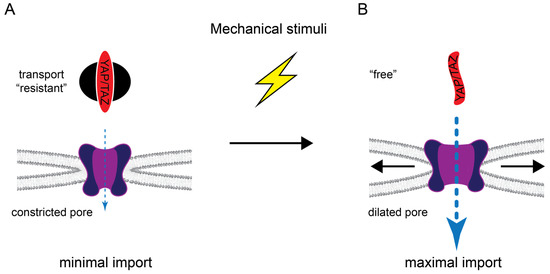
Figure 3.
Regulation of YAP/TAZ nuclear transport by mechanical stimuli. (A) In the basal state, YAP/TAZ are transport “resistant” (see Figure 1) and the NPC is in a more constricted conformation, resulting in minimal/no import. (B) Mechanical stimuli can convert YAP/TAZ into a free state, for example, by reducing Hippo pathway activity (and thereby reducing YAP/TAZ phosphorylation) and/or by inducing cytoskeletal rearrangements that reduce cytoplasmic retention via angiomotins and MRTF. Mechanical forces are also transmitted to the nuclear envelope to increase tension which provokes dilation of the NPC and nuclear import.
Furthermore, the mechanical forces enhanced both Ran-dependent YAP import and passive influx of control constructs. These changes in import were sensitive to the molecular weight and mechanical stability of the cargoes themselves [93,178]. The relatively small YAP (≈55 kD), being mainly composed of intrinsically disordered regions, displays low mechanical stability. Hence, YAP is perfectly fit for the mechanosensitive import. This work compellingly revealed that the cytoskeleton, in addition to its impact on the Hippo pathway activity (see Section 3.4) and direct cytoplasmic retention via angiomotins and MRTF, also modulated YAP transport itself and suggested pore dilation as the underlying mechanism. It remains to be seen how these findings can be integrated with the importin 7-mediated, mechanosensitive YAP import [116], as well as the role of identified NLS sequences.
Moreover, we could also show that mediated import of TAZ, which depended on the novel NLS we identified, was increased upon stimulation of Rho activity [80]. Unpublished work indicates that the TAZ NLS-mediated import is importin 7-independent, suggesting that nuclear deformation might stimulate import through this pathway as well.
Lastly, mathematical modeling of substrate stiffness, engagement, and dimensionality (2D vs. 3D cultures) was used to explain nuclear-cytoplasmic distribution of YAP/TAZ under different conditions [179]. This work laid out the complex regulation of YAP/TAZ localization by mechanical and geometrical properties of the cell environment as well as cell and nuclear shape. For example, the clear differences in localization in 2D vs. 3D cultures at the same substrate stiffness underlines the need to study dynamics of YAP/TAZ localization in vivo.
4. Nuclear Export of Yorkie, YAP, and TAZ
4.1. CRM1-Mediated Export
Strong support for a CRM1-mediated export pathway of YAP/TAZ stems from their observed nuclear accumulation upon treatment of cells with the CRM1-specific inhibitor LMB [80,180,181]. Using a rapamycin-induced cytoplasmic retention system, we verified the LMB-sensitive export, and identified the corresponding NES involving residues 35–38 within the TEAD binding domain of TAZ [80]. These experiments also revealed that LMB directly affected efflux dynamics, rather than affecting localization via redistribution of retention factors from the nucleus to the cytoplasm. The localization of the NES within the TEAD binding domain suggested that TEAD binding masks the NES. Moreover, binding studies revealed that 14-3-3 binding reduced TEAD association, and this could consequently unmask the NES, establishing an export-prone state of the molecule [80].
4.2. Piggyback Export
In addition to this direct nuclear export, a piggyback-based mechanism involving the NES of Merlin has been proposed [182]. In epithelial cells, Furukawa et al. found that the association of Merlin with the adherens junction component E-cadherin is regulated by tension in the underlying circumferential actin belt. In contrast to other parts of the cytoskeleton, this actomyosin-generated tension led to YAP suppression (reduced nuclear localization and transcriptional activity), rather than activation. Actin belt tension induced the dissociation of Merlin from the adherens junctions, increased YAP–Merlin interaction and enhanced nuclear efflux, whereas import was unaltered, as determined by LMB-time course experiments.
4.3. Regulation of Export
It is plausible that the nuclear export of YAP/TAZ is a highly regulated process. Using quantitative analysis of fluorescence photobleaching experiments, coupled with sophisticated mathematical modeling of transport kinetics, Ege et al. determined rates for global association and dissociation of YAP with nuclear and cytoplasmic retention factors and rates for import and export [183]. The detailed analysis identified the major regulatory pathways impinging on YAP/TAZ phosphorylation, such as the Hippo pathway and Src-family kinase activity downstream of the cytoskeleton, to regulate primarily YAP export. These are intriguing insights, adding details to the predominant import- and retention-centric views of the regulation of YAP/TAZ localization. While hugely informative, it can be added that, given the large number of known factors affecting retention and transport in a context-dependent manner, care should be taken when interpreting modeling-based analysis with its inherent simplifications.
5. YAP and TAZ in the Focus of Drug Therapy
5.1. Brief Overview of the Current State
Given their role in tumorigenesis, YAP and TAZ are attractive drug targets. However, because they mainly consist of inherently disordered regions and lack typical “druggable pockets” or enzymatic activity, their direct pharmacological inhibition is extremely challenging [184]. Therefore, apart from the use of antisense oligonucleotides directed against YAP/TAZ transcripts, either their upstream regulators or downstream effectors have been targeted. Notable examples in the first category include statins, which are widely used inhibitors of the Rho/Rho kinase pathway [73,185,186], the antidiabetic drug Metformin, which acts primarily by inducing AMPK-mediated inhibitory phosphorylation of YAP [187,188], and various tyrosine kinase (e.g., Src, Abl) inhibitors [189,190,191], which downregulate YAP/TAZ expression, stability, and nuclear localization. However, the real breakthrough has come with the development of drugs that inhibit YAP/TAZ–TEAD interaction and TEAD activity. The first compound recognized to interfere with YAP/TAZ-dependent TEAD activation was Verteporfin, a photosensitizer used in the treatment of macular degeneration [192,193]. It is, however, rather unspecific with poor pharmacokinetics and toxicity profile. Recently, several companies developed new TEAD inhibitors with high selectivity, efficacy, and improved pharmacokinetics. These drugs either target with high affinity YAP/TAZ binding sites (interface 3 and 2) in TEAD (protein–protein interaction (PPI) inhibitors), or an internal hydrophobic pocket, where TEADs are (auto)palmitoylated (allosteric palmitoylation inhibitors). Palmitoylation is necessary for TEAD activity [184,194,195]. Excellent recent reviews summarize the progress in this evolving field, describing the various drugs and their actions in different preclinical models [18,184,195,196,197,198]. For example, palmitoylation inhibitors (by Vivace Therapeutics and Dana Farber) were shown to suppress tumor growth in strictly YAP/TAZ-dependent NF2-deficient tumors, such as mesothelioma, schwannoma, and meningioma [199,200,201]. Importantly, three TEAD inhibitors, VT3989 (Vivace Therapeutics), IK-930 (Ikena Oncology), and IAG933 (Novartis) entered Phase I clinical trials [198].
5.2. Inhibition of Nuclear Transport in Cancer: General Mechanisms and YAP/TAZ Specific Perspectives
With the above advances in mind, the question arises whether the potential inhibition of nuclear entry of YAP/TAZ signifies a viable option. Several karyopherins, including importin β1, various importin α subtypes, and CRM1 (XPO1) are overexpressed in different malignancies [202,203,204]. Moreover, inhibitors of nuclear protein transport exert strong anti-tumor effects. For example, nuclear import (importin β1 or α/β1) inhibitors, such as ivermectin, importazole, INI-43, diaminoazole derivates, and diterpenoids show potent tumor growth-inhibitory effects in ovarian, cervical, esophageal, colorectal, and prostate cancer as well as glioblastomas, schwannomas, and hematological malignancies [204,205,206,207]. Conversely, selective inhibitors of nuclear export (SINE), e.g., selinexor that targets CRM1, are clinically used drugs in the treatment of multiple myeloma, leukemias, and sarcomas [205,207,208,209]. Perturbation of nuclear traffic interferes with tumor growth through several mechanisms, encompassing altered cycling of transcription factors, induction of cell cycle arrest, and apoptosis. Thus, in principle, targeting nuclear traffic is a viable option for cancer therapy. However, two remarks of caution must be added. First, given the large number of cargoes using these karyopherins, the selectivity of these compounds is bound to be limited, and the potential toxicity is an important caveat. In this regard, cargo-specific karyopherin inhibitors represent a major advance, and are emerging as potent antiviral agents. Second, the impact of these drugs on YAP/TAZ traffic, and the potential contribution of this to their overall anti-tumor effect remains to be elucidated. Nonetheless, the recent identification of several YAP/TAZ sequences critical for import, as well as specific karyopherins and NPC components involved in their nuclear uptake, holds realistic promise for the development of selective YAP/TAZ transport inhibitors. Indeed, some hits in high-throughput screens seem to act via such mechanisms. Thus, a better understanding of YAP/TAZ nuclear import and export may lead to the discovery of a completely new class of drugs, suppressing the action of these proteins in an orthogonal way.
6. Conclusions and Future Directions
Our understanding of the nuclear traffic of YAP/TAZ is in a nascent state. The emerging picture suggests that the nucleocytoplasmic shuttling of these critical regulators is brought about by multiple mediated mechanisms, which involve different sequences within the cargo itself, various NTRs, other ancillary transport partners, and distinct components of the NPC (Figure 4, Table 2). However, the sought-after synthesis is still missing. Future studies should clarify the exact relationship between proposed (and not yet identified) NLS and the relevant karyopherins, as well as the presence and significance of direct interactions of YAP/TAZ (or their novel binding partners) with the critical nucleoporins. The regulation of import and export—in terms of the availability/affinity of the NLS and NES, with respect to the posttranslational modifications of YAP/TAZ and the NTRs and via mechanical or chemical modulation of the resistance/permeability of the NPC itself, are fertile grounds for future research. Distinction between sequestration and import/export per se should also be further elucidated. However, it is important to realize that these inputs do not act in isolation; in fact, the same sequences or molecular players may affect both transport, retention, and intrinsic transcriptional activity (e.g., the NLS identified by us also contributes to transactivation, while the availability of the NES is regulated by sequestration). Finally, deciphering the transport machinery holds real promise for selectively targeting YAP/TAZ import and export. This would provide new tools to modify the activity of these important regulators and might one day contribute to the treatment of fibrosis and cancer.
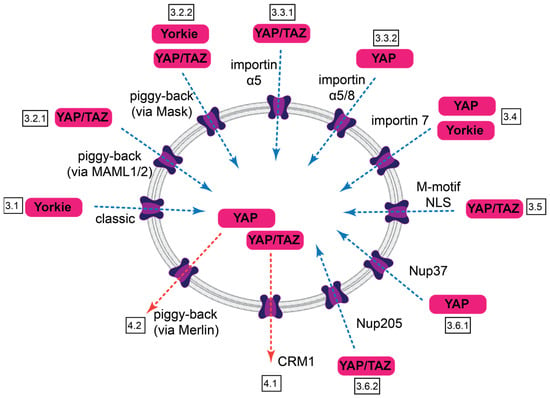
Figure 4.
Summary of nuclear import and export pathways for Yorkie, YAP, and TAZ. Import routes are indicated by blue dotted arrows, export routes by red ones. Identified features of the transport and boxed section numbers are indicated.

Table 2.
Transport characteristics for Yorkie, YAP, and TAZ.
Author Contributions
M.K. and A.K. both contributed to the conceptualization, design, and writing of the manuscript. M.K. generated the figures. All authors have read and agreed to the published version of the manuscript.
Funding
Experimental work reviewed here and performed in the authors’ laboratory was funded by grants from the Canadian Institutes of Health Research (CIHR, PJT 148608, PJT 162360), the Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2019-05222), and the Kidney Foundation of Canada to A.K. A.K. is the Thor Eaton Professor of Fibrosis Research, jointly appointed by the University of Toronto and St. Michael’s Hospital.
Acknowledgments
We are indebted to all member of the Kapus and Szászi laboratories who contributed to our related studies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Kim, J.; Jho, E.H. Role of the Hippo pathway and mechanisms for controlling cellular localization of YAP/TAZ. FEBS J. 2022, 289, 5798–5818. [Google Scholar] [CrossRef] [PubMed]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef]
- Dasgupta, I.; McCollum, D. Control of cellular responses to mechanical cues through YAP/TAZ regulation. J. Biol. Chem. 2019, 294, 17693–17706. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, K.C.; Meng, Z. Mechanoregulation of YAP and TAZ in Cellular Homeostasis and Disease Progression. Front. Cell Dev. Biol. 2021, 9, 673599. [Google Scholar] [CrossRef] [PubMed]
- Totaro, A.; Panciera, T.; Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018, 20, 888–899. [Google Scholar] [CrossRef]
- Pocaterra, A.; Romani, P.; Dupont, S. YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 2020, 133, jcs230425. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.R.; Tapon, N. Hippo signalling during development. Development 2019, 146, dev167106. [Google Scholar] [CrossRef]
- Jeong, M.G.; Kim, H.K.; Hwang, E.S. The essential role of TAZ in normal tissue homeostasis. Arch. Pharm. Res. 2021, 44, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Driskill, J.H.; Pan, D. Control of stem cell renewal and fate by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lagares, D.; Choi, K.M.; Stopfer, L.; Marinkovic, A.; Vrbanac, V.; Probst, C.K.; Hiemer, S.E.; Sisson, T.H.; Horowitz, J.C.; et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L344–L357. [Google Scholar] [CrossRef] [PubMed]
- Speight, P.; Kofler, M.; Szaszi, K.; Kapus, A. Context-dependent switch in chemo/mechanotransduction via multilevel crosstalk among cytoskeleton-regulated MRTF and TAZ and TGFbeta-regulated Smad3. Nat. Commun. 2016, 7, 11642. [Google Scholar] [CrossRef] [PubMed]
- Bialik, J.F.; Ding, M.; Speight, P.; Dan, Q.; Miranda, M.Z.; Di Ciano-Oliveira, C.; Kofler, M.M.; Rotstein, O.D.; Pedersen, S.F.; Szaszi, K.; et al. Profibrotic epithelial phenotype: A central role for MRTF and TAZ. Sci. Rep. 2019, 9, 4323. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Varelas, X.; Guan, K.L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef] [PubMed]
- Mia, M.M.; Singh, M.K. New Insights into Hippo/YAP Signaling in Fibrotic Diseases. Cells 2022, 11, 2065. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, R.; Hansen, C.G. The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clin. Sci. 2022, 136, 197–222. [Google Scholar] [CrossRef]
- Piccolo, S.; Panciera, T.; Contessotto, P.; Cordenonsi, M. YAP/TAZ as master regulators in cancer: Modulation, function and therapeutic approaches. Nat. Cancer 2023, 4, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Franklin, J.M.; Wu, Z.; Guan, K.L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat. Rev. Cancer 2023, 23, 512–525. [Google Scholar] [CrossRef]
- Van Haele, M.; Moya, I.M.; Karaman, R.; Rens, G.; Snoeck, J.; Govaere, O.; Nevens, F.; Verslype, C.; Topal, B.; Monbaliu, D.; et al. YAP and TAZ Heterogeneity in Primary Liver Cancer: An Analysis of Its Prognostic and Diagnostic Role. Int. J. Mol. Sci. 2019, 20, 638. [Google Scholar] [CrossRef]
- Thompson, B.J. YAP/TAZ: Drivers of Tumor Growth, Metastasis, and Resistance to Therapy. Bioessays 2020, 42, e1900162. [Google Scholar] [CrossRef]
- Wang, D.; He, J.; Dong, J.; Meyer, T.F.; Xu, T. The HIPPO pathway in gynecological malignancies. Am. J. Cancer Res. 2020, 10, 610–629. [Google Scholar] [PubMed]
- Kovar, H.; Bierbaumer, L.; Radic-Sarikas, B. The YAP/TAZ Pathway in Osteogenesis and Bone Sarcoma Pathogenesis. Cells 2020, 9, 972. [Google Scholar] [CrossRef] [PubMed]
- Masliantsev, K.; Karayan-Tapon, L.; Guichet, P.O. Hippo Signaling Pathway in Gliomas. Cells 2021, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, N.J.; Konstantinou, E.K.; Gragoudas, E.S.; Marinkovic, M.; Luyten, G.P.M.; Kim, I.K.; Jager, M.J.; Vavvas, D.G. Targeting the YAP/TAZ Pathway in Uveal and Conjunctival Melanoma With Verteporfin. Investig. Ophthalmol. Vis. Sci. 2021, 62, 3. [Google Scholar] [CrossRef]
- Della Chiara, G.; Gervasoni, F.; Fakiola, M.; Godano, C.; D’Oria, C.; Azzolin, L.; Bonnal, R.J.P.; Moreni, G.; Drufuca, L.; Rossetti, G.; et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nat. Commun. 2021, 12, 2340. [Google Scholar] [CrossRef]
- Yin, F.; Dong, J.; Kang, L.I.; Liu, X. Hippo-YAP signaling in digestive system tumors. Am. J. Cancer Res. 2021, 11, 2495–2507. [Google Scholar] [PubMed]
- Castellan, M.; Guarnieri, A.; Fujimura, A.; Zanconato, F.; Battilana, G.; Panciera, T.; Sladitschek, H.L.; Contessotto, P.; Citron, A.; Grilli, A.; et al. Single-cell analyses reveal YAP/TAZ as regulators of stemness and cell plasticity in Glioblastoma. Nat. Cancer 2021, 2, 174–188. [Google Scholar] [CrossRef]
- Koinis, F.; Chantzara, E.; Samarinas, M.; Xagara, A.; Kratiras, Z.; Leontopoulou, V.; Kotsakis, A. Emerging Role of YAP and the Hippo Pathway in Prostate Cancer. Biomedicines 2022, 10, 2834. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zou, H.; Guo, Y.; Tong, T.; Chen, Y.; Xiao, Y.; Pan, Y.; Li, P. The oncogenic roles and clinical implications of YAP/TAZ in breast cancer. Br. J. Cancer 2023, 128, 1611–1624. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018, 25, 1304–1317.e5. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Deng, L.; Zou, H.; Guo, Y.; Tong, T.; Huang, M.; Ling, G.; Li, P. New insights into the ambivalent role of YAP/TAZ in human cancers. J. Exp. Clin. Cancer Res. 2023, 42, 130. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.; Shevde, L.A. Merlin regulates signaling events at the nexus of development and cancer. Cell Commun. Signal. 2020, 18, 63. [Google Scholar] [CrossRef]
- Bianchi, A.B.; Mitsunaga, S.I.; Cheng, J.Q.; Klein, W.M.; Jhanwar, S.C.; Seizinger, B.; Kley, N.; Klein-Szanto, A.J.; Testa, J.R. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc. Natl. Acad. Sci. USA 1995, 92, 10854–10858. [Google Scholar] [CrossRef] [PubMed]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef]
- Hacking, S.M.; Pavlick, D.; Wang, Y.; Carneiro, B.A.; Mullally, M.; Lu, S.; Canepa, M.; Bratslavsky, G.; Jacob, J.; Necchi, A.; et al. Comprehensive Genomic Profiling of NF2-Mutated Kidney Tumors Reveals Potential Targets for Therapy. Oncologist 2023, 28, e508–e519. [Google Scholar] [CrossRef] [PubMed]
- Sekido, Y.; Sato, T. NF2 alteration in mesothelioma. Front. Toxicol. 2023, 5, 1161995. [Google Scholar] [CrossRef] [PubMed]
- Calvo, F.; Ege, N.; Grande-Garcia, A.; Hooper, S.; Jenkins, R.P.; Chaudhry, S.I.; Harrington, K.; Williamson, P.; Moeendarbary, E.; Charras, G.; et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 2013, 15, 637–646. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP and TAZ: A signalling hub of the tumour microenvironment. Nat. Rev. Cancer 2019, 19, 454–464. [Google Scholar] [CrossRef]
- Hooglugt, A.; van der Stoel, M.M.; Boon, R.A.; Huveneers, S. Endothelial YAP/TAZ Signaling in Angiogenesis and Tumor Vasculature. Front. Oncol. 2020, 10, 612802. [Google Scholar] [CrossRef]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal. Transduct. Target. Ther. 2021, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Riehl, B.D.; Kim, E.; Bouzid, T.; Lim, J.Y. The Role of Microenvironmental Cues and Mechanical Loading Milieus in Breast Cancer Cell Progression and Metastasis. Front. Bioeng. Biotechnol. 2020, 8, 608526. [Google Scholar] [CrossRef]
- Hassan, M.S.; Cwidak, N.; Awasthi, N.; von Holzen, U. Cytokine Interaction With Cancer-Associated Fibroblasts in Esophageal Cancer. Cancer Control 2022, 29, 10732748221078470. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Nie, P.; Chen, M.; Tang, Y.; Zhang, H.; Guan, J.; Cao, Z.; Hou, C.; Wang, W.; Zhao, Y.; et al. MST4 kinase suppresses gastric tumorigenesis by limiting YAP activation via a non-canonical pathway. J. Exp. Med. 2020, 217, jem20191817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, Y.; Li, F.; Tong, X.; Ren, Y.; Han, X.; Yao, S.; Long, F.; Yang, Z.; Fan, H.; et al. YAP promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of survivin. Cancer Res. 2015, 75, 4450–4457. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Maitra, A.; Anders, R.A.; Taketo, M.M.; Pan, D. beta-Catenin destruction complex-independent regulation of Hippo-YAP signaling by APC in intestinal tumorigenesis. Genes. Dev. 2015, 29, 1493–1506. [Google Scholar] [CrossRef]
- Tiffon, C.; Giraud, J.; Molina-Castro, S.E.; Peru, S.; Seeneevassen, L.; Sifre, E.; Staedel, C.; Bessede, E.; Dubus, P.; Megraud, F.; et al. TAZ Controls Helicobacter pylori-Induced Epithelial-Mesenchymal Transition and Cancer Stem Cell-Like Invasive and Tumorigenic Properties. Cells 2020, 9, 1462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Panciera, T.; Citron, A.; Di Biagio, D.; Battilana, G.; Gandin, A.; Giulitti, S.; Forcato, M.; Bicciato, S.; Panzetta, V.; Fusco, S.; et al. Reprogramming normal cells into tumour precursors requires ECM stiffness and oncogene-mediated changes of cell mechanical properties. Nat. Mater. 2020, 19, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Debaugnies, M.; Sanchez-Danes, A.; Rorive, S.; Raphael, M.; Liagre, M.; Parent, M.A.; Brisebarre, A.; Salmon, I.; Blanpain, C. YAP and TAZ are essential for basal and squamous cell carcinoma initiation. EMBO Rep. 2018, 19, e45809. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Jeong, S.H.; Jang, C.; Bae, H.; Kim, Y.H.; Park, I.; Kim, S.K.; Koh, G.Y. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 2019, 363, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Eisinger-Mathason, T.S.; Mucaj, V.; Biju, K.M.; Nakazawa, M.S.; Gohil, M.; Cash, T.P.; Yoon, S.S.; Skuli, N.; Park, K.M.; Gerecht, S.; et al. Deregulation of the Hippo pathway in soft-tissue sarcoma promotes FOXM1 expression and tumorigenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E3402–E3411. [Google Scholar] [CrossRef]
- Chen, H.Y.; Yu, S.L.; Ho, B.C.; Su, K.Y.; Hsu, Y.C.; Chang, C.S.; Li, Y.C.; Yang, S.Y.; Hsu, P.Y.; Ho, H.; et al. R331W Missense Mutation of Oncogene YAP1 Is a Germline Risk Allele for Lung Adenocarcinoma With Medical Actionability. J. Clin. Oncol. 2015, 33, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, J.Z.; Vergara, I.A.; Zhang, Y.; Szeto, P.; Yang, L.; Mintoff, C.; Colebatch, A.; McIntosh, L.; Mitchell, K.A.; et al. Somatic Hypermutation of the YAP Oncogene in a Human Cutaneous Melanoma. Mol. Cancer Res. 2019, 17, 1435–1449. [Google Scholar] [CrossRef]
- Garcia, K.; Gingras, A.C.; Harvey, K.F.; Tanas, M.R. TAZ/YAP fusion proteins: Mechanistic insights and therapeutic opportunities. Trends Cancer 2022, 8, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Szulzewsky, F.; Holland, E.C.; Vasioukhin, V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev. Biol. 2021, 475, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Seavey, C.N.; Pobbati, A.V.; Rubin, B.P. Unraveling the Biology of Epithelioid Hemangioendothelioma, a TAZ-CAMTA1 Fusion Driven Sarcoma. Cancers 2022, 14, 2980. [Google Scholar] [CrossRef] [PubMed]
- Hiemer, S.E.; Zhang, L.; Kartha, V.K.; Packer, T.S.; Almershed, M.; Noonan, V.; Kukuruzinska, M.; Bais, M.V.; Monti, S.; Varelas, X. A YAP/TAZ-Regulated Molecular Signature Is Associated with Oral Squamous Cell Carcinoma. Mol. Cancer Res. 2015, 13, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Finch-Edmondson, M.; Cognart, H.; Zhu, B.; Song, H.; Low, B.C.; Sudol, M. Common and Unique Transcription Signatures of YAP and TAZ in Gastric Cancer Cells. Cancers 2020, 12, 3667. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, C.Y.; Zha, Z.Y.; Zhao, B.; Yao, J.; Zhao, S.; Xiong, Y.; Lei, Q.Y.; Guan, K.L. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 2009, 284, 13355–13362. [Google Scholar] [CrossRef] [PubMed]
- Huh, H.D.; Kim, D.H.; Jeong, H.S.; Park, H.W. Regulation of TEAD Transcription Factors in Cancer Biology. Cells 2019, 8, 600. [Google Scholar] [CrossRef]
- Currey, L.; Thor, S.; Piper, M. TEAD family transcription factors in development and disease. Development 2021, 148, dev196675. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.S.A.; Xiao, Y.; Lamar, J.M. YAP/TAZ Activation as a Target for Treating Metastatic Cancer. Cancers 2018, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes. Dev. 2007, 21, 2747–2761. [Google Scholar] [CrossRef] [PubMed]
- Freeman, A.K.; Morrison, D.K. 14-3-3 Proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011, 22, 681–687. [Google Scholar] [CrossRef]
- Karaman, R.; Halder, G. Cell Junctions in Hippo Signaling. Cold Spring Harb. Perspect. Biol. 2018, 10, a028753. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.J.; McCaffrey, L. Emerging role of cell polarity proteins in breast cancer progression and metastasis. Breast Cancer 2014, 6, 15–27. [Google Scholar] [CrossRef]
- Mana-Capelli, S.; Paramasivam, M.; Dutta, S.; McCollum, D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol. Biol. Cell 2014, 25, 1676–1685. [Google Scholar] [CrossRef]
- Ahmad, U.S.; Uttagomol, J.; Wan, H. The Regulation of the Hippo Pathway by Intercellular Junction Proteins. Life 2022, 12, 1792. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, G.; Ruggeri, N.; Specchia, V.; Cordenonsi, M.; Mano, M.; Dupont, S.; Manfrin, A.; Ingallina, E.; Sommaggio, R.; Piazza, S.; et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 2014, 16, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Santinon, G.; Pocaterra, A.; Dupont, S. Control of YAP/TAZ Activity by Metabolic and Nutrient-Sensing Pathways. Trends Cell Biol. 2016, 26, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Halder, G.; Dupont, S.; Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012, 13, 591–600. [Google Scholar] [CrossRef]
- Chan, S.W.; Lim, C.J.; Loo, L.S.; Chong, Y.F.; Huang, C.; Hong, W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J. Biol. Chem. 2009, 284, 14347–14358. [Google Scholar] [CrossRef]
- Sansores-Garcia, L.; Atkins, M.; Moya, I.M.; Shahmoradgoli, M.; Tao, C.; Mills, G.B.; Halder, G. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr. Biol. 2013, 23, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Manning, S.A.; Dent, L.G.; Kondo, S.; Zhao, Z.W.; Plachta, N.; Harvey, K.F. Dynamic Fluctuations in Subcellular Localization of the Hippo Pathway Effector Yorkie In Vivo. Curr. Biol. 2018, 28, 1651–1660 e1654. [Google Scholar] [CrossRef]
- Kofler, M.; Speight, P.; Little, D.; Di Ciano-Oliveira, C.; Szaszi, K.; Kapus, A. Mediated nuclear import and export of TAZ and the underlying molecular requirements. Nat. Commun. 2018, 9, 4966. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Jiang, J. Hippo-Independent Regulation of Yki/Yap/Taz: A Non-canonical View. Front. Cell Dev. Biol. 2021, 9, 658481. [Google Scholar] [CrossRef] [PubMed]
- Kodaka, M.; Hata, Y. The mammalian Hippo pathway: Regulation and function of YAP1 and TAZ. Cell. Mol. Life Sci. 2015, 72, 285–306. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, Z.; Shah, A.A.; Hong, Y.; Chen, Q.; Wan, Y. New insights into posttranslational modifications of Hippo pathway in carcinogenesis and therapeutics. Cell Div. 2016, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.S.; Meng, Z.; Kim, Y.C.; Park, H.W.; Hansen, C.G.; Kim, S.; Lim, D.S.; Guan, K.L. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 2015, 17, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, V.; Gudmundsdottir, K.; Luong, P.; Leung, K.Y.; Knebel, A.; Basu, S. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis. 2010, 1, e29. [Google Scholar] [CrossRef] [PubMed]
- Sudol, M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 1994, 9, 2145–2152. [Google Scholar]
- Tamm, C.; Bower, N.; Anneren, C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J. Cell Sci. 2011, 124, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Wu, L.W.; Grivennikov, S.I.; de Jong, P.R.; Lian, I.; Yu, F.X.; Wang, K.; Ho, S.B.; Boland, B.S.; Chang, J.T.; et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 2015, 519, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Hoj, J.P.; Mayro, B.; Pendergast, A.M. A TAZ-AXL-ABL2 Feed-Forward Signaling Axis Promotes Lung Adenocarcinoma Brain Metastasis. Cell Rep. 2019, 29, 3421–3434 e3428. [Google Scholar] [CrossRef]
- Hata, S.; Hirayama, J.; Kajiho, H.; Nakagawa, K.; Hata, Y.; Katada, T.; Furutani-Seiki, M.; Nishina, H. A novel acetylation cycle of transcription co-activator Yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents. J. Biol. Chem. 2012, 287, 22089–22098. [Google Scholar] [CrossRef]
- Oudhoff, M.J.; Freeman, S.A.; Couzens, A.L.; Antignano, F.; Kuznetsova, E.; Min, P.H.; Northrop, J.P.; Lehnertz, B.; Barsyte-Lovejoy, D.; Vedadi, M.; et al. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell 2013, 26, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Lapi, E.; Di Agostino, S.; Donzelli, S.; Gal, H.; Domany, E.; Rechavi, G.; Pandolfi, P.P.; Givol, D.; Strano, S.; Lu, X.; et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol. Cell 2008, 32, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Elosegui-Artola, A.; Andreu, I.; Beedle, A.E.M.; Lezamiz, A.; Uroz, M.; Kosmalska, A.J.; Oria, R.; Kechagia, J.Z.; Rico-Lastres, P.; Le Roux, A.L.; et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 2017, 171, 1397–1410 e1314. [Google Scholar] [CrossRef]
- Lin, D.H.; Hoelz, A. The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem. 2019, 88, 725–783. [Google Scholar] [CrossRef] [PubMed]
- Strambio-De-Castillia, C.; Niepel, M.; Rout, M.P. The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 490–501. [Google Scholar] [CrossRef]
- Jamali, T.; Jamali, Y.; Mehrbod, M.; Mofrad, M.R. Nuclear pore complex: Biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int. Rev. Cell Mol. Biol. 2011, 287, 233–286. [Google Scholar] [CrossRef] [PubMed]
- Dultz, E.; Wojtynek, M.; Medalia, O.; Onischenko, E. The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth. Cells 2022, 11, 1456. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Popken, P.; Ghavami, A.; Onck, P.R.; Poolman, B.; Veenhoff, L.M. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell 2015, 26, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Seibel, N.M.; Eljouni, J.; Nalaskowski, M.M.; Hampe, W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal. Biochem. 2007, 368, 95–99. [Google Scholar] [CrossRef]
- Cowburn, D.; Rout, M. Improving the hole picture: Towards a consensus on the mechanism of nuclear transport. Biochem. Soc. Trans. 2023, 51, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.B.; Gorlich, D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem. Sci. 2016, 41, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D. Transport into and out of the cell nucleus. EMBO J. 1998, 17, 2721–2727. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef]
- Lu, J.; Wu, T.; Zhang, B.; Liu, S.; Song, W.; Qiao, J.; Ruan, H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021, 19, 60. [Google Scholar] [CrossRef]
- Wing, C.E.; Fung, H.Y.J.; Chook, Y.M. Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 2022, 23, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Yoneda, Y. Importin alpha: Functions as a nuclear transport factor and beyond. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 259–274. [Google Scholar] [CrossRef]
- Chook, Y.M.; Blobel, G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001, 11, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, A.; Mofrad, M.R.K. On the nuclear pore complex and its emerging role in cellular mechanotransduction. APL Bioeng. 2022, 6, 011504. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.B.; Gorlich, D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 2015, 4, e04251. [Google Scholar] [CrossRef] [PubMed]
- Christie, M.; Chang, C.W.; Rona, G.; Smith, K.M.; Stewart, A.G.; Takeda, A.A.; Fontes, M.R.; Stewart, M.; Vertessy, B.G.; Forwood, J.K.; et al. Structural Biology and Regulation of Protein Import into the Nucleus. J. Mol. Biol. 2016, 428, 2060–2090. [Google Scholar] [CrossRef] [PubMed]
- Moroianu, J.; Blobel, G.; Radu, A. Nuclear protein import: Ran-GTP dissociates the karyopherin alphabeta heterodimer by displacing alpha from an overlapping binding site on beta. Proc. Natl. Acad. Sci. USA 1996, 93, 7059–7062. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Ribbeck, K.; Lipowsky, G.; Kent, H.M.; Stewart, M.; Gorlich, D. NTF2 mediates nuclear import of Ran. EMBO J. 1998, 17, 6587–6598. [Google Scholar] [CrossRef]
- Marfori, M.; Mynott, A.; Ellis, J.J.; Mehdi, A.M.; Saunders, N.F.; Curmi, P.M.; Forwood, J.K.; Boden, M.; Kobe, B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 2011, 1813, 1562–1577. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, M.; Sanchez-Perales, S.; Jarabo, P.; Calvo, E.; Huyton, T.; Fu, L.; Ng, S.C.; Sotodosos-Alonso, L.; Vazquez, J.; Casas-Tinto, S.; et al. Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP. Nat. Commun. 2022, 13, 1174. [Google Scholar] [CrossRef]
- Lu, M.; Zak, J.; Chen, S.; Sanchez-Pulido, L.; Severson, D.T.; Endicott, J.; Ponting, C.P.; Schofield, C.J.; Lu, X. A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell 2014, 157, 1130–1145. [Google Scholar] [CrossRef] [PubMed]
- Soniat, M.; Chook, Y.M. Karyopherin-beta2 Recognition of a PY-NLS Variant that Lacks the Proline-Tyrosine Motif. Structure 2016, 24, 1802–1809. [Google Scholar] [CrossRef]
- Fischer, U.; Schauble, N.; Schutz, S.; Altvater, M.; Chang, Y.; Faza, M.B.; Panse, V.G. A non-canonical mechanism for Crm1-export cargo complex assembly. eLife 2015, 4, e05745. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, Y.; Feng, W.; Lei, L.; Du, Y.; Wu, J.; Wang, S. NUP37, a positive regulator of YAP/TEAD signaling, promotes the progression of hepatocellular carcinoma. Oncotarget 2017, 8, 98004–98013. [Google Scholar] [CrossRef]
- Ester, L.; Cabrita, I.; Ventzke, M.; Kieckhofer, E.; Christodoulou, M.; Mandel, A.M.; Diefenhardt, P.; Fabretti, F.; Benzing, T.; Habbig, S.; et al. The role of the FSGS disease gene product and nuclear pore protein NUP205 in regulating nuclear localization and activity of transcriptional regulators YAP and TAZ. Hum. Mol. Genet. 2023, ddad135. [Google Scholar] [CrossRef] [PubMed]
- Marg, A.; Shan, Y.; Meyer, T.; Meissner, T.; Brandenburg, M.; Vinkemeier, U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 2004, 165, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.R.; Onischenko, E.; Tang, J.H.; Kumar, P.; Chen, J.Z.; Ulrich, A.; Liphardt, J.T.; Weis, K.; Schwartz, T.U. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2013, 2, e00745. [Google Scholar] [CrossRef] [PubMed]
- Kotera, I.; Sekimoto, T.; Miyamoto, Y.; Saiwaki, T.; Nagoshi, E.; Sakagami, H.; Kondo, H.; Yoneda, Y. Importin alpha transports CaMKIV to the nucleus without utilizing importin beta. EMBO J. 2005, 24, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Tessier, T.M.; MacNeil, K.M.; Mymryk, J.S. Piggybacking on Classical Import and Other Non-Classical Mechanisms of Nuclear Import Appear Highly Prevalent within the Human Proteome. Biology 2020, 9, 188. [Google Scholar] [CrossRef]
- Wang, S.; Lu, Y.; Yin, M.X.; Wang, C.; Wu, W.; Li, J.; Wu, W.; Ge, L.; Hu, L.; Zhao, Y.; et al. Importin alpha1 Mediates Yorkie Nuclear Import via an N-terminal Non-canonical Nuclear Localization Signal. J. Biol. Chem. 2016, 291, 7926–7937. [Google Scholar] [CrossRef] [PubMed]
- Reggiani, F.; Gobbi, G.; Ciarrocchi, A.; Sancisi, V. YAP and TAZ Are Not Identical Twins. Trends Biochem. Sci. 2021, 46, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Rebay, I.; Fleming, R.J.; Scottgale, T.N.; Artavanis-Tsakonas, S. The Notch locus and the genetic circuitry involved in early Drosophila neurogenesis. Genes Dev. 1990, 4, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Harigaya, K.; Sasaki, N.; Okamura, Y.; Kokubo, H.; Saga, Y.; Hozumi, K.; Suganami, A.; Tamura, Y.; Nagase, T.; et al. Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo. Development 2011, 138, 5235–5246. [Google Scholar] [CrossRef] [PubMed]
- Zema, S.; Pelullo, M.; Nardozza, F.; Felli, M.P.; Screpanti, I.; Bellavia, D. A Dynamic Role of Mastermind-Like 1: A Journey Through the Main (Path)ways Between Development and Cancer. Front. Cell Dev. Biol. 2020, 8, 613557. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. WNT antagonist, DKK2, is a Notch signaling target in intestinal stem cells: Augmentation of a negative regulation system for canonical WNT signaling pathway by the Notch-DKK2 signaling loop in primates. Int. J. Mol. Med. 2007, 19, 197–201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alves-Guerra, M.C.; Ronchini, C.; Capobianco, A.J. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007, 67, 8690–8698. [Google Scholar] [CrossRef]
- Quaranta, R.; Pelullo, M.; Zema, S.; Nardozza, F.; Checquolo, S.; Lauer, D.M.; Bufalieri, F.; Palermo, R.; Felli, M.P.; Vacca, A.; et al. Maml1 acts cooperatively with Gli proteins to regulate sonic hedgehog signaling pathway. Cell Death Dis. 2017, 8, e2942. [Google Scholar] [CrossRef]
- Jin, B.; Shen, H.; Lin, S.; Li, J.L.; Chen, Z.; Griffin, J.D.; Wu, L. The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J. Biol. Chem. 2010, 285, 14356–14365. [Google Scholar] [CrossRef] [PubMed]
- Heynen, G.J.; Nevedomskaya, E.; Palit, S.; Jagalur Basheer, N.; Lieftink, C.; Schlicker, A.; Zwart, W.; Bernards, R.; Bajpe, P.K. Mastermind-Like 3 Controls Proliferation and Differentiation in Neuroblastoma. Mol. Cancer Res. 2016, 14, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Kiyono, T.; Ryo, E.; Ogawa, R.; Wakai, S.; Ichikawa, H.; Suzuki, K.; Arai, S.; Tsuta, K.; Ishida, M.; et al. Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J. Clin. Investig. 2019, 129, 3827–3832. [Google Scholar] [CrossRef]
- Kim, J.; Kwon, H.; Shin, Y.K.; Song, G.; Lee, T.; Kim, Y.; Jeong, W.; Lee, U.; Zhang, X.; Nam, G.; et al. MAML1/2 promote YAP/TAZ nuclear localization and tumorigenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 13529–13540. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.K.; Carroll, P.M.; Allard, J.D.; Simon, M.A. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development 2002, 129, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Almeida, B.O.; Machado-Neto, J.A. Emerging functions for ANKHD1 in cancer-related signaling pathways and cellular processes. BMB Rep. 2020, 53, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Hollenbeck, J.J.; Danner, D.J.; Landgren, R.M.; Rainbolt, T.K.; Roberts, D.S. Designed ankyrin repeat proteins as scaffolds for multivalent recognition. Biomacromolecules 2012, 13, 1996–2002. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.; Nagampalli, R.S.K.; Fatima, M.T.; Ashraf, G.M. New paradigm in ankyrin repeats: Beyond protein-protein interaction module. Int. J. Biol. Macromol. 2018, 109, 1164–1173. [Google Scholar] [CrossRef]
- Nicastro, G.; Taylor, I.A.; Ramos, A. KH-RNA interactions: Back in the groove. Curr. Opin. Struct. Biol. 2015, 30, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Dhyani, A.; Duarte, A.S.; Machado-Neto, J.A.; Favaro, P.; Ortega, M.M.; Olalla Saad, S.T. ANKHD1 regulates cell cycle progression and proliferation in multiple myeloma cells. FEBS Lett. 2012, 586, 4311–4318. [Google Scholar] [CrossRef]
- Machado-Neto, J.A.; Lazarini, M.; Favaro, P.; de Melo Campos, P.; Scopim-Ribeiro, R.; Franchi Junior, G.C.; Nowill, A.E.; Lima, P.R.; Costa, F.F.; Benichou, S.; et al. ANKHD1 silencing inhibits Stathmin 1 activity, cell proliferation and migration of leukemia cells. Biochim. Biophys. Acta 2015, 1853, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Zhu, M.; Guidry, J.J.; Majeste, N.; Mao, H.; Yanofsky, S.T.; Tian, X.; Wu, C. Mask, the Drosophila ankyrin repeat and KH domain-containing protein, affects microtubule stability. J. Cell Sci. 2021, 134, jcs258512. [Google Scholar] [CrossRef] [PubMed]
- Traina, F.; Favaro, P.M.; Medina Sde, S.; Duarte Ada, S.; Winnischofer, S.M.; Costa, F.F.; Saad, S.T. ANKHD1, ankyrin repeat and KH domain containing 1, is overexpressed in acute leukemias and is associated with SHP2 in K562 cells. Biochim. Biophys. Acta 2006, 1762, 828–834. [Google Scholar] [CrossRef]
- de Almeida, B.O.; de Almeida, L.C.; Costa-Lotufo, L.V.; Machado-Neto, J.A. ANKHD1 contributes to the malignant phenotype of triple-negative breast cancer cells. Cell Biol. Int. 2022, 46, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Han, Q.; Rong, X.Z.; Yang, M.; Han, Y.C.; Yu, J.H.; Lin, X.Y. ANKHD1 promotes proliferation and invasion of non-small-cell lung cancer cells via regulating YAP oncoprotein expression and inactivating the Hippo pathway. Int. J. Oncol. 2020, 56, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Sidor, C.M.; Brain, R.; Thompson, B.J. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr. Biol. 2013, 23, 223–228. [Google Scholar] [CrossRef]
- Sidor, C.; Borreguero-Munoz, N.; Fletcher, G.C.; Elbediwy, A.; Guillermin, O.; Thompson, B.J. Mask family proteins ANKHD1 and ANKRD17 regulate YAP nuclear import and stability. eLife 2019, 8, e48601. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tu, K.; Liu, D.; Guo, L.; Chen, Y.; Li, Q.; Maiers, J.L.; Liu, Z.; Shah, V.H.; Dou, C.; et al. p300 Acetyltransferase Is a Cytoplasm-to-Nucleus Shuttle for SMAD2/3 and TAZ Nuclear Transport in Transforming Growth Factor beta-Stimulated Hepatic Stellate Cells. Hepatology 2019, 70, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Posern, G.; Treisman, R. Actin’ together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006, 16, 588–596. [Google Scholar] [CrossRef]
- Olson, E.N.; Nordheim, A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010, 11, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.Z.; Lichner, Z.; Szaszi, K.; Kapus, A. MRTF: Basic Biology and Role in Kidney Disease. Int. J. Mol. Sci. 2021, 22, 6040. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.M.; Miyamoto, S.; Brown, J.H. Myocardin-Related Transcription Factor A and Yes-Associated Protein Exert Dual Control in G Protein-Coupled Receptor- and RhoA-Mediated Transcriptional Regulation and Cell Proliferation. Mol. Cell. Biol. 2016, 36, 39–49. [Google Scholar] [CrossRef]
- Foster, C.T.; Gualdrini, F.; Treisman, R. Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes. Dev. 2017, 31, 2361–2375. [Google Scholar] [CrossRef]
- Nagashima, S.; Maruyama, J.; Honda, K.; Kondoh, Y.; Osada, H.; Nawa, M.; Nakahama, K.I.; Ishigami-Yuasa, M.; Kagechika, H.; Sugimura, H.; et al. CSE1L promotes nuclear accumulation of transcriptional coactivator TAZ and enhances invasiveness of human cancer cells. J. Biol. Chem. 2021, 297, 100803. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Hieda, M.; Harreman, M.T.; Fukumoto, M.; Saiwaki, T.; Hodel, A.E.; Corbett, A.H.; Yoneda, Y. Importin alpha can migrate into the nucleus in an importin beta- and Ran-independent manner. EMBO J. 2002, 21, 5833–5842. [Google Scholar] [CrossRef]
- Moreno-Vicente, R.; Pavon, D.M.; Martin-Padura, I.; Catala-Montoro, M.; Diez-Sanchez, A.; Quilez-Alvarez, A.; Lopez, J.A.; Sanchez-Alvarez, M.; Vazquez, J.; Strippoli, R.; et al. Caveolin-1 Modulates Mechanotransduction Responses to Substrate Stiffness through Actin-Dependent Control of YAP. Cell Rep. 2018, 25, 1622–1635 e1626. [Google Scholar] [CrossRef]
- Panagiotopoulos, A.A.; Polioudaki, C.; Ntallis, S.G.; Dellis, D.; Notas, G.; Panagiotidis, C.A.; Theodoropoulos, P.A.; Castanas, E.; Kampa, M. The sequence [EKRKI(E/R)(K/L/R/S/T)] is a nuclear localization signal for importin 7 binding (NLS7). Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129851. [Google Scholar] [CrossRef]
- Kanai, F.; Marignani, P.A.; Sarbassova, D.; Yagi, R.; Hall, R.A.; Donowitz, M.; Hisaminato, A.; Fujiwara, T.; Ito, Y.; Cantley, L.C.; et al. TAZ: A novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000, 19, 6778–6791. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Sudol, M. Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells 2009, 14, 607–615. [Google Scholar] [CrossRef]
- Kofler, M.; Kapus, A. Nucleocytoplasmic Shuttling of the Mechanosensitive Transcription Factors MRTF and YAP/TAZ. Methods Mol. Biol. 2021, 2299, 197–216. [Google Scholar] [CrossRef]
- Bilokapic, S.; Schwartz, T.U. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2012, 109, 15241–15246. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Sadowski, C.E.; Kohl, S.; Lovric, S.; Astrinidis, S.A.; Pabst, W.L.; Gee, H.Y.; Ashraf, S.; Lawson, J.A.; Shril, S.; et al. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat. Genet. 2016, 48, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.A.; Donato, D.M.; Balcioglu, H.E.; Schmidt, T.; Danen, E.H.; Koenderink, G.H. A guide to mechanobiology: Where biology and physics meet. Biochim. Biophys. Acta 2015, 1853, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Jung-Garcia, Y.; Maiques, O.; Monger, J.; Rodriguez-Hernandez, I.; Fanshawe, B.; Domart, M.C.; Renshaw, M.J.; Marti, R.M.; Matias-Guiu, X.; Collinson, L.M.; et al. LAP1 supports nuclear adaptability during constrained melanoma cell migration and invasion. Nat. Cell Biol. 2023, 25, 108–119. [Google Scholar] [CrossRef]
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017, 216, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Major, L.G.; Holle, A.W.; Young, J.L.; Hepburn, M.S.; Jeong, K.; Chin, I.L.; Sanderson, R.W.; Jeong, J.H.; Aman, Z.M.; Kennedy, B.F.; et al. Volume Adaptation Controls Stem Cell Mechanotransduction. ACS Appl. Mater. Interfaces 2019, 11, 45520–45530. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.M.; Smith, M.A.; Jensen, C.C.; Yoshigi, M.; Blankman, E.; Ullman, K.S.; Beckerle, M.C. Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 2020, 31, 1774–1787. [Google Scholar] [CrossRef]
- Urciuoli, E.; Petrini, S.; D’Oria, V.; Leopizzi, M.; Rocca, C.D.; Peruzzi, B. Nuclear Lamins and Emerin Are Differentially Expressed in Osteosarcoma Cells and Scale with Tumor Aggressiveness. Cancers 2020, 12, 443. [Google Scholar] [CrossRef]
- Takata, T.; Matsumura, M. The LINC Complex Assists the Nuclear Import of Mechanosensitive Transcriptional Regulators. Results Probl. Cell Differ. 2022, 70, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Lovett, D.B.; Shekhar, N.; Nickerson, J.A.; Roux, K.J.; Lele, T.P. Modulation of Nuclear Shape by Substrate Rigidity. Cell Mol. Bioeng. 2013, 6, 230–238. [Google Scholar] [CrossRef]
- Zimmerli, C.E.; Allegretti, M.; Rantos, V.; Goetz, S.K.; Obarska-Kosinska, A.; Zagoriy, I.; Halavatyi, A.; Hummer, G.; Mahamid, J.; Kosinski, J.; et al. Nuclear pores dilate and constrict in cellulo. Science 2021, 374, eabd9776. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, A.T.; Ziaei, S.; Ehret, C.; Duchemin, H.; Mamchaoui, K.; Bigot, A.; Mayer, M.; Quijano-Roy, S.; Desguerre, I.; Laine, J.; et al. Cellular microenvironments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J. Cell Sci. 2014, 127, 2873–2884. [Google Scholar] [CrossRef]
- Shiu, J.Y.; Aires, L.; Lin, Z.; Vogel, V. Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nat. Cell Biol. 2018, 20, 262–271. [Google Scholar] [CrossRef]
- Koushki, N.; Ghagre, A.; Srivastava, L.K.; Molter, C.; Ehrlicher, A.J. Nuclear compression regulates YAP spatiotemporal fluctuations in living cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2301285120. [Google Scholar] [CrossRef]
- Infante, E.; Stannard, A.; Board, S.J.; Rico-Lastres, P.; Panagaki, F.; Beedle, A.E.M.; Rajan, V.S.; Rostkova, E.; Lezamiz, A.; Wang, Y.J.; et al. The mechanical stability of proteins regulates their translocation rate into the cell nucleus. Nat. Phys. 2019, 15, 973–981. [Google Scholar] [CrossRef]
- Scott, K.E.; Fraley, S.I.; Rangamani, P. A spatial model of YAP/TAZ signaling reveals how stiffness, dimensionality, and shape contribute to emergent outcomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2021571118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.Y.; Yu, J.; Guan, K.L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes. Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Zhang, L.; Jiang, J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev. Biol. 2010, 337, 303–312. [Google Scholar] [CrossRef]
- Furukawa, K.T.; Yamashita, K.; Sakurai, N.; Ohno, S. The Epithelial Circumferential Actin Belt Regulates YAP/TAZ through Nucleocytoplasmic Shuttling of Merlin. Cell Rep. 2017, 20, 1435–1447. [Google Scholar] [CrossRef]
- Ege, N.; Dowbaj, A.M.; Jiang, M.; Howell, M.; Hooper, S.; Foster, C.; Jenkins, R.P.; Sahai, E. Quantitative Analysis Reveals that Actin and Src-Family Kinases Regulate Nuclear YAP1 and Its Export. Cell Syst. 2018, 6, 692–708 e613. [Google Scholar] [CrossRef] [PubMed]
- Barry, E.R.; Simov, V.; Valtingojer, I.; Venier, O. Recent Therapeutic Approaches to Modulate the Hippo Pathway in Oncology and Regenerative Medicine. Cells 2021, 10, 2715. [Google Scholar] [CrossRef]
- Mi, W.; Lin, Q.; Childress, C.; Sudol, M.; Robishaw, J.; Berlot, C.H.; Shabahang, M.; Yang, W. Geranylgeranylation signals to the Hippo pathway for breast cancer cell proliferation and migration. Oncogene 2015, 34, 3095–3106. [Google Scholar] [CrossRef] [PubMed]
- Vigneau, A.L.; Rico, C.; Boerboom, D.; Paquet, M. Statins downregulate YAP and TAZ and exert anti-cancer effects in canine mammary tumour cells. Vet. Comp. Oncol. 2022, 20, 437–448. [Google Scholar] [CrossRef]
- Messina, B.; Lo Sardo, F.; Scalera, S.; Memeo, L.; Colarossi, C.; Mare, M.; Blandino, G.; Ciliberto, G.; Maugeri-Sacca, M.; Bon, G. Hippo pathway dysregulation in gastric cancer: From Helicobacter pylori infection to tumor promotion and progression. Cell Death Dis. 2023, 14, 21. [Google Scholar] [CrossRef]
- Kang, L.; Yi, J.; Lau, C.W.; He, L.; Chen, Q.; Xu, S.; Li, J.; Xia, Y.; Zhang, Y.; Huang, Y.; et al. AMPK-Dependent YAP Inhibition Mediates the Protective Effect of Metformin against Obesity-Associated Endothelial Dysfunction and Inflammation. Antioxidants 2023, 12, 1681. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.J.; Rouse, C.; Xu, X.; Wang, J.; Onaitis, M.W.; Pendergast, A.M. Inactivation of ABL kinases suppresses non-small cell lung cancer metastasis. JCI Insight 2016, 1, e89647. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rao, G.; Kim, I.K.; Conforti, F.; Liu, J.; Zhang, Y.W.; Giaccone, G. Dasatinib sensitises KRAS-mutant cancer cells to mitogen-activated protein kinase kinase inhibitor via inhibition of TAZ activity. Eur. J. Cancer 2018, 99, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Martellucci, S.; Clementi, L.; Sabetta, S.; Mattei, V.; Botta, L.; Angelucci, A. Src Family Kinases as Therapeutic Targets in Advanced Solid Tumors: What We Have Learned so Far. Cancers 2020, 12, 1448. [Google Scholar] [CrossRef]
- Liu-Chittenden, Y.; Huang, B.; Shim, J.S.; Chen, Q.; Lee, S.J.; Anders, R.A.; Liu, J.O.; Pan, D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012, 26, 1300–1305. [Google Scholar] [CrossRef]
- Yu, F.X.; Luo, J.; Mo, J.S.; Liu, G.; Kim, Y.C.; Meng, Z.; Zhao, L.; Peyman, G.; Ouyang, H.; Jiang, W.; et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 2014, 25, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.; Han, X.; Zheng, B.; DeRan, M.; Yu, J.; Jarugumilli, G.K.; Deng, H.; Pan, D.; Luo, X.; Wu, X. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat. Chem. Biol. 2016, 12, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Zagiel, B.; Melnyk, P.; Cotelle, P. Progress with YAP/TAZ-TEAD inhibitors: A patent review (2018-present). Expert Opin. Ther. Pat. 2022, 32, 899–912. [Google Scholar] [CrossRef]
- Lou, J.; Lu, Y.; Cheng, J.; Zhou, F.; Yan, Z.; Zhang, D.; Meng, X.; Zhao, Y. A chemical perspective on the modulation of TEAD transcriptional activities: Recent progress, challenges, and opportunities. Eur. J. Med. Chem. 2022, 243, 114684. [Google Scholar] [CrossRef]
- Pobbati, A.V.; Kumar, R.; Rubin, B.P.; Hong, W. Therapeutic targeting of TEAD transcription factors in cancer. Trends Biochem. Sci. 2023, 48, 450–462. [Google Scholar] [CrossRef]
- Zhao, B.; Pobbati, A.V.; Rubin, B.P.; Stauffer, S. Leveraging Hot Spots of TEAD-Coregulator Interactions in the Design of Direct Small Molecule Protein-Protein Interaction Disruptors Targeting Hippo Pathway Signaling. Pharmaceuticals 2023, 16, 583. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.; Konradi, A.W.; Feng, Y.; Peng, X.; Ma, M.; Li, J.; Yu, F.X.; Guan, K.L.; Post, L. Small Molecule Inhibitors of TEAD Auto-palmitoylation Selectively Inhibit Proliferation and Tumor Growth of NF2-deficient Mesothelioma. Mol. Cancer Ther. 2021, 20, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Lu, W.; Che, J.; Kwiatkowski, N.P.; Gao, Y.; Seo, H.S.; Ficarro, S.B.; Gokhale, P.C.; Liu, Y.; Geffken, E.A.; et al. Covalent disruptor of YAP-TEAD association suppresses defective Hippo signaling. eLife 2022, 11, e78810. [Google Scholar] [CrossRef]
- Laraba, L.; Hillson, L.; de Guibert, J.G.; Hewitt, A.; Jaques, M.R.; Tang, T.T.; Post, L.; Ercolano, E.; Rai, G.; Yang, S.M.; et al. Inhibition of YAP/TAZ-driven TEAD activity prevents growth of NF2-null schwannoma and meningioma. Brain 2023, 146, 1697–1713. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, T.; Chook, Y.M. Karyopherins in cancer. Curr. Opin. Cell Biol. 2018, 52, 30–42. [Google Scholar] [CrossRef]
- Han, Y.; Wang, X. The emerging roles of KPNA2 in cancer. Life Sci. 2020, 241, 117140. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Lin, M.; Cheng, X.; Zhang, Z.; Deng, S.; Lang, K.; Yang, Z.; Sun, X. KPNB1-mediated nuclear import in cancer. Eur. J. Pharmacol. 2023, 955, 175925. [Google Scholar] [CrossRef] [PubMed]
- Jans, D.A.; Martin, A.J.; Wagstaff, K.M. Inhibitors of nuclear transport. Curr. Opin. Cell Biol. 2019, 58, 50–60. [Google Scholar] [CrossRef]
- Huang, J.L.; Yan, X.L.; Li, W.; Fan, R.Z.; Li, S.; Chen, J.; Zhang, Z.; Sang, J.; Gan, L.; Tang, G.H.; et al. Discovery of Highly Potent Daphnane Diterpenoids Uncovers Importin-beta1 as a Druggable Vulnerability in Castration-Resistant Prostate Cancer. J. Am. Chem. Soc. 2022, 144, 17522–17532. [Google Scholar] [CrossRef]
- Nachmias, B.; Schimmer, A.D. Targeting nuclear import and export in hematological malignancies. Leukemia 2020, 34, 2875–2886. [Google Scholar] [CrossRef]
- Podar, K.; Shah, J.; Chari, A.; Richardson, P.G.; Jagannath, S. Selinexor for the treatment of multiple myeloma. Expert Opin. Pharmacother. 2020, 21, 399–408. [Google Scholar] [CrossRef]
- Mo, C.C.; Yee, A.J.; Midha, S.; Hartley-Brown, M.A.; Nadeem, O.; O’Donnell, E.K.; Bianchi, G.; Sperling, A.S.; Laubach, J.P.; Richardson, P.G. Selinexor: Targeting a novel pathway in multiple myeloma. eJHaem 2023, 4, 792–810. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).