E3 Ubiquitin Ligase TRIP12 Controls Exit from Mitosis via Positive Regulation of MCL-1 in Response to Taxol
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
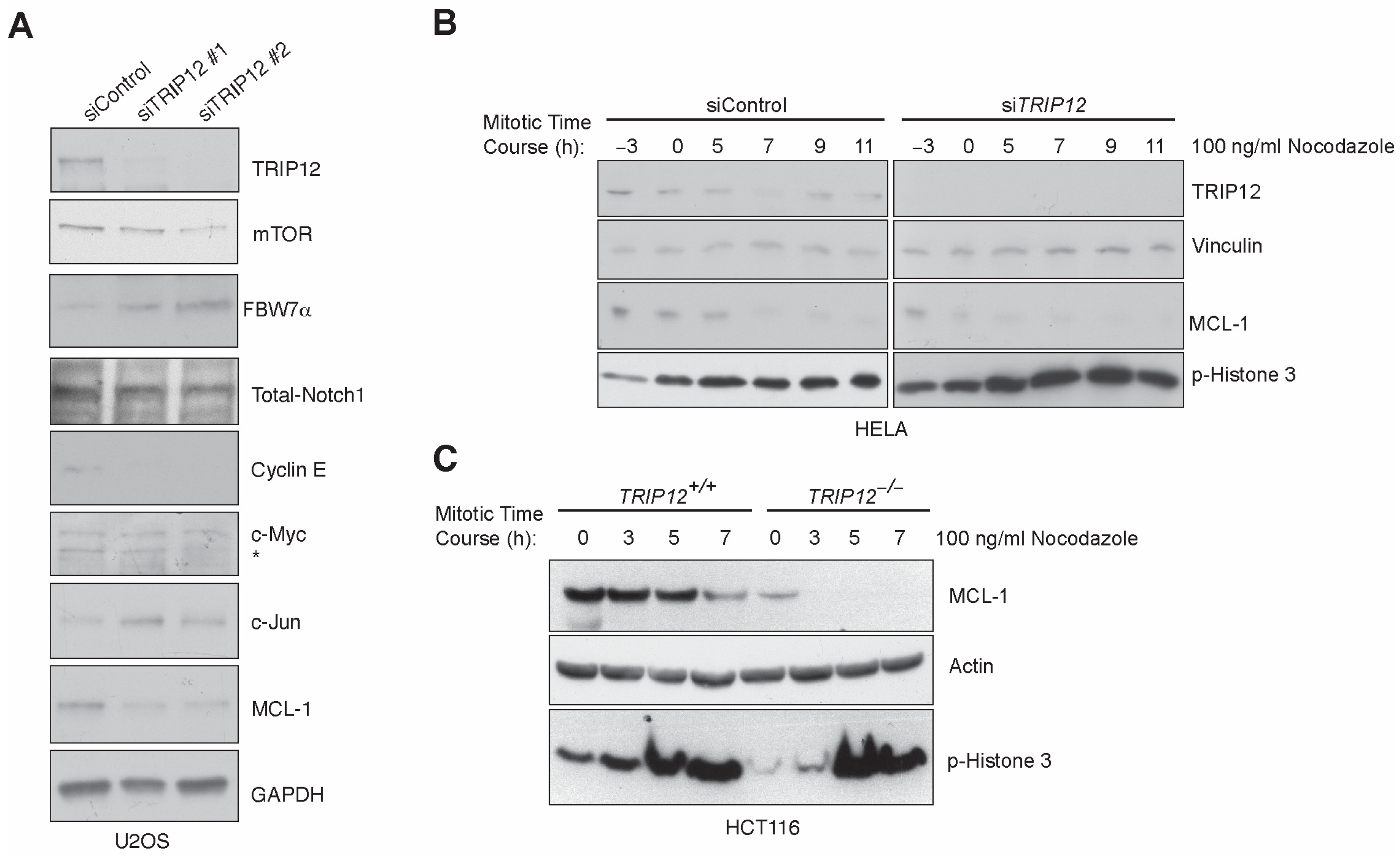
2.1. Enhanced Mitotic Degradation of MCL-1 in the Absence of TRIP12
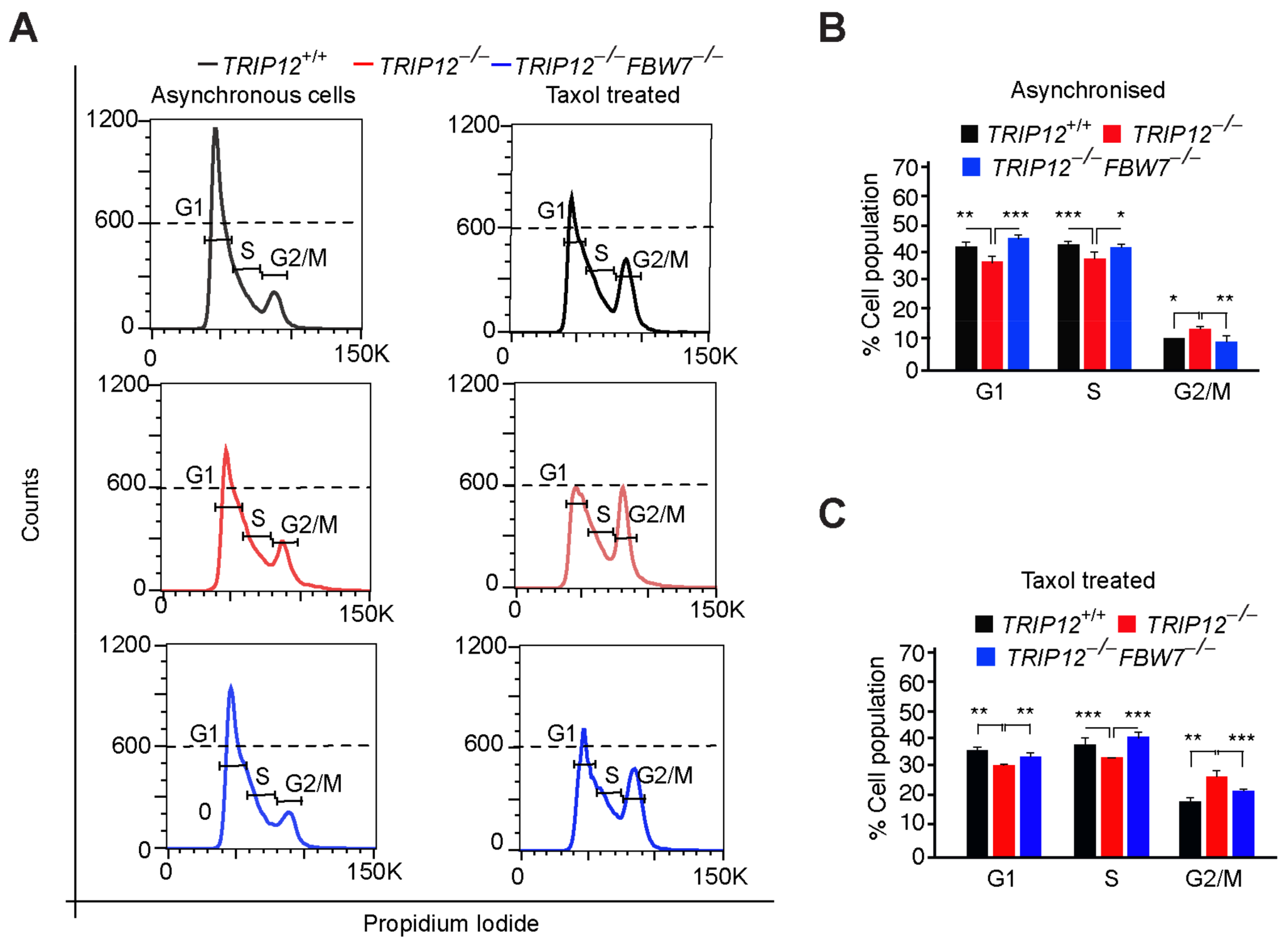
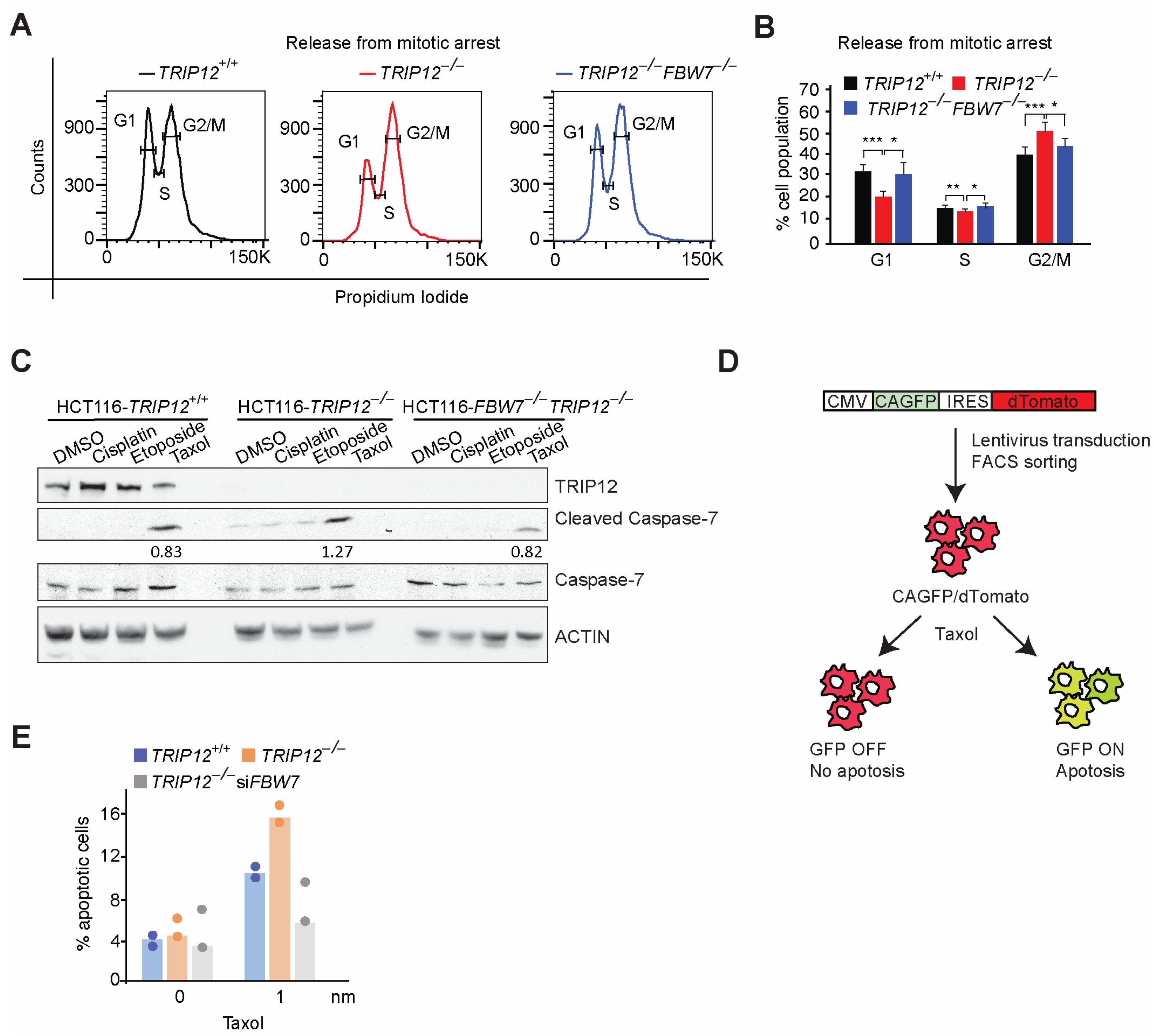
2.2. TRIP12 Controls Exit from Mitosis via Negative Regulation of FBW7
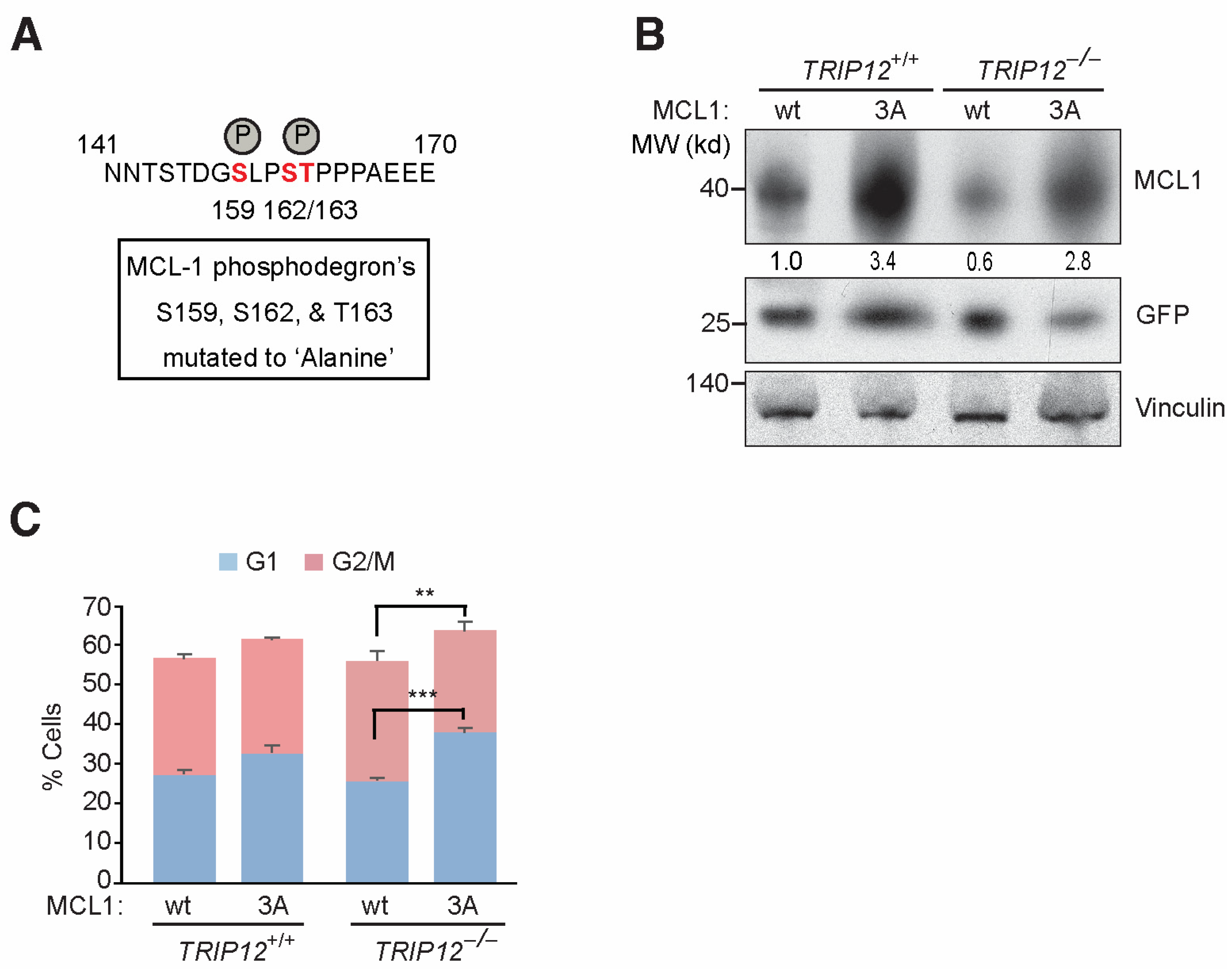
2.3. Mutating FBW7 Recognizable GSK3β Phosphodegron on MCL-1 Reverses Mitotic Arrest in TRIP12−/− Cells
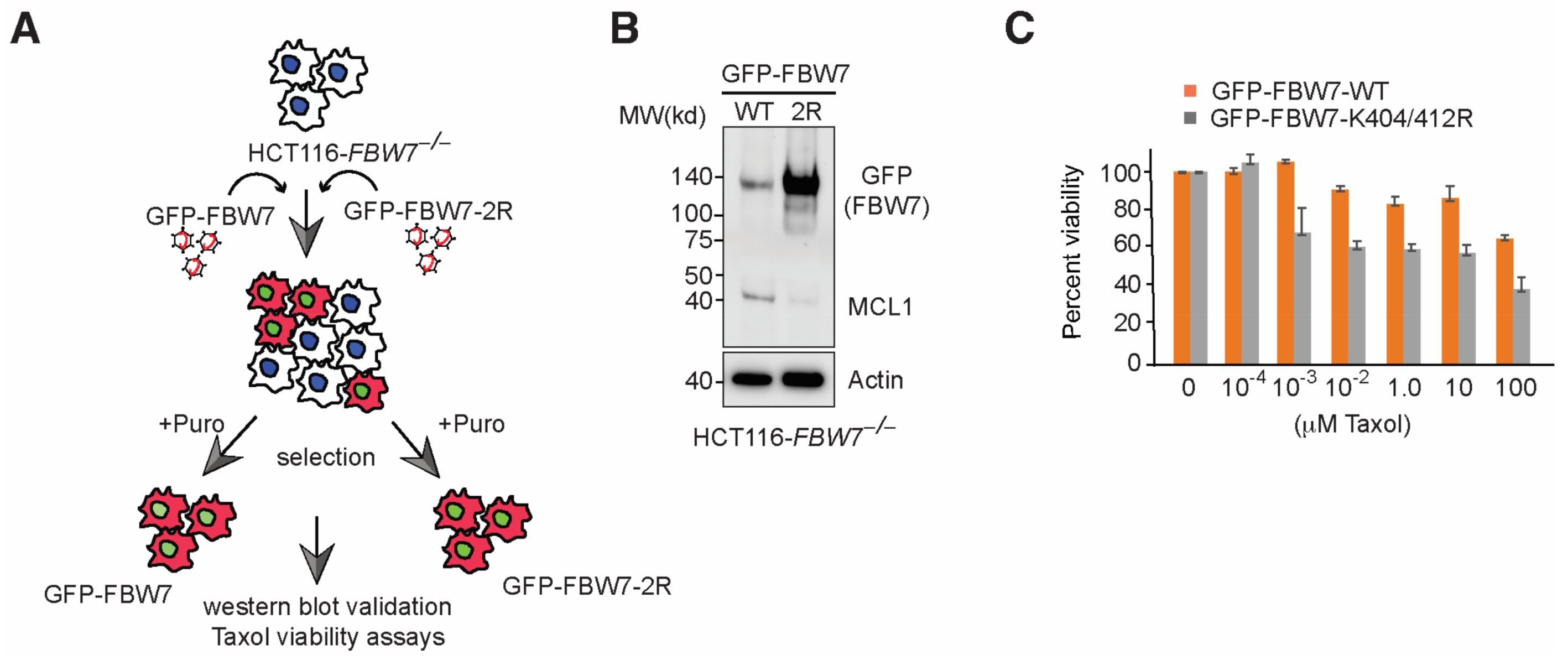
2.4. FBW7 Ubiquitylation Resistant Mutant Reduces MCL-1 Protein Levels and Sensitize HCT116 Cells to Taxol
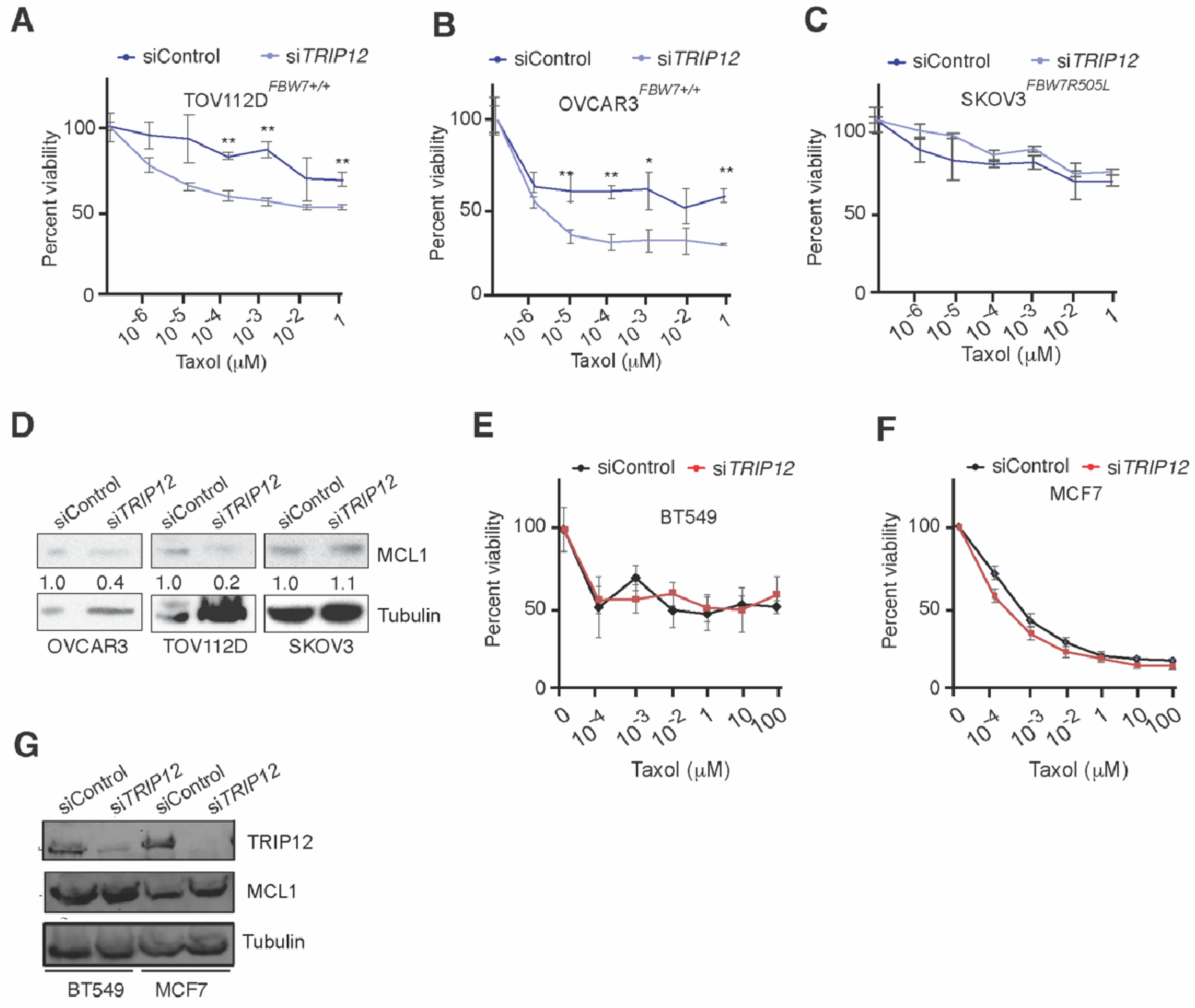
2.5. Targeting TRIP12 Sensitizes Ovarian but Not Breast Cancer Cells to Taxol-Induced Cell Death
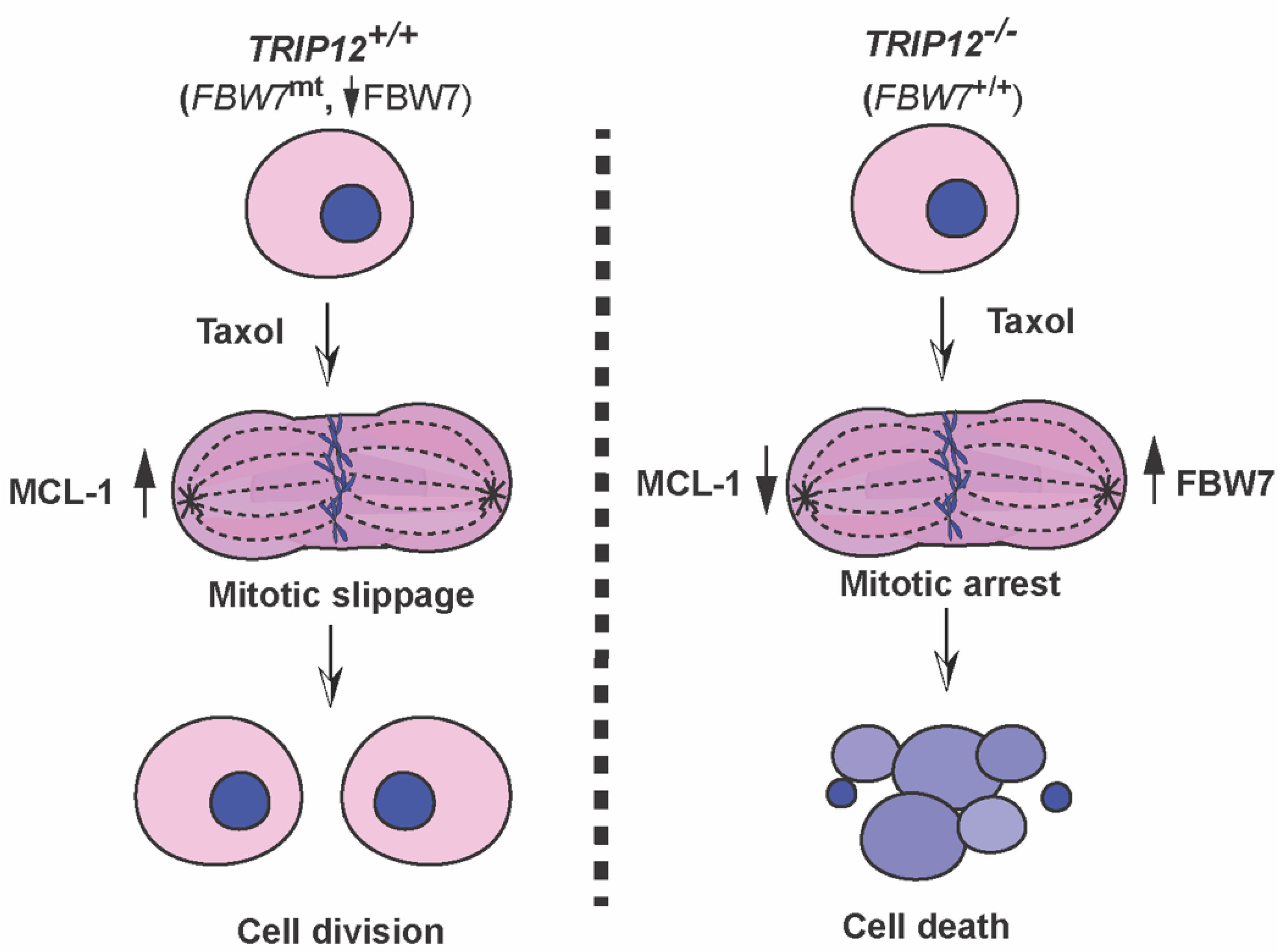
3. Discussion
4. Methods and Materials
4.1. Cell Lines
4.2. Site Directed Mutagenesis
4.3. Western Blot Assays
4.4. Antibodies
4.5. Cell Viability Assays
4.6. LDH Cell Cytotoxicity Assays
4.7. Cell Cycle Analysis
4.8. Generation CA-GFP Apoptosis Reported Plasmid and Cell Lines
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nagai, H.; Kim, Y.H. Cancer Prevention from the Perspective of Global Cancer Burden Patterns. J. Thorac. Dis. 2017, 9, 448–451. [Google Scholar] [CrossRef]
- Ojima, I.; Lichtenthal, B.; Lee, S.; Wang, C.; Wang, X. Taxane Anticancer Agents: A Patent Perspective. Expert Opin. Ther. Pat. 2016, 26, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hüsemann, L.C.; Reese, A.; Radine, C.; Piekorz, R.P.; Budach, W.; Sohn, D.; Jänicke, R.U. The Microtubule Targeting Agents Eribulin and Paclitaxel Activate Similar Signaling Pathways and Induce Cell Death Predominantly in a Caspase-Independent Manner. Cell Cycle 2020, 19, 464–478. [Google Scholar] [CrossRef]
- Choi, Y.H.; Yoo, Y.H. Taxol-Induced Growth Arrest and Apoptosis Is Associated with the Upregulation of the Cdk Inhibitor, P21waf1/Cip1, in Human Breast Cancer Cells. Oncol. Rep. 2012, 28, 2163–2169. [Google Scholar] [CrossRef]
- Welcker, M.; Clurman, B.E. FBW7 Ubiquitin Ligase: A Tumour Suppressor at the Crossroads of Cell Division, Growth and Differentiation. Nat. Rev. Cancer 2008, 8, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; Kusam, S.; Lam, C.; Okamoto, T.; Sandoval, W.; Anderson, D.J.; Helgason, E.; Ernst, J.A.; Eby, M.; Liu, J.; et al. Sensitivity to Antitubulin Chemotherapeutics Is Regulated by MCL-1 and FBW7. Nature 2011, 471, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, H.; Shaik, S.; Onoyama, I.; Gao, D.; Tseng, A.; Maser, R.S.; Zhai, B.; Wan, L.; Gutierrez, A.; Lau, A.W.; et al. Scf(FBW7) Regulates Cellular Apoptosis by Targeting MCL-1 for Ubiquitylation and Destruction. Nature 2011, 471, 104–109. [Google Scholar] [CrossRef]
- Khan, O.M.; Carvalho, J.; Spencer-Dene, B.; Mitter, R.; Frith, D.; Snijders, A.P.; Wood, S.A.; Behrens, A. The Deubiquitinase Usp9x Regulates FBW7 Stability and Suppresses Colorectal Cancer. J. Clin. Investig. 2018, 128, 1326–1337. [Google Scholar] [CrossRef]
- Min, S.-H.; Lau, A.W.; Lee, T.H.; Inuzuka, H.; Wei, S.; Huang, P.; Shaik, S.; Lee, D.Y.; Finn, G.; Balastik, M.; et al. Negative Regulation of the Stability and Tumor Suppressor Function of FBW7 by the Pin1 Prolyl Isomerase. Mol. Cell 2012, 46, 771–783. [Google Scholar] [CrossRef]
- Johnson, E.S.; Ma, P.C.; Ota, I.M.; Varshavsky, A. A Proteolytic Pathway That Recognizes Ubiquitin as a Degradation Signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Taki, T.; Oda, M.; Watanabe, T.; Yumura-Yagi, K.; Kobayashi, R.; Suzuki, N.; Hara, J.; Horibe, K.; Hayashi, Y. Fbxw7 and Notch1 Mutations in Childhood T Cell Acute Lymphoblastic Leukaemia and T Cell Non-Hodgkin Lymphoma. Br. J. Haematol. 2009, 145, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Gudjonsson, T.; Altmeyer, M.; Savic, V.; Toledo, L.; Dinant, C.; Grøfte, M.; Bartkova, J.; Poulsen, M.; Oka, Y.; Bekker-Jensen, S.; et al. TRIP12 and Ubr5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell 2012, 150, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Larrieu, D.; Brunet, M.; Vargas, C.; Hanoun, N.; Ligat, L.; Dagnon, L.; Lulka, H.; Pommier, R.M.; Selves, J.; Jády, B.E.; et al. The E3 Ubiquitin Ligase TRIP12 Participates in Cell Cycle Progression and Chromosome Stability. Sci. Rep. 2020, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.A.; Kim, D.; Hwang, H.; Kim, M.S.; Ma, S.-X.; Kwon, S.-H.; Kweon, S.H.; Yoo, J.M.; Choi, S.; Kwon, S.H.; et al. TRIP12 Ubiquitination of Glucocerebrosidase Contributes to Neurodegeneration in Parkinson’s Disease. Neuron 2021, 109, 3758–3774.e11. [Google Scholar] [CrossRef]
- Chen, D.; Yoon, J.B.; Gu, W. Reactivating the Arf-P53 Axis in Aml Cells by Targeting Ulf. Cell Cycle 2010, 9, 2946–2951. [Google Scholar] [CrossRef]
- Gatti, M.; Imhof, R.; Huang, Q.; Baudis, M.; Altmeyer, M. The Ubiquitin Ligase TRIP12 Limits Parp1 Trapping and Constrains Parp Inhibitor Efficiency. Cell Rep. 2020, 32, 107985. [Google Scholar] [CrossRef]
- Khan, O.M.; Almagro, J.; Nelson, J.K.; Horswell, S.; Encheva, V.; Keyan, K.S.; Clurman, B.E.; Snijders, A.P.; Behrens, A. Proteasomal Degradation of the Tumour Suppressor FBW7 Requires Branched Ubiquitylation by TRIP12. Nat. Commun. 2021, 12, 2043. [Google Scholar] [CrossRef]
- Grim, J.E.; Gustafson, M.P.; Hirata, R.K.; Hagar, A.C.; Swanger, J.; Welcker, M.; Hwang, H.C.; Ericsson, J.; Russell, D.W.; Clurman, B.E. Isoform- and Cell Cycle-Dependent Substrate Degradation by the FBW7 Ubiquitin Ligase. J. Cell Biol. 2008, 181, 913–920. [Google Scholar] [CrossRef]
- Nicholls, S.B.; Chu, J.; Abbruzzese, G.; Tremblay, K.D.; Hardy, J.A. Mechanism of a Genetically Encoded Dark-to-Bright Reporter for Caspase Activity. J. Biol. Chem. 2011, 286, 24977–24986. [Google Scholar] [CrossRef]
- Campbell, K.J.; Mason, S.M.; Winder, M.L.; Willemsen, R.B.E.; Cloix, C.; Lawson, H.; Rooney, N.; Dhayade, S.; Sims, A.H.; Blyth, K.; et al. Breast Cancer Dependence on MCL-1 Is Due to Its Canonical Anti-Apoptotic Function. Cell Death Differ. 2021, 28, 2589–2600. [Google Scholar] [CrossRef]
- Verma, M. Personalized Medicine and Cancer. J. Pers. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Bellon, M.; Ju, M.; Zhao, L.; Wei, M.; Fu, L.; Nicot, C. Clinical Significance of Fbxw7 Loss of Function in Human Cancers. Mol. Cancer 2022, 21, 87. [Google Scholar] [CrossRef] [PubMed]
- Bolomsky, A.; Vogler, M.; Köse, M.C.; Heckman, C.A.; Ehx, G.; Ludwig, H.; Caers, J. MCL-1 Inhibitors, Fast-Lane Development of a New Class of Anti-Cancer Agents. J. Hematol. Oncol. 2020, 13, 173. [Google Scholar] [CrossRef]
- Akula, M.K.; Ibrahim, M.X.; Ivarsson, E.G.; Khan, O.M.; Kumar, I.T.; Erlandsson, M.; Karlsson, C.; Xu, X.; Brisslert, M.; Brakebusch, C.; et al. Protein Prenylation Restrains Innate Immunity by Inhibiting Rac1 Effector Interactions. Nat. Commun. 2019, 10, 3975. [Google Scholar] [CrossRef] [PubMed]
- Aldaalis, A.; Bengoechea-Alonso, M.T.; Ericsson, J. The Srebp-Dependent Regulation of Cyclin D1 Coordinates Cell Proliferation and Lipid Synthesis. Front. Oncol. 2022, 12, 942386. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Aldaalis, A.; Ericsson, J. Loss of the FBW7 Tumor Suppressor Rewires Cholesterol Metabolism in Cancer Cells Leading to Activation of the Pi3k-Akt Signalling Axis. Front. Oncol. 2022, 12, 990672. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keyan, K.S.; Alanany, R.; Kohil, A.; Khan, O.M. E3 Ubiquitin Ligase TRIP12 Controls Exit from Mitosis via Positive Regulation of MCL-1 in Response to Taxol. Cancers 2023, 15, 505. https://doi.org/10.3390/cancers15020505
Keyan KS, Alanany R, Kohil A, Khan OM. E3 Ubiquitin Ligase TRIP12 Controls Exit from Mitosis via Positive Regulation of MCL-1 in Response to Taxol. Cancers. 2023; 15(2):505. https://doi.org/10.3390/cancers15020505
Chicago/Turabian StyleKeyan, Kripa S., Rania Alanany, Amira Kohil, and Omar M. Khan. 2023. "E3 Ubiquitin Ligase TRIP12 Controls Exit from Mitosis via Positive Regulation of MCL-1 in Response to Taxol" Cancers 15, no. 2: 505. https://doi.org/10.3390/cancers15020505
APA StyleKeyan, K. S., Alanany, R., Kohil, A., & Khan, O. M. (2023). E3 Ubiquitin Ligase TRIP12 Controls Exit from Mitosis via Positive Regulation of MCL-1 in Response to Taxol. Cancers, 15(2), 505. https://doi.org/10.3390/cancers15020505





