Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Dataset
2.3. Image Acquisition and Processing
2.4. Input Data
2.5. ResNet Architecture Description and Model Development
2.6. Evaluation of the Classification Performance
3. Results
3.1. IDH and 1p/19q Status Prediction in Lower Grade Gliomas
3.2. IDH Status and Three Molecular Subtypes Prediction in All Glioma Grades
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Metellus, P.; Coulibaly, B.; Colin, C.; de Paula, A.M.; Vasiljevic, A.; Taieb, D.; Barlier, A.; Boisselier, B.; Mokhtari, K.; Wang, X.W.; et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010, 120, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Soffietti, R.; Baumert, B.; Bello, L.; Von Deimling, A.; Duffau, H.; Frénay, M.; Grisold, W.; Grant, R.; Graus, F.; Hoang-Xuan, K. Guidelines on management of low-grade gliomas: Report of an EFNS–EANO Task Force. Eur. J. Neurol. 2010, 17, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; Van Den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Bangalore Yogananda, C.G.; Shah, B.R.; Vejdani-Jahromi, M.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Mickey, B.; Patel, T.R.; et al. A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas. Neuro-Oncology 2020, 22, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.-G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.-K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro-Oncology 2021, 23, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Jian, A.; Jang, K.; Manuguerra, M.; Liu, S.; Magnussen, J.; Di Ieva, A. Machine Learning for the Prediction of Molecular Markers in Glioma on Magnetic Resonance Imaging: A Systematic Review and Meta-Analysis. Neurosurgery 2021, 89, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Alhasan, A.S. Clinical Applications of Artificial Intelligence, Machine Learning, and Deep Learning in the Imaging of Gliomas: A Systematic Review. Cureus 2021, 13, e19580. [Google Scholar] [CrossRef]
- Smits, M. Imaging of oligodendroglioma. Br. J. Radiol. 2016, 89, 20150857. [Google Scholar] [CrossRef] [PubMed]
- Rudà, R.; Bruno, F.; Ius, T.; Silvani, A.; Minniti, G.; Pace, A.; Lombardi, G.; Bertero, L.; Pizzolitto, S.; Pollo, B.; et al. IDH wild-type grade 2 diffuse astrocytomas: Prognostic factors and impact of treatments within molecular subgroups. Neuro-Oncology 2022, 24, 809–820. [Google Scholar] [CrossRef]
- Michiwaki, Y.; Hata, N.; Mizoguchi, M.; Hiwatashi, A.; Kuga, D.; Hatae, R.; Akagi, Y.; Amemiya, T.; Fujioka, Y.; Togao, O.; et al. Relevance of calcification and contrast enhancement pattern for molecular diagnosis and survival prediction of gliomas based on the 2016 World Health Organization Classification. Clin. Neurol. Neurosurg. 2019, 187, 105556. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, C.; Barritault, M.; Poncet, D.; Cartalat, S.; Joubert, B.; Bruna, J.; Jouanneau, E.; Guyotat, J.; Vasiljevic, A.; Fenouil, T.; et al. Radiological characteristics and natural history of adult IDH-wildtype astrocytomas with TERT promoter mutations. Neurosurgery 2019, 85, E448–E456. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, B.; Zhang, S.; Cheng, J.; Liu, X.; Wang, W.; Dong, Y.; Zhang, L.; Mo, X.; Chen, Q.; et al. Quantitative MRI-based radiomics for noninvasively predicting molecular subtypes and survival in glioma patients. NPJ Precis. Oncol. 2021, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Lu, C.; Li, G.; Li, Z.; Sun, S.; Zhang, Y.; Hou, Z.; Xie, J. Prediction of lower grade insular glioma molecular pathology using diffusion tensor imaging metric-based histogram parameters. Front. Oncol. 2021, 11, 627202. [Google Scholar] [CrossRef] [PubMed]
- Aliotta, E.; Nourzadeh, H.; Batchala, P.; Schiff, D.; Lopes, M.; Druzgal, J.; Mukherjee, S.; Patel, S. Molecular subtype classification in lower-grade glioma with accelerated DTI. Am. J. Neuroradiol. 2019, 40, 1458–1463. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Han, K.; Ahn, S.; Choi, Y.; Chang, J.; Kim, S.; Kang, S.-G.; Kim, E.; Lee, S.-K. Whole-tumor histogram and texture analyses of DTI for evaluation of IDH1-mutation and 1p/19q-codeletion status in World Health Organization grade II gliomas. Am. J. Neuroradiol. 2018, 39, 693–698. [Google Scholar] [CrossRef]
- Chu, J.-P.; Song, Y.-K.; Tian, Y.-S.; Qiu, H.-S.; Huang, X.-H.; Wang, Y.-L.; Huang, Y.-Q.; Zhao, J. Diffusion kurtosis imaging in evaluating gliomas: Different region of interest selection methods on time efficiency, measurement repeatability, and diagnostic ability. Eur. Radiol. 2021, 31, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Choi, S.H.; Kim, Y.-J.; Kim, K.G.; Sohn, C.-H.; Kim, J.-H.; Yun, T.J.; Chang, K.-H. Gliomas: Histogram analysis of apparent diffusion coefficient maps with standard-or high-b-value diffusion-weighted MR imaging—Correlation with tumor grade. Radiology 2011, 261, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, W. Quantitative evaluation of diffusion tensor imaging for clinical management of glioma. Neurosurg. Rev. 2020, 43, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Bisdas, S.; Shen, H.; Thust, S.; Katsaros, V.; Stranjalis, G.; Boskos, C.; Brandner, S.; Zhang, J. Texture analysis-and support vector machine-assisted diffusional kurtosis imaging may allow in vivo gliomas grading and IDH-mutation status prediction: A preliminary study. Sci. Rep. 2018, 8, 6108. [Google Scholar] [CrossRef]
- Zhang, H.; Schneider, T.; Wheeler-Kingshott, C.A.; Alexander, D.C. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012, 61, 1000–1016. [Google Scholar] [CrossRef]
- Figini, M.; Riva, M.; Graham, M.; Castelli, G.M.; Fernandes, B.; Grimaldi, M.; Baselli, G.; Pessina, F.; Bello, L.; Zhang, H.; et al. Prediction of isocitrate dehydrogenase genotype in brain gliomas with MRI: Single-shell versus multishell diffusion models. Radiology 2018, 289, 788–796. [Google Scholar] [CrossRef]
- Jiang, S.; Zanazzi, G.J.; Hassanpour, S. Predicting prognosis and IDH mutation status for patients with lower-grade gliomas using whole slide images. Sci. Rep. 2021, 11, 16849. [Google Scholar] [CrossRef]
- Pei, L.; Jones, K.A.; Shboul, Z.A.; Chen, J.Y.; Iftekharuddin, K.M. Deep neural network analysis of pathology images with integrated molecular data for enhanced glioma classification and grading. Front. Oncol. 2021, 11, 668694. [Google Scholar] [CrossRef]
- Ali, M.B.; Gu, I.Y.-H.; Berger, M.S.; Pallud, J.; Southwell, D.; Widhalm, G.; Roux, A.; Vecchio, T.G.; Jakola, A.S. Domain mapping and deep learning from multiple MRI clinical datasets for prediction of molecular subtypes in low grade gliomas. Brain Sci. 2020, 10, 463. [Google Scholar] [CrossRef]
- Li, Z.-C.; Yan, J.; Zhang, S.; Liang, C.; Lv, X.; Zou, Y.; Zhang, H.; Liang, D.; Zhang, Z.; Chen, Y. Glioma survival prediction from whole-brain MRI without tumor segmentation using deep attention network: A multicenter study. Eur. Radiol. 2022, 32, 5719–5729. [Google Scholar] [CrossRef]
- Pasquini, L.; Napolitano, A.; Tagliente, E.; Dellepiane, F.; Lucignani, M.; Vidiri, A.; Ranazzi, G.; Stoppacciaro, A.; Moltoni, G.; Nicolai, M.; et al. Deep learning can differentiate IDH-mutant from IDH-wild GBM. J. Pers. Med. 2021, 11, 290. [Google Scholar] [CrossRef]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Nandoe Tewarie, R.; Lycklama, G.J.; De Witt Hamer, P.C.; Eijgelaar, R.S.; et al. Combined molecular subtyping, grading, and segmentation of glioma using multi-task deep learning. Neuro-Oncology 2022. [Google Scholar] [CrossRef]
- Lu, Z.; Bai, Y.; Chen, Y.; Su, C.; Lu, S.; Zhan, T.; Hong, X.; Wang, S. The classification of gliomas based on a pyramid dilated convolution resnet model. Pattern Recognit. Lett. 2020, 133, 173–179. [Google Scholar] [CrossRef]
- Gao, Y.; Xiao, X.; Han, B.; Li, G.; Ning, X.; Wang, D.; Cai, W.; Kikinis, R.; Berkovsky, S.; Di Ieva, A.; et al. Deep learning methodology for differentiating glioma recurrence from radiation necrosis using multimodal magnetic resonance imaging: Algorithm development and validation. JMIR Med. Inform. 2020, 8, e19805. [Google Scholar] [CrossRef]
- Liu, S.; Shah, Z.; Sav, A.; Russo, C.; Berkovsky, S.; Qian, Y.; Coiera, E.; Di Ieva, A. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep. 2020, 10, 7733. [Google Scholar] [CrossRef]
- Zeineldin, R.A.; Karar, M.E.; Coburger, J.; Wirtz, C.R.; Burgert, O. DeepSeg: Deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 909–920. [Google Scholar] [CrossRef]
- Matsui, Y.; Maruyama, T.; Nitta, M.; Saito, T.; Tsuzuki, S.; Tamura, M.; Kusuda, K.; Fukuya, Y.; Asano, H.; Kawamata, T.; et al. Prediction of lower-grade glioma molecular subtypes using deep learning. J. Neuro-Oncol. 2020, 146, 321–327. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A. Residual Convolutional Neural Network for the Determination of IDH Status in Low-and High-Grade Gliomas from MR ImagingNeural Network for Determination of IDH Status in Gliomas. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Cluceru, J.; Interian, Y.; Phillips, J.J.; Molinaro, A.M.; Luks, T.L.; Alcaide-Leon, P.; Olson, M.P.; Nair, D.; LaFontaine, M.; Shai, A.; et al. Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion-weighted imaging. Neuro-Oncology 2022, 24, 639–652. [Google Scholar] [CrossRef]
- Chicco, D.; Tötsch, N.; Jurman, G. The Matthews correlation coefficient (MCC) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. BioData Min. 2021, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Huang, W.; Yin, B.; Xiong, J.; Wu, J.; Geng, D. Can diffusion tensor imaging noninvasively detect IDH1 gene mutations in astrogliomas? A retrospective study of 112 cases. Am. J. Neuroradiol. 2014, 35, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Tan, W.L.; Pan, J.W.; Wang, Y.; Yin, B.; Zhang, J.; Geng, D.Y. Detecting isocitrate dehydrogenase gene mutations in oligodendroglial tumors using diffusion tensor imaging metrics and their correlations with proliferation and microvascular density. J. Magn. Reson. Imaging 2016, 43, 45–54. [Google Scholar] [CrossRef]
- Xiong, J.; Tan, W.; Wen, J.; Pan, J.; Wang, Y.; Zhang, J.; Geng, D. Combination of diffusion tensor imaging and conventional MRI correlates with isocitrate dehydrogenase 1/2 mutations but not 1p/19q genotyping in oligodendroglial tumours. Eur. Radiol. 2016, 26, 1705–1715. [Google Scholar] [CrossRef]
- Maynard, J.; Okuchi, S.; Wastling, S.; Busaidi, A.A.; Almossawi, O.; Mbatha, W.; Brandner, S.; Jaunmuktane, Z.; Koc, A.M.; Mancini, L.; et al. World Health Organization grade II/III glioma molecular status: Prediction by MRI morphologic features and apparent diffusion coefficient. Radiology 2020, 296, 111–121. [Google Scholar] [CrossRef]
- Gao, A.; Zhang, H.; Yan, X.; Wang, S.; Chen, Q.; Gao, E.; Qi, J.; Bai, J.; Zhang, Y.; Cheng, J. Whole-Tumor Histogram Analysis of Multiple Diffusion Metrics for Glioma Genotyping. Radiology 2022, 302, 652–661. [Google Scholar] [CrossRef]
- Aliotta, E.; Dutta, S.W.; Feng, X.; Tustison, N.J.; Batchala, P.P.; Schiff, D.; Lopes, M.B.; Jain, R.; Druzgal, T.J.; Mukherjee, S.; et al. Automated apparent diffusion coefficient analysis for genotype prediction in lower grade glioma: Association with the T2-FLAIR mismatch sign. J. Neuro-Oncol. 2020, 149, 325–335. [Google Scholar] [CrossRef]
- Wu, C.-C.; Jain, R.; Radmanesh, A.; Poisson, L.M.; Guo, W.-Y.; Zagzag, D.; Snuderl, M.; Placantonakis, D.; Golfinos, J.; Chi, A. Predicting genotype and survival in glioma using standard clinical MR imaging apparent diffusion coefficient images: A pilot study from the cancer genome atlas. Am. J. Neuroradiol. 2018, 39, 1814–1820. [Google Scholar] [CrossRef]
- Du, N.; Zhou, X.; Mao, R.; Shu, W.; Xiao, L.; Ye, Y.; Xu, X.; Shen, Y.; Lin, G.; Fang, X.; et al. Preoperative and Noninvasive Prediction of Gliomas Histopathological Grades and IDH Molecular Types Using Multiple MRI Characteristics. Front. Oncol. 2022, 12, 873839. [Google Scholar] [CrossRef]
- Xie, Y.; Li, S.; Shen, N.; Gan, T.; Zhang, S.; Liu, W.V.; Zhu, W. Assessment of Isocitrate Dehydrogenase 1 Genotype and Cell Proliferation in Gliomas Using Multiple Diffusion Magnetic Resonance Imaging. Front. Neurosci. 2021, 15, 783361. [Google Scholar] [CrossRef]
- Thust, S.; Maynard, J.; Benenati, M.; Wastling, S.; Mancini, L.; Jaunmuktane, Z.; Brandner, S.; Jäger, H. Regional and volumetric parameters for diffusion-weighted WHO Grade II and III glioma genotyping: A method comparison. Am. J. Neuroradiol. 2021, 42, 441–447. [Google Scholar] [CrossRef]
- Guo, H.; Liu, J.; Hu, J.; Zhang, H.; Zhao, W.; Gao, M.; Zhang, Y.; Yang, G.; Cui, Y. Diagnostic performance of gliomas grading and IDH status decoding A comparison between 3D amide proton transfer APT and four diffusion-weighted MRI models. J. Magn. Reson. Imaging 2022, 56, 1834–1844. [Google Scholar] [CrossRef]
- Zhou, H.; Chang, K.; Bai, H.X.; Xiao, B.; Su, C.; Bi, W.L.; Zhang, P.J.; Senders, J.T.; Vallières, M.; Kavouridis, V.K.; et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low-and high-grade gliomas. J. Neuro-Oncol. 2019, 142, 299–307. [Google Scholar] [CrossRef]
- Han, Y.; Xie, Z.; Zang, Y.; Zhang, S.; Gu, D.; Zhou, M.; Gevaert, O.; Wei, J.; Li, C.; Chen, H.; et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J. Neuro-Oncol. 2018, 140, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tian, Q.; Wang, L.; Liu, Y.; Li, B.; Liang, Z.; Gao, P.; Zheng, K.; Zhao, B.; Lu, H. Radiomics strategy for molecular subtype stratification of lower-grade glioma: Detecting IDH and TP53 mutations based on multimodal MRI. J. Magn. Reson. Imaging 2018, 48, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Kihira, S.; Tsankova, N.M.; Bauer, A.; Sakai, Y.; Mahmoudi, K.; Zubizarreta, N.; Houldsworth, J.; Khan, F.; Salamon, N.; Hormigo, A.; et al. Multiparametric MRI texture analysis in prediction of glioma biomarker status: Added value of MR diffusion. Neuro-Oncol. Adv. 2021, 3, vdab051. [Google Scholar] [CrossRef]
- He, J.; Ren, J.; Niu, G.; Liu, A.; Wu, Q.; Xie, S.; Ma, X.; Li, B.; Wang, P.; Shen, J.; et al. Multiparametric MR radiomics in brain glioma: Models comparation to predict biomarker status. BMC Med. Imaging 2022, 22, 137. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion-and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur. Radiol. 2020, 30, 2142–2151. [Google Scholar] [CrossRef]
- Park, C.J.; Choi, Y.S.; Park, Y.W.; Ahn, S.S.; Kang, S.-G.; Chang, J.-H.; Kim, S.H.; Lee, S.-K. Diffusion tensor imaging radiomics in lower-grade glioma: Improving subtyping of isocitrate dehydrogenase mutation status. Neuroradiology 2020, 62, 319–326. [Google Scholar] [CrossRef]
- Tan, Y.; Mu, W.; Wang, X.-C.; Yang, G.-Q.; Gillies, R.J.; Zhang, H. Whole-tumor radiomics analysis of DKI and DTI may improve the prediction of genotypes for astrocytomas: A preliminary study. Eur. J. Radiol. 2020, 124, 108785. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, S.-T.; Wei, J.-W.; Dong, D.; Wang, X.-C.; Yang, G.-Q.; Tian, J.; Zhang, H. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur. Radiol. 2019, 29, 3325–3337. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, X.; Rui, W.; Sheng, Y.; Yu, Y.; Zhang, Y.; Yao, Z.; Qiu, T.; Ren, Y. A nomogram strategy for identifying the subclassification of IDH mutation and ATRX expression loss in lower-grade gliomas. Eur. Radiol. 2022, 32, 3187–3198. [Google Scholar] [CrossRef]
- Shen, N.; Zhao, L.; Jiang, J.; Jiang, R.; Su, C.; Zhang, S.; Tang, X.; Zhu, W. Intravoxel incoherent motion diffusion-weighted imaging analysis of diffusion and microperfusion in grading gliomas and comparison with arterial spin labeling for evaluation of tumor perfusion. J. Magn. Reson. Imaging 2016, 44, 620–632. [Google Scholar] [CrossRef]
- Panagiotaki, E.; Walker-Samuel, S.; Siow, B.; Johnson, S.P.; Rajkumar, V.; Pedley, R.B.; Lythgoe, M.F.; Alexander, D.C. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res. 2014, 74, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Szczepankiewicz, F.; van Westen, D.; Englund, E.; Westin, C.-F.; Ståhlberg, F.; Lätt, J.; Sundgren, P.C.; Nilsson, M. The link between diffusion MRI and tumor heterogeneity: Mapping cell eccentricity and density by diffusional variance decomposition (DIVIDE). Neuroimage 2016, 142, 522–532. [Google Scholar] [CrossRef] [PubMed]
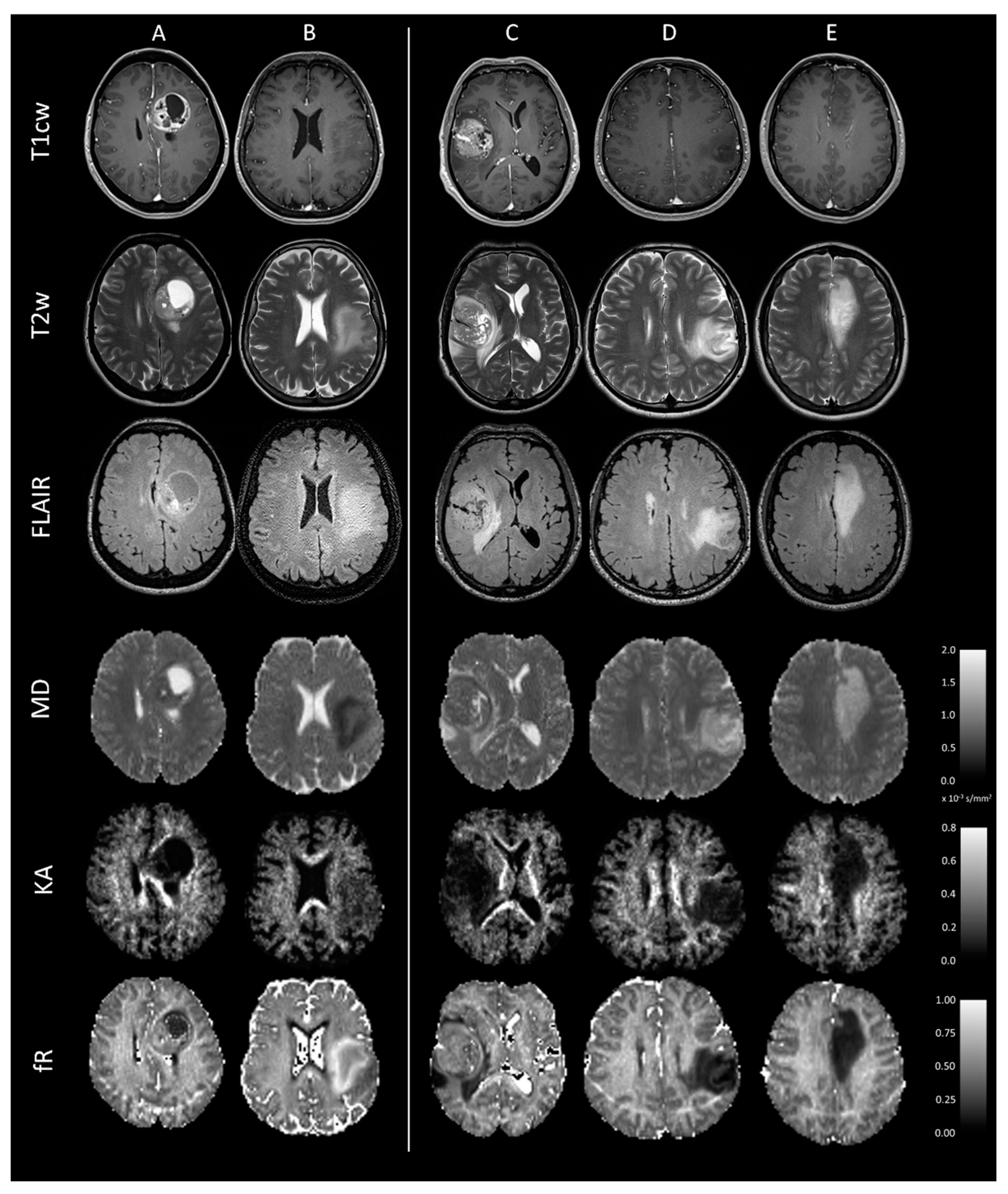

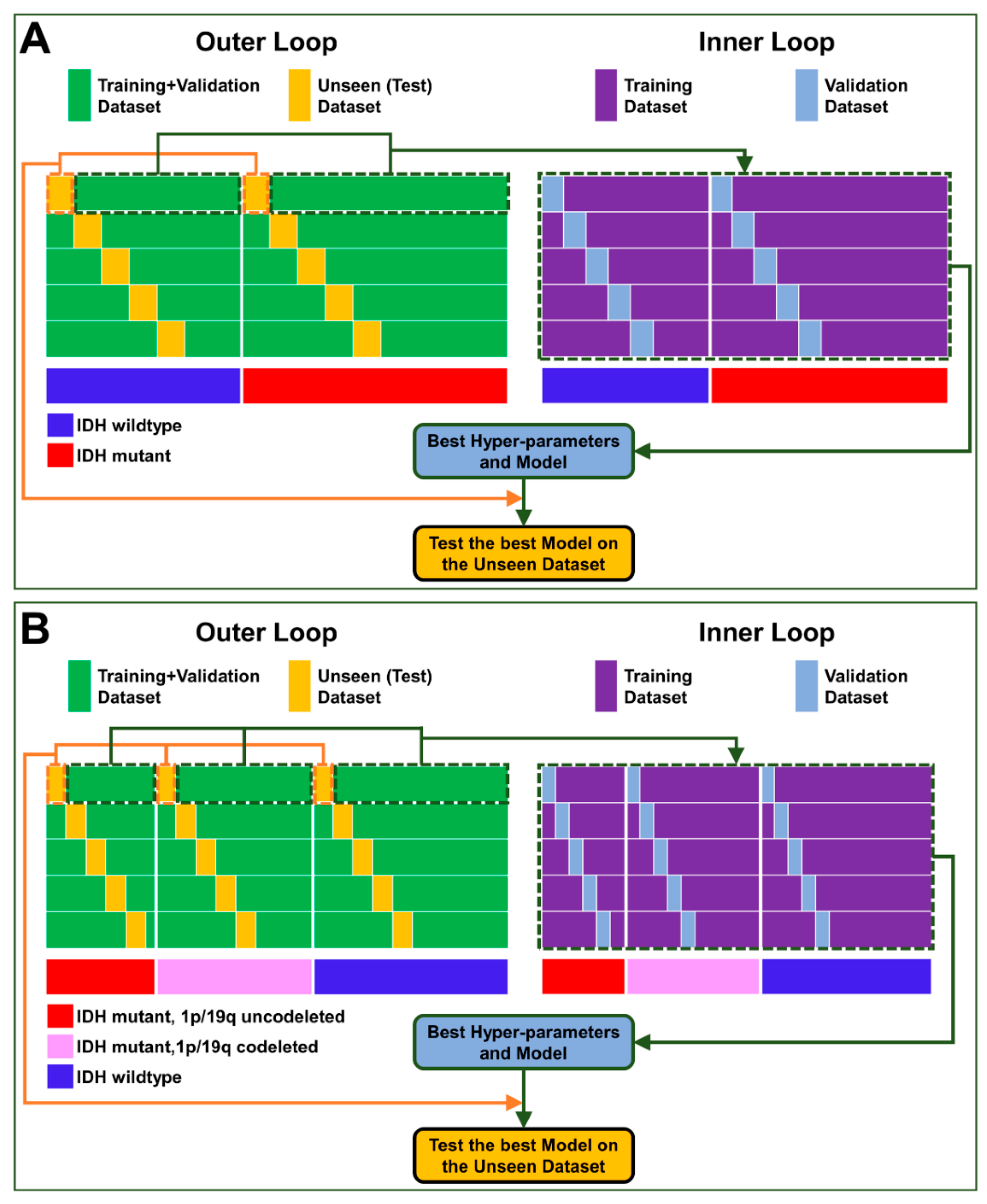

| Clinical Characteristics | Values |
|---|---|
| Number of subjects, N (%) | 146 (100) |
| Age, mean (range) years | 46.2 (14–77) |
| Sex | |
| Male, N (%) | 88 (60) |
| Female, N (%) | 58 (40) |
| Hemisphere | |
| Left, N (%) | 95 (65) |
| Right, N (%) | 45 (31) |
| Bilateral, N (%) | 6 (4) |
| WHO grade | |
| 2, N (%) | 50 (34) |
| 3, N (%) | 50 (34) |
| 4, N (%) | 46 (32) |
| Molecular subtype | |
| Grade 2–3 diffuse astrocytoma, IDH-mutant, N (%) | 28 (19.2) |
| Oligodendroglioma, IDH-mutant, 1p/19q codeleted, N (%) | 50 (34.2) |
| Molecular glioblastoma, IDH-wildtype, N (%) | 22 (15) |
| Grade 4 diffuse astrocytoma, IDH-mutant, N (%) | 6 (4.1) |
| Glioblastoma, IDH-wildtype, N (%) | 40 (27.5) |
| Target Population | Classification Task | Training 1 | Validation 1 | Test 2 |
|---|---|---|---|---|
| Only patients with grade 2 and 3 gliomas (n = 100) | IDH-mutant (n = 78) vs. IDH-wildtype (n = 22) | n = 72 vs. n = 16 | n = 2 vs. n = 2 | n = 4 vs. n = 4 |
| 1p/19q-codeleted (n = 50) vs. 1p/19q-uncodeleted (n = 50) | n = 41 vs. n = 41 | n = 3 vs. n = 3 | n = 6 vs. n = 6 | |
| Patients with grade 2, 3, and 4 gliomas (n = 146) | IDH-mutant (n = 84) vs. IDH-wildtype (n = 62) | n = 71 vs. n = 49 | n = 4 vs. n = 4 | n = 9 vs. n = 9 |
| IDH-mutant and 1p/19q-codeleted (n = 50) vs. IDH-mutant and 1p/19q-uncodeleted (n = 34) vs. IDH-wildtype (n = 62) | n = 40 vs. n = 24 vs. n = 52 | n = 4 vs. n = 4 vs. n = 4 | n = 6 vs. n = 6 vs. n = 6 |
| Classification Metrics | cMRI | Multi-Shell dMRI | cMRI and Multi-Shell dMRI |
|---|---|---|---|
| Overall performance | |||
| Accuracy | 70% ± 7% | 70% ± 7% | 75% ± 9% |
| Precision 1 | 0.81 ± 0.03 | 0.78 ± 0.03 | 0.82 ± 0.06 |
| MCC | 0.50 ± 0.11 | 0.47 ± 0.09 | 0.56 ± 0.14 |
| Individual group sensitivity | |||
| IDH mutant | 100% ± 0% | 75% ± 31% | 95% ± 11% |
| IDH wildtype | 40% ± 14% | 65% ± 28% | 55% ± 21% |
| Classification Metrics | cMRI | Multi-Shell dMRI | cMRI and Multi-Shell dMRI |
|---|---|---|---|
| Overall performance | |||
| Accuracy | 65% ± 6% | 66% ± 9% | 72% ± 4% |
| Precision 1 | 0.66 ± 0.06 | 0.73 ± 0.10 | 0.75 ± 0.05 |
| MCC | 0.30 ± 0.10 | 0.38 ± 0.20 | 0.45 ± 0.08 |
| Individual group sensitivity | |||
| 1p/19q codeleted | 70% ± 10% | 60% ± 10% | 60% ± 10% |
| 1p/19q uncodeleted | 61% ± 20% | 70% ± 20% | 83% ± 10% |
| Classification Metrics | cMRI | Multi-Shell dMRI | cMRI and Multi-Shell dMRI |
|---|---|---|---|
| Overall performance | |||
| Accuracy | 74 ± 5% | 73 ± 6% | 81 ± 5% |
| Precision 1 | 0.77 ± 0.04 | 0.77 ± 0.09 | 0.83 ± 0.04 |
| MCC | 0.52 ± 0.08 | 0.52 ± 0.18 | 0.64 ± 0.06 |
| Individual group sensitivity | |||
| IDH mutant | 84 ± 6% | 84 ± 6% | 89 ± 8% |
| IDH wildtype | 64 ± 9% | 62 ± 6% | 73 ± 13% |
| Classification Metrics | cMRI | Multi-Shell dMRI | cMRI and Multi-Shell dMRI |
|---|---|---|---|
| Overall performance | |||
| Accuracy | 57 ± 8% | 56 ± 7% | 60 ± 5% |
| Precision 1 | 0.60 ± 0.14 | 0.59 ± 0.07 | 0.65 ± 0.05 |
| MCC | 0.37 ± 0.14 | 0.35 ± 0.10 | 0.43 ± 0.06 |
| Individual group sensitivity | |||
| IDH-mutant 1p/19q-codeleted | 60 ± 19% | 53 ± 18% | 63 ± 18% |
| IDH-mutant 1p/19q-uncodeleted | 37 ± 22% | 43 ± 9% | 43 ± 22% |
| IDH wildtype | 73 ± 19% | 70 ± 7% | 73 ± 15% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karami, G.; Pascuzzo, R.; Figini, M.; Del Gratta, C.; Zhang, H.; Bizzi, A. Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning. Cancers 2023, 15, 482. https://doi.org/10.3390/cancers15020482
Karami G, Pascuzzo R, Figini M, Del Gratta C, Zhang H, Bizzi A. Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning. Cancers. 2023; 15(2):482. https://doi.org/10.3390/cancers15020482
Chicago/Turabian StyleKarami, Golestan, Riccardo Pascuzzo, Matteo Figini, Cosimo Del Gratta, Hui Zhang, and Alberto Bizzi. 2023. "Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning" Cancers 15, no. 2: 482. https://doi.org/10.3390/cancers15020482
APA StyleKarami, G., Pascuzzo, R., Figini, M., Del Gratta, C., Zhang, H., & Bizzi, A. (2023). Combining Multi-Shell Diffusion with Conventional MRI Improves Molecular Diagnosis of Diffuse Gliomas with Deep Learning. Cancers, 15(2), 482. https://doi.org/10.3390/cancers15020482







