Development of Radiomic-Based Model to Predict Clinical Outcomes in Non-Small Cell Lung Cancer Patients Treated with Immunotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Image Acquisition, Segmentation, Pre-Processing and Feature Extraction
2.3. Patient Outcomes
2.3.1. Follow-Up and Response Assessment
2.3.2. Survival and Durable Clinical Benefit (DCB)
2.4. Statistical Analysis
3. Results
3.1. Patients Characteristics
3.2. Patient Outcomes
3.3. Radiomics and Prediction of Progression, DCB and Survival
3.3.1. Univariate Analysis
At Baseline (PET/CT0)
At Month 2 (PET/CT1)
Delta-Radiomics
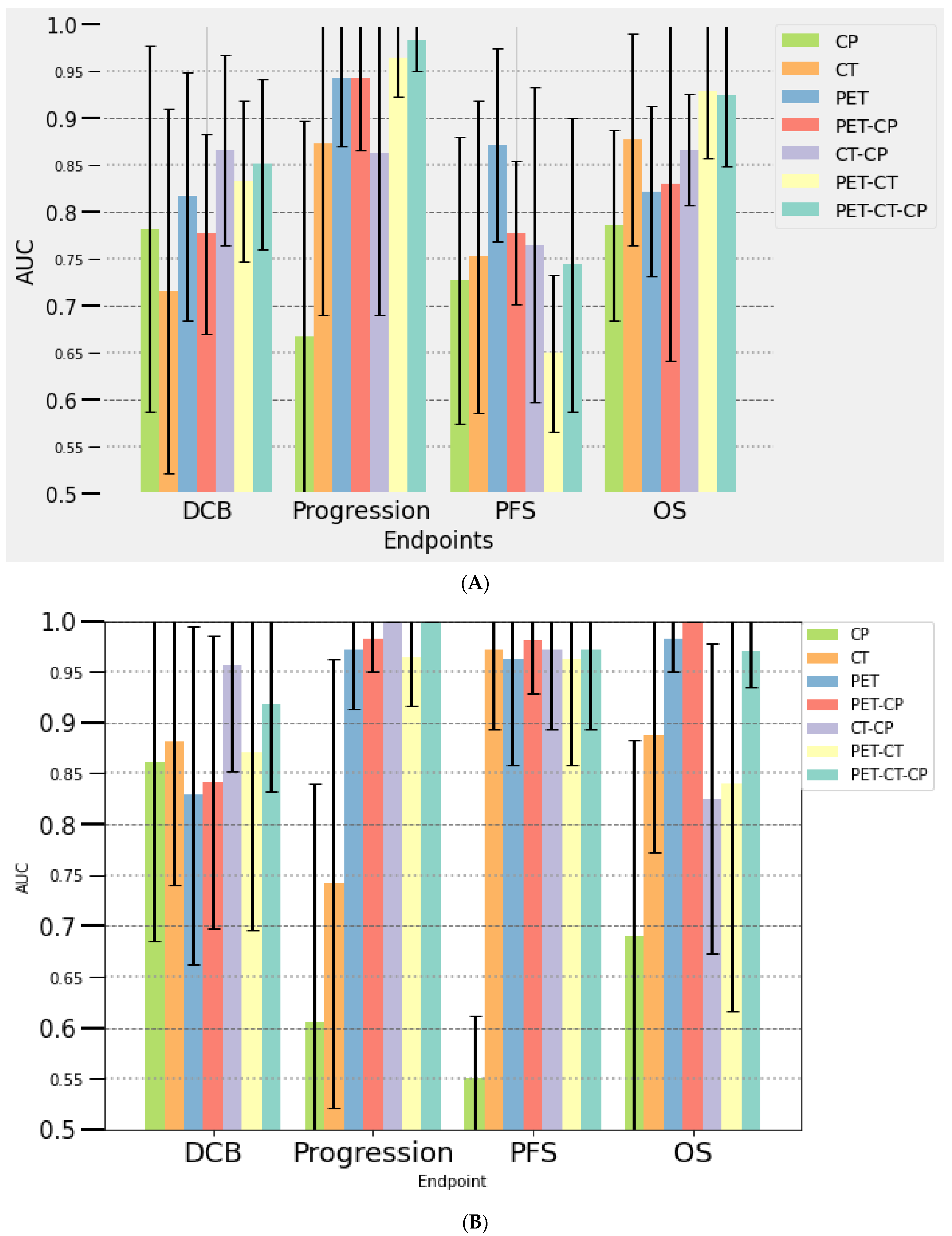
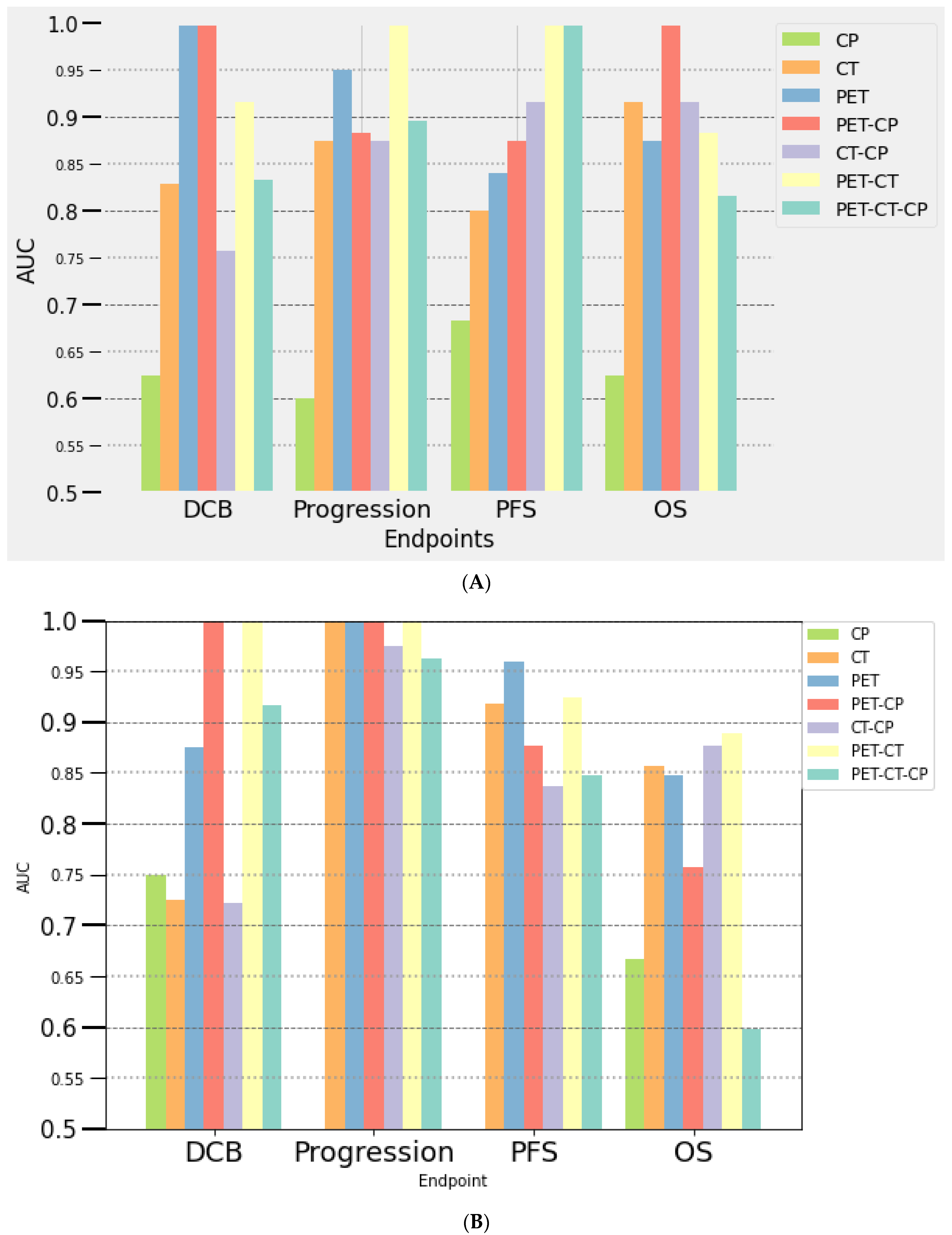
3.3.2. Multivariate Analysis
At Baseline (PET/CT0)
At Month 2 (PET/CT1)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Ahn, M.J.; Girard, N.; Johnson, M.; Kim, D.W.; Lopes, G.; Pillai, R.N.; Solomon, B.; Villacampa, G.; Zhou, Q. Advanced-Stage Non-Small Cell Lung Cancer: Advances in Thoracic Oncology 2018. J. Thorac. Oncol. 2019, 14, 1134–1155. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Champiat, S.; Dercle, L.; Ammari, S.; Massard, C.; Hollebecque, A.; Postel-Vinay, S.; Chaput, N.; Eggermont, A.; Marabelle, A.; Soria, J.C.; et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clin. Cancer Res. 2017, 23, 1920–1928. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crino, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Updated Analysis of KEYNOTE-024: Pembrolizumab Versus Platinum-Based Chemotherapy for Advanced Non-Small-Cell Lung Cancer With PD-L1 Tumor Proportion Score of 50% or Greater. J. Clin. Oncol. 2019, 37, 537–546. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, L.; Cai, Z.; Jiang, W.; Li, J.; Yang, C.; Yu, C.; Jiang, B.; Wang, W.; Xu, W.; et al. Potential feature exploration and model development based on 18F-FDG PET/CT images for differentiating benign and malignant lung lesions. Eur. J. Radiol. 2019, 121, 108735. [Google Scholar] [CrossRef]
- Kim, S.K.; Allen-Auerbach, M.; Goldin, J.; Fueger, B.J.; Dahlbom, M.; Brown, M.; Czernin, J.; Schiepers, C. Accuracy of PET/CT in characterization of solitary pulmonary lesions. J. Nucl. Med. 2007, 48, 214–220. [Google Scholar] [PubMed]
- Balagurunathan, Y.; Schabath, M.B.; Wang, H.; Liu, Y.; Gillies, R.J. Quantitative Imaging features Improve Discrimination of Malignancy in Pulmonary nodules. Sci. Rep. 2019, 9, 8528. [Google Scholar] [CrossRef] [PubMed]
- Hyun, S.H.; Ahn, M.S.; Koh, Y.W.; Lee, S.J. A Machine-Learning Approach Using PET-Based Radiomics to Predict the Histological Subtypes of Lung Cancer. Clin. Nucl. Med. 2019, 44, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, M.; Cozzi, L.; Antunovic, L.; Lozza, L.; Fogliata, A.; Voulaz, E.; Rossi, A.; Chiti, A.; Sollini, M. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 207–217. [Google Scholar] [CrossRef]
- Dissaux, G.; Visvikis, D.; Da-Ano, R.; Pradier, O.; Chajon, E.; Barillot, I.; Duverge, L.; Masson, I.; Abgral, R.; Santiago Ribeiro, M.J.; et al. Pretreatment (18)F-FDG PET/CT Radiomics Predict Local Recurrence in Patients Treated with Stereotactic Body Radiotherapy for Early-Stage Non-Small Cell Lung Cancer: A Multicentric Study. J. Nucl. Med. 2020, 61, 814–820. [Google Scholar] [CrossRef]
- Baek, S.; He, Y.; Allen, B.G.; Buatti, J.; Smith, B.; Tong, L.; Sun, Z.; Wu, J.; Diehn, M.; Loo, B.; et al. Deep segmentation networks predict survival of non-small cell lung cancer. Sci. Rep. 2019, 9, 1286. [Google Scholar] [CrossRef]
- Khorrami, M.; Prasanna, P.; Gupta, A.; Patil, P.; Velu, P.D.; Thawani, R.; Corredor, G.; Alilou, M.; Bera, K.; Fu, P.; et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 108–119. [Google Scholar] [CrossRef]
- Barabino, E.; Rossi, G.; Pamparino, S.; Fiannacca, M.; Caprioli, S.; Fedeli, A.; Zullo, L.; Vagge, S.; Cittadini, G.; Genova, C. Exploring Response to Immunotherapy in Non-Small Cell Lung Cancer Using Delta-Radiomics. Cancers 2022, 14, 350. [Google Scholar] [CrossRef]
- Mu, W.; Tunali, I.; Gray, J.E.; Qi, J.; Schabath, M.B.; Gillies, R.J. Radiomics of (18)F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1168–1182. [Google Scholar] [CrossRef]
- Detterbeck, F.C.; Boffa, D.J.; Kim, A.W.; Tanoue, L.T. The Eighth Edition Lung Cancer Stage Classification. Chest 2017, 151, 193–203. [Google Scholar] [CrossRef]
- Grootjans, W.; Usmanij, E.; Oyen, W.; van der Heijden, E.; Visser, E.; Visvikis, D.; Hatt, M.; Bussink, J.; de Geus-Oei, L. Performance of automatic image segmentation algorithms for calculating total lesion glycolysis for early response monitoring in non-small cell lung cancer patients during concomitant chemoradiotherapy. Radiother. Oncol. 2016, 119, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Hatt, M.; Cheze Le Rest, C.; Albarghach, N.; Pradier, O.; Visvikis, D. PET functional volume delineation: A robustness and repeatability study. Eur. J. Nucl. Med. Mol. Imaging 2011, 38, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Hatt, M.; Laurent, B.; Ouahabi, A.; Fayad, H.; Tan, S.; Li, L.; Lu, W.; Jaouen, V.; Tauber, C.; Czakon, J.; et al. The first MICCAI challenge on PET tumor segmentation. Med. Image Anal. 2018, 44, 177–195. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Wahl, R.L.; Jacene, H.; Kasamon, Y.; Lodge, M.A. From RECIST to PERCIST: Evolving Considerations for PET response criteria in solid tumors. J. Nucl. Med. 2009, 50 (Suppl. 1), 122S–150S. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, L.; Duchemann, B.; Chouahnia, K.; Zelek, L.; Soussan, M. Monitoring anti-PD-1-based immunotherapy in non-small cell lung cancer with FDG PET: Introduction of iPERCIST. EJNMMI Res. 2019, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Humbert, O.; Cadour, N.; Paquet, M.; Schiappa, R.; Poudenx, M.; Chardin, D.; Borchiellini, D.; Benisvy, D.; Ouvrier, M.J.; Zwarthoed, C.; et al. (18)FDG PET/CT in the early assessment of non-small cell lung cancer response to immunotherapy: Frequency and clinical significance of atypical evolutive patterns. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Tazdait, M.; Mezquita, L.; Lahmar, J.; Ferrara, R.; Bidault, F.; Ammari, S.; Balleyguier, C.; Planchard, D.; Gazzah, A.; Soria, J.C.; et al. Patterns of responses in metastatic NSCLC during PD-1 or PDL-1 inhibitor therapy: Comparison of RECIST 1.1, irRECIST and iRECIST criteria. Eur. J. Cancer 2018, 88, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Genova, C.; Bassanelli, M.; De Giglio, A.; Brambilla, M.; Metro, G.; Baglivo, S.; Dal Bello, M.G.; Ceribelli, A.; Grossi, F.; et al. Safety and Efficacy of Nivolumab in Patients With Advanced Non-small-cell Lung Cancer Treated Beyond Progression. Clin. Lung Cancer 2019, 20, 178–185 e172. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Toschi, L.; Rossi, S.; Mazziotti, E.; Lopci, E. The immune-metabolic-prognostic index and clinical outcomes in patients with non-small cell lung carcinoma under checkpoint inhibitors. J. Cancer Res. Clin. Oncol. 2020, 146, 1235–1243. [Google Scholar] [CrossRef]
- Seban, R.D.; Mezquita, L.; Berenbaum, A.; Dercle, L.; Botticella, A.; Le Pechoux, C.; Caramella, C.; Deutsch, E.; Grimaldi, S.; Adam, J.; et al. Baseline metabolic tumor burden on FDG PET/CT scans predicts outcome in advanced NSCLC patients treated with immune checkpoint inhibitors. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1147–1157. [Google Scholar] [CrossRef]
- Nardone, V.; Tini, P.; Pastina, P.; Botta, C.; Reginelli, A.; Carbone, S.F.; Giannicola, R.; Calabrese, G.; Tebala, C.; Guida, C.; et al. Radiomics predicts survival of patients with advanced non-small cell lung cancer undergoing PD-1 blockade using Nivolumab. Oncol. Lett. 2020, 19, 1559–1566. [Google Scholar] [CrossRef]
- Zhao, W.; Jiang, W.; Wang, H.; He, J.; Su, C.; Yu, Q. Impact of Smoking History on Response to Immunotherapy in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 703143. [Google Scholar] [CrossRef]
- Polverari, G.; Ceci, F.; Bertaglia, V.; Reale, M.L.; Rampado, O.; Gallio, E.; Passera, R.; Liberini, V.; Scapoli, P.; Arena, V.; et al. (18)F-FDG Pet Parameters and Radiomics Features Analysis in Advanced Nsclc Treated with Immunotherapy as Predictors of Therapy Response and Survival. Cancers 2020, 12, 1163. [Google Scholar] [CrossRef]
- Kaira, K.; Higuchi, T.; Naruse, I.; Arisaka, Y.; Tokue, A.; Altan, B.; Suda, S.; Mogi, A.; Shimizu, K.; Sunaga, N.; et al. Metabolic activity by (18)F-FDG-PET/CT is predictive of early response after nivolumab in previously treated NSCLC. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 56–66. [Google Scholar] [CrossRef]
- Nishino, M.; Dahlberg, S.E.; Adeni, A.E.; Lydon, C.A.; Hatabu, H.; Janne, P.A.; Hodi, F.S.; Awad, M.M. Tumor Response Dynamics of Advanced Non-small Cell Lung Cancer Patients Treated with PD-1 Inhibitors: Imaging Markers for Treatment Outcome. Clin. Cancer Res. 2017, 23, 5737–5744. [Google Scholar] [CrossRef]
- Valentini, V.; Lambin, P.; Myerson, R.J. Is it time for tailored treatment of rectal cancer? From prescribing by consensus to prescribing by numbers. Radiother. Oncol. 2012, 102, 1–3. [Google Scholar] [CrossRef]
- Ahn, H.K.; Lee, H.; Kim, S.G.; Hyun, S.H. Pre-treatment (18)F-FDG PET-based radiomics predict survival in resected non-small cell lung cancer. Clin. Radiol. 2019, 74, 467–473. [Google Scholar] [CrossRef]
- Saeed-Vafa, D.; Bravo, R.; Dean, J.A.; El-Kenawi, A.; Mon Père, N.; Strobl, M.; Daniels, C.; Stringfield, O.; Damaghi, M.; Tunali, I.; et al. Combining radiomics and mathematical modeling to elucidate mechanisms of resistance to immune checkpoint blockade in non-small cell lung cancer. bioRxiv 2017. [Google Scholar] [CrossRef]
- Dercle, L.; Fronheiser, M.; Lu, L.; Du, S.; Hayes, W.; Leung, D.K.; Roy, A.; Wilkerson, J.; Guo, P.; Fojo, A.T.; et al. Identification of Non–Small Cell Lung Cancer Sensitive to Systemic Cancer Therapies Using Radiomics. Clin. Cancer Res. 2020, 26, 2151–2162. [Google Scholar] [CrossRef] [PubMed]
- Majdoub, M.; Hoeben, B.; Troost, E.G.C.; Oyen, W.; Kaanders, J.; Cheze Le Rest, C.; Visser, E.; Visvikis, D.; Hatt, M. Prognostic Value of Head and Neck Tumor Proliferative Sphericity From 3′-Deoxy-3′-[<sup>18</sup>F] Fluorothymidine Positron Emission Tomography. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 2, 33–40. [Google Scholar] [CrossRef]
- Grove, O.; Berglund, A.E.; Schabath, M.B.; Aerts, H.; Dekker, A.; Wang, H.; Velazquez, E.R.; Lambin, P.; Gu, Y.; Balagurunathan, Y.; et al. Correction: Quantitative Computed Tomographic Descriptors Associate Tumor Shape Complexity and Intratumor Heterogeneity with Prognosis in Lung Adenocarcinoma. PLoS ONE 2021, 16, e0248541. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallieres, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Grossmann, P.; Stringfield, O.; El-Hachem, N.; Bui, M.M.; Rios Velazquez, E.; Parmar, C.; Leijenaar, R.T.; Haibe-Kains, B.; Lambin, P.; Gillies, R.J.; et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017, 6, e23421. [Google Scholar] [CrossRef]
- Xu, C.J.; van der Schaaf, A.; Schilstra, C.; Langendijk, J.A.; van’t Veld, A.A. Impact of statistical learning methods on the predictive power of multivariate normal tissue complication probability models. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e677–e684. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedlander, M. The Elements of Statistical Learning; Springer: New York, NY, USA, 2009. [Google Scholar]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallieres, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Overall Population (n = 83) | Responders (n = 40) | Non-Responders (n = 31) | p Value |
|---|---|---|---|---|
| Age (years), median (range) | 63.5 (38–85) | 61.5 (47–82) | 65 (43–85) | 0.28 |
| Sex, n (%) | 0.78 | |||
| Men | 59 (71) | 31 (77) | 22 (71) | |
| Women | 24 (29) | 9 (23) | 9 (29) | |
| Smoking, n (%) | 0.09 | |||
| Smoker | 79 (95) | 40 (100) | 29 (94) | |
| Non-smoker | 4 (5) | 0 | 2 (6) | |
| ECOG PS, n (%) | 0.3 | |||
| 0 | 35 (42) | 19 (47,5) | 12 (39) | |
| 1 | 44 (53) | 19 (47,5) | 18 (58) | |
| ≥2 | 4 (5) | 2 (5) | 1 (3) | |
| Histology, n (%) | 0.13 | |||
| Adenocarcinoma | 48 (58) | 26 (65) | 15 | |
| Squamous cell carcinoma | 22 (26) | 8 (20) | 11 | |
| Other type | 13 (16) | 6 (15) | 5 | |
| PD-L1, n (%) | - | |||
| <1% | 20 (24) | 12 (30) | 9 (29) | |
| 1–49% | 18 (22) | 8 (20) | 2 (6) | |
| ≥50% | 15 (18) | 11 (28) | 3 (10) | |
| Not performed | 30 (36) | 9 (22) | 17 (55) | |
| Stage before immunotherapy, n (%) | 0.27 | |||
| IIB | 1 (1) | 1 (2) | 0 | |
| III | 11 (13) | 6 (15) | 3 (10) | |
| IV | 71 (86) | 33 (83) | 28 (90) | |
| Previous treatment, n (%) | ||||
| Surgery | 19 (23) | 11 (28) | 7 (23) | |
| Radiotherapy | 14 (17) | 10 (25) | 10 (32) | |
| Chemotherapy | 58 (70) | 22 (55) | 26 (84) | |
| Immunotherapy, n (%) | ||||
| First line | 23 (28) | 24 (60) | 26 (84) | |
| Second or >line | 60 (72) | 16 (40) | 5 (16) | |
| Nivolumab | 41 (49) | 13 (33) | 24 (78) | |
| Pembrolizumab | 37 (45) | 26 (65) | 6 (19) | |
| Atezolizumab | 5 (6) | 1 (2) | 1 (3) | |
| PFS, median (days) | 156 | 250 | 52 | <0.001 |
| DCB, n (%) | 35 (42) | 25 (63) | 3 (10) | <0.001 |
| OS, median (days) | 256 | 351 | 194 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tankyevych, O.; Trousset, F.; Latappy, C.; Berraho, M.; Dutilh, J.; Tasu, J.P.; Lamour, C.; Cheze Le Rest, C. Development of Radiomic-Based Model to Predict Clinical Outcomes in Non-Small Cell Lung Cancer Patients Treated with Immunotherapy. Cancers 2022, 14, 5931. https://doi.org/10.3390/cancers14235931
Tankyevych O, Trousset F, Latappy C, Berraho M, Dutilh J, Tasu JP, Lamour C, Cheze Le Rest C. Development of Radiomic-Based Model to Predict Clinical Outcomes in Non-Small Cell Lung Cancer Patients Treated with Immunotherapy. Cancers. 2022; 14(23):5931. https://doi.org/10.3390/cancers14235931
Chicago/Turabian StyleTankyevych, Olena, Flora Trousset, Claire Latappy, Moran Berraho, Julien Dutilh, Jean Pierre Tasu, Corinne Lamour, and Catherine Cheze Le Rest. 2022. "Development of Radiomic-Based Model to Predict Clinical Outcomes in Non-Small Cell Lung Cancer Patients Treated with Immunotherapy" Cancers 14, no. 23: 5931. https://doi.org/10.3390/cancers14235931
APA StyleTankyevych, O., Trousset, F., Latappy, C., Berraho, M., Dutilh, J., Tasu, J. P., Lamour, C., & Cheze Le Rest, C. (2022). Development of Radiomic-Based Model to Predict Clinical Outcomes in Non-Small Cell Lung Cancer Patients Treated with Immunotherapy. Cancers, 14(23), 5931. https://doi.org/10.3390/cancers14235931







