Laminin-332 γ2 Monomeric Chain Promotes Adhesion and Migration of Hepatocellular Carcinoma Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
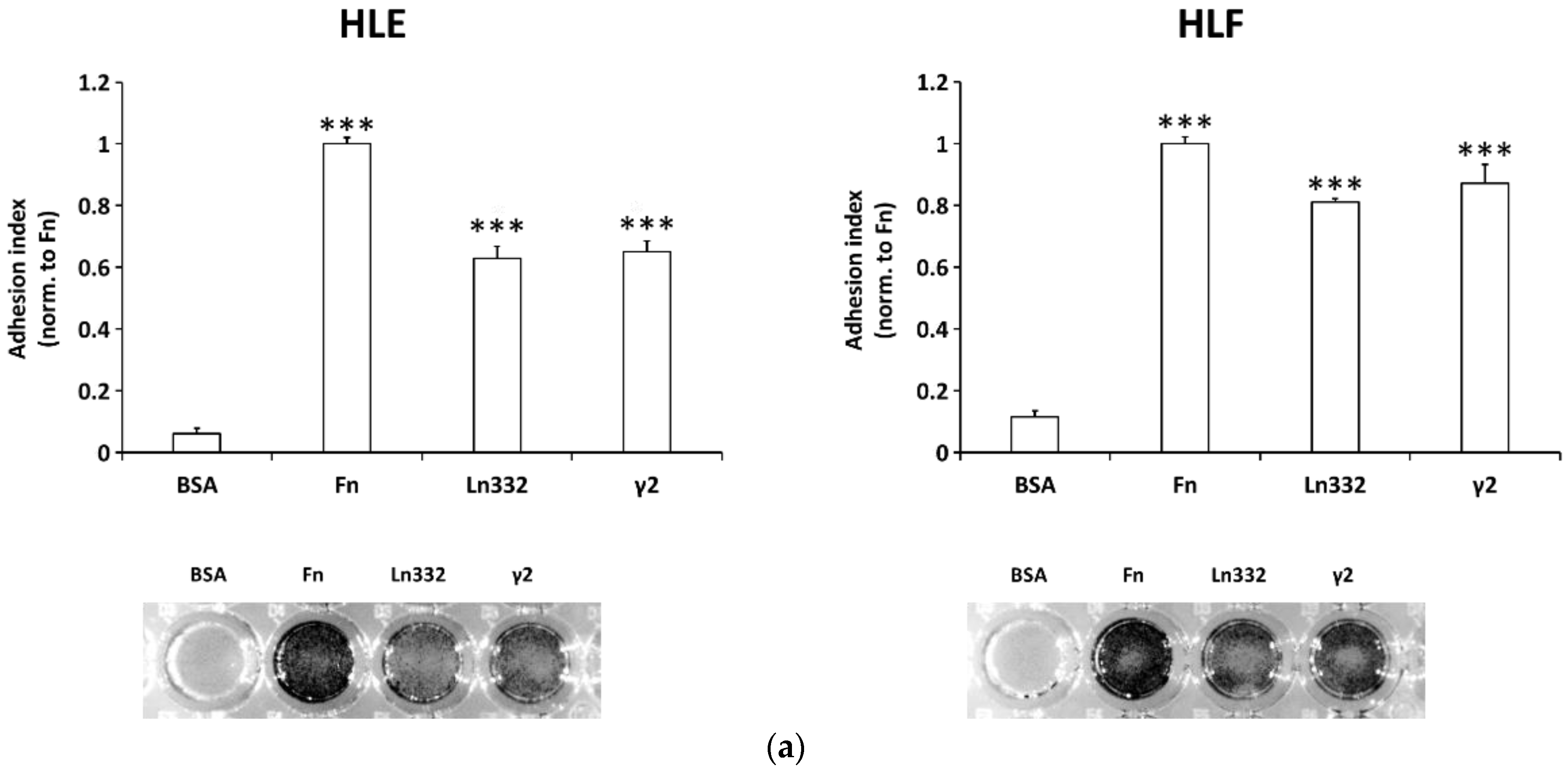
2.2. Adhesion Assay
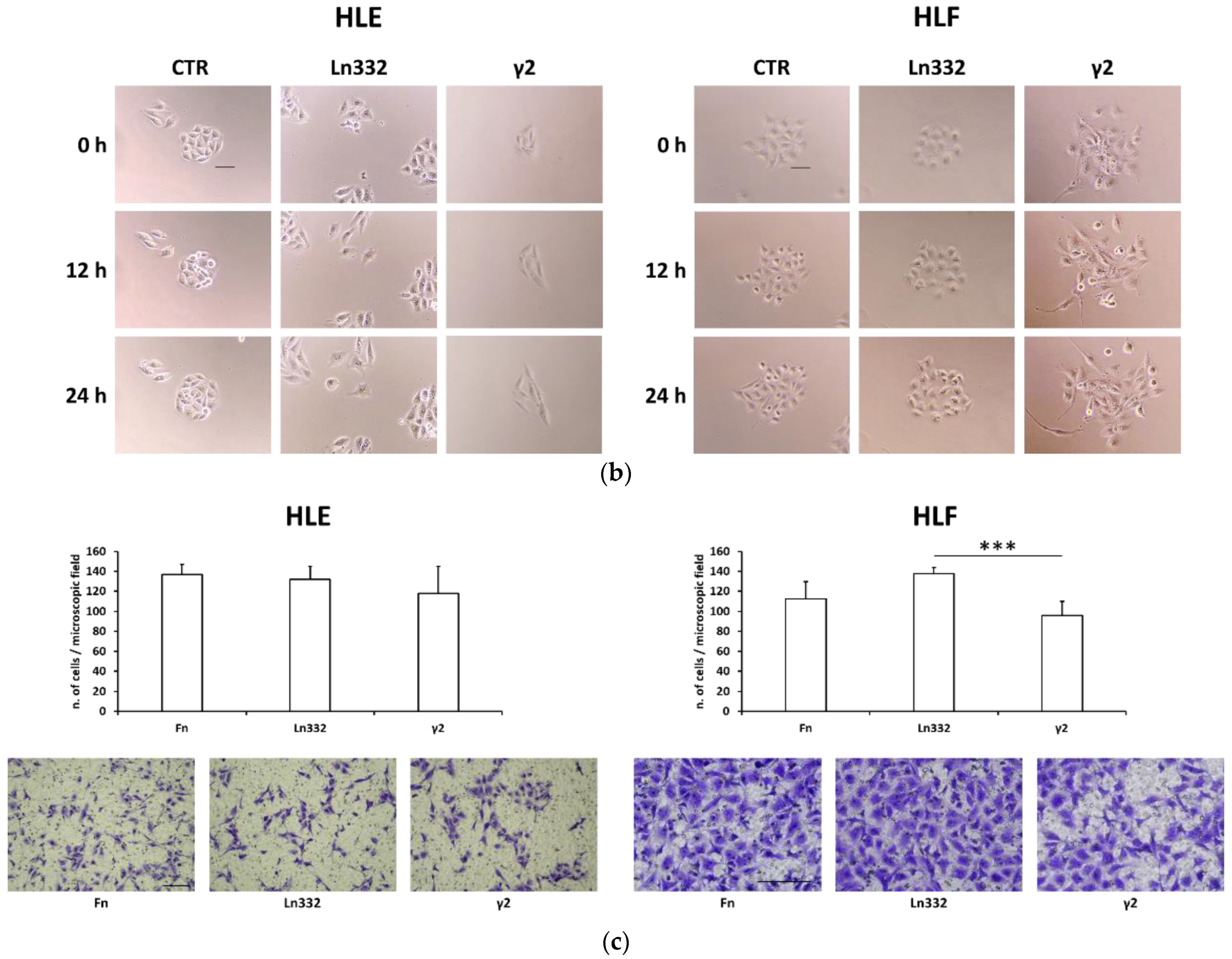
2.3. Transwell Migration Assay
2.4. Scattering Assay
2.5. Microscopy Analyses
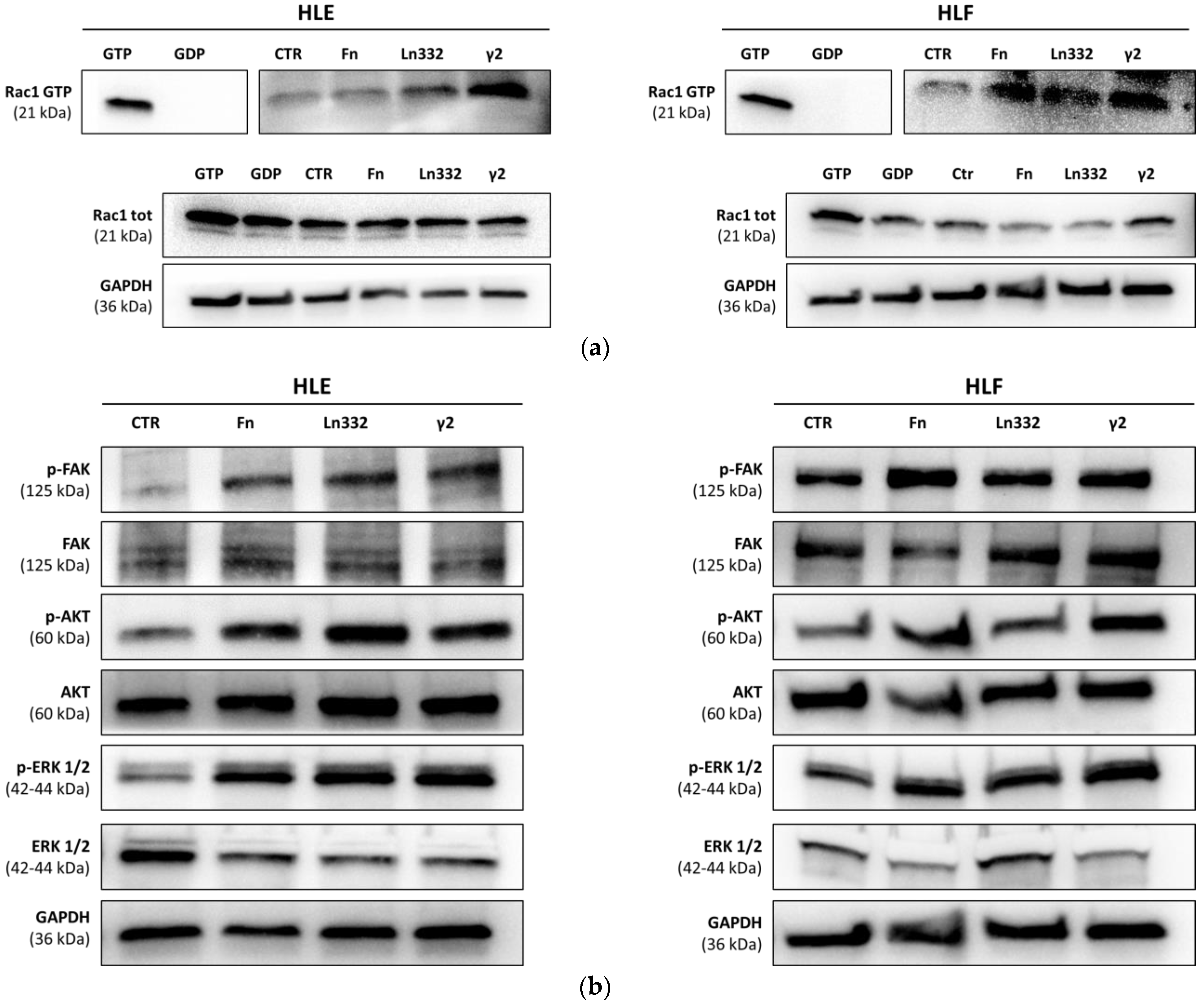
2.6. Western Blot
2.7. Active Rac1 Pull-Down Assays
2.8. Statistical Analysis
3. Results
3.1. Soluble γ2 Chain of Ln332 Promotes HCC Cell Adhesion, Scattering, and Migration
3.2. Ln332 γ2 Chain Activates Pro-Migratory Signaling Pathway in HCC Cells
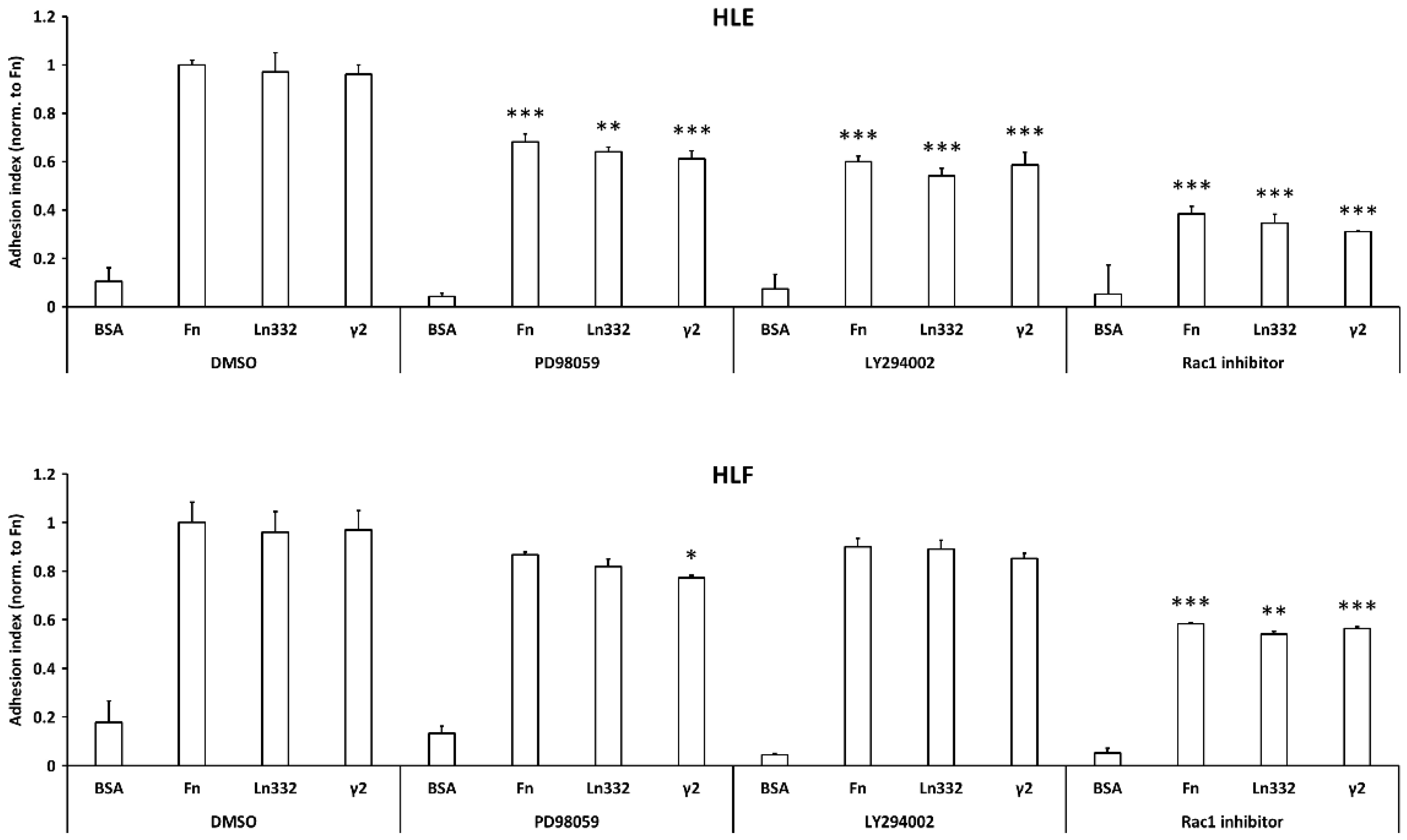
3.3. Adhesion and Spreading of HCC Cells on γ2 Chain of Laminin-332 depends on ERKs and Rac1 Activation
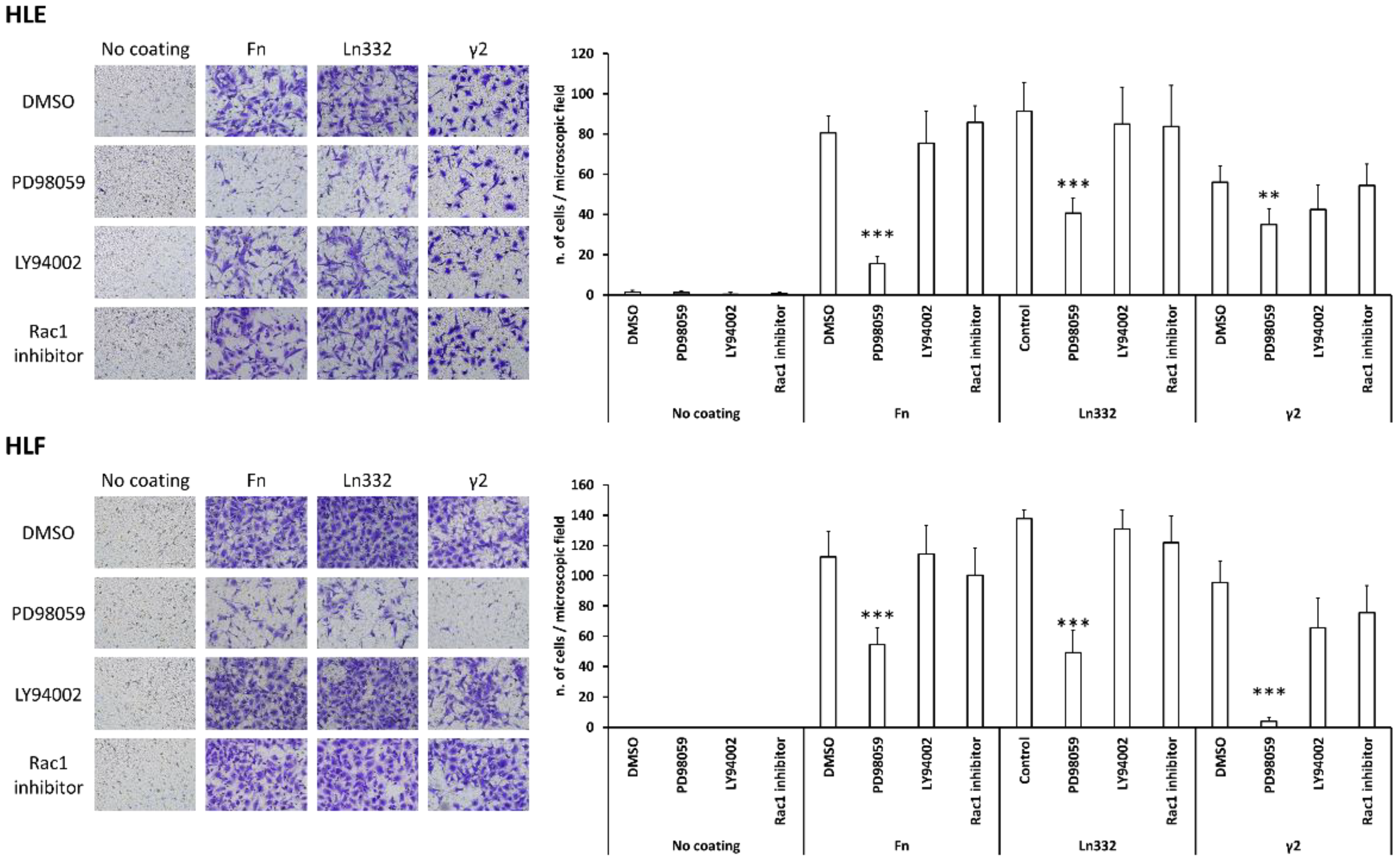
3.4. Pro-Migratory Activity of Ln332 γ2 depends upon MEK-ERK1/2 Pathway Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Rani, B. Role of the tissue microenvironment as a therapeutic target in hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 4128. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, M.; Macias-Barragan, J. Update on the pathophysiology of liver fibrosis. Expert Rev. Gastroenterol. Hepatol. 2010, 4, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Santamato, A.; Fransvea, E.; Dituri, F.; Caligiuri, A.; Quaranta, M.; Niimi, T.; Pinzani, M.; Antonaci, S.; Giannelli, G. Hepatic stellate cells stimulate HCC cell migration via laminin-5 production. Clin. Sci. 2011, 121, 159–168. [Google Scholar] [CrossRef]
- Burgeson, R.E.; Chiquet, M.; Deutzmann, R.; Ekblom, P.; Engel, J.; Kleinman, H.; Martin, G.R.; Meneguzzi, G.; Paulsson, M.; Sanes, J. A new nomenclature for the laminins. Matrix Biol. 1994, 14, 209–211. [Google Scholar] [CrossRef]
- Giannelli, G.; Fransvea, E.; Bergamini, C.; Marinosci, F.; Antonaci, S. Laminin-5 chains are expressed differentially in metastatic and nonmetastatic hepatocellular carcinoma. Clin. Cancer Res. 2003, 9, 3684–3691. [Google Scholar]
- Bergamini, C.; Sgarra, C.; Trerotoli, P.; Lupo, L.; Azzariti, A.; Antonaci, S.; Giannelli, G. Laminin-5 stimulates hepatocellular carcinoma growth through a different function of α6β4 and α3β1 integrins. Hepatology 2007, 46, 1801–1809. [Google Scholar] [CrossRef]
- Korkut, A.; Zaidi, S.; Kanchi, R.S.; Rao, S.; Gough, N.R.; Schultz, A.; Li, X.; Lorenzi, P.L.; Berger, A.C.; Robertson, G.; et al. A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily. Cell Syst. 2018, 7, 422–437.e7. [Google Scholar] [CrossRef]
- Eke, I.; Cordes, N. Focal adhesion signaling and therapy resistance in cancer. Semin. Cancer Biol. 2015, 31, 65–75. [Google Scholar] [CrossRef]
- Giannelli, G.; Bergamini, C.; Fransvea, E.; Sgarra, C.; Antonaci, S. Laminin-5 with Transforming Growth Factor-β1 Induces Epithelial to Mesenchymal Transition in Hepatocellular Carcinoma. Gastroenterology 2005, 129, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Koshikawa, N.; Moriyama, K.; Takamura, H.; Mizushima, H.; Nagashima, Y.; Yanoma, S.; Miyazaki, K. Overexpression of laminin gamma2 chain monomer in invading gastric carcinoma cells. Cancer Res. 1999, 59, 5596–5601. [Google Scholar] [PubMed]
- Chen, J.; Zhang, H.; Luo, J.; Wu, X.; Li, X.; Zhao, X.; Zhou, D.; Yu, S. Overexpression of α3, β3 and γ2 chains of laminin-332 is associated with poor prognosis in pancreatic ductal adenocarcinoma. Oncol. Lett. 2018, 16, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Tsubota, Y.; Hashimoto, J.; Kariya, Y.; Miyazaki, K. The short arm of laminin gamma2 chain of laminin-5 (laminin-332) binds syndecan-1 and regulates cellular adhesion and migration by suppressing phosphorylation of integrin beta4 chain. Mol. Biol. Cell 2007, 18, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Falk-Marzillier, J.; Schiraldi, O.; Stetler-Stevenson, W.G.; Quaranta, V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science 1997, 277, 225–228. [Google Scholar] [CrossRef]
- Koshikawa, N.; Giannelli, G.; Cirulli, V.; Miyazaki, K.; Quaranta, V. Role of Cell Surface Metalloprotease Mt1-Mmp in Epithelial Cell Migration over Laminin-5. J. Cell Biol. 2000, 148, 615–624. [Google Scholar] [CrossRef]
- Koshikawa, N.; Schenk, S.; Moeckel, G.; Sharabi, A.; Miyazaki, K.; Gardner, H.; Zent, R.; Quaranta, V. Proteolytic processing of laminin-5 by MT1-MMP in tissues and its effects on epithelial cell morphology. FASEB J. 2004, 18, 1–22. [Google Scholar] [CrossRef]
- Dituri, F.; Scialpi, R.; Schmidt, T.A.; Frusciante, M.; Mancarella, S.; Lupo, L.G.; Villa, E.; Giannelli, G. Proteoglycan-4 is correlated with longer survival in HCC patients and enhances sorafenib and regorafenib effectiveness via CD44 in vitro. Cell Death Dis. 2020, 11, 984. [Google Scholar] [CrossRef]
- Fransvea, E.; Angelotti, U.; Antonaci, S.; Giannelli, G. Blocking transforming growth factor-beta up-regulates E-cadherin and reduces migration and invasion of hepatocellular carcinoma cells. Hepatology 2008, 47, 1557–1566. [Google Scholar] [CrossRef]
- Vilchez Mercedes, S.A.; Bocci, F.; Levine, H.; Onuchic, J.N.; Jolly, M.K.; Wong, P.K. Decoding leader cells in collective cancer invasion. Nat. Rev. Cancer 2021, 21, 592–604. [Google Scholar] [CrossRef]
- Mackay, D.J.G.; Esch, F.; Furthmayr, H.; Hall, A. Rho- and Rac-dependent Assembly of Focal Adhesion Complexes and Actin Filaments in Permeabilized Fibroblasts: An Essential Role for Ezrin/Radixin/Moesin Proteins. J. Cell Biol. 1997, 138, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Nethe, M.; Hordijk, P.L. The role of ubiquitylation and degradation in RhoGTPase signalling. J. Cell Sci. 2010, 123, 4011–4018. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Barton, M.J.; Nguyen, N.-T. Biophysical properties of cells for cancer diagnosis. J. Biomech. 2019, 86, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kazanietz, M.G.; Caloca, M.J. The Rac GTPase in Cancer: From Old Concepts to New Paradigms. Cancer Res. 2017, 77, 5445–5451. [Google Scholar] [CrossRef]
- Bustelo, X.R. RHO GTPases in cancer: Known facts, open questions, and therapeutic challenges. Biochem. Soc. Trans. 2018, 46, 741–760. [Google Scholar] [CrossRef]
- Schenk, S.; Hintermann, E.; Bilban, M.; Koshikawa, N.; Hojilla, C.; Khokha, R.; Quaranta, V. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J. Cell Biol. 2003, 161, 197–209. [Google Scholar] [CrossRef]
- Schenk, S.; Quaranta, V. Tales from the crypt[ic] sites of the extracellular matrix. Trends Cell Biol. 2003, 13, 366–375. [Google Scholar] [CrossRef]
- Gagnoux-Palacios, L.; Allegra, M.; Spirito, F.; Pommeret, O.; Romero, C.; Ortonne, J.; Meneguzzi, G. The Short Arm of the Laminin γ2 Chain Plays a Pivotal Role in the Incorporation of Laminin 5 into the Extracellular Matrix and in Cell Adhesion. J. Cell Biol. 2001, 153, 835–850. [Google Scholar] [CrossRef]
- Pyke, C.; Rømer, J.; Kallunki, P.; Lund, L.R.; Ralfkiaer, E.; Danø, K.; Tryggvason, K. The gamma 2 chain of kalinin/laminin 5 is preferentially expressed in invading malignant cells in human cancers. Am. J. Pathol. 1994, 145, 782–791. [Google Scholar]
- Pyke, C.; Salo, S.; Ralfkiaer, E.; Rømer, J.; Danø, K.; Tryggvason, K. Laminin-5 is a marker of invading cancer cells in some human carcinomas and is coexpressed with the receptor for urokinase plasminogen activator in budding cancer cells in colon adenocarcinomas. Cancer Res. 1995, 55, 4132–4139. [Google Scholar]
- Shen, X.-M.; Wu, Y.-P.; Feng, Y.-B.; Luo, M.-L.; Du, X.-L.; Zhang, Y.; Cai, Y.; Xu, X.; Han, Y.-L.; Zhang, X.; et al. Interaction of MT1-MMP and laminin-5gamma2 chain correlates with metastasis and invasiveness in human esophageal squamous cell carcinoma. Clin. Exp. Metastasis 2007, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Katayama, M.; Funakoshi, A.; Sumii, T.; Sanzen, N.; Sekiguchi, K. Laminin gamma2-chain fragment circulating level increases in patients with metastatic pancreatic ductal cell adenocarcinomas. Cancer Lett. 2005, 225, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef]
- Hall, A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu. Rev. Cell Biol. 1994, 10, 31–54. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.B.; Nabha, S.M.; Atanaskova, N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003, 22, 395–403. [Google Scholar] [CrossRef]
- Shih, Y.-W.; Chen, P.-S.; Wu, C.-H.; Jeng, Y.-F.; Wang, C.-J. Alpha-chaconine-reduced metastasis involves a PI3K/Akt signaling pathway with downregulation of NF-kappaB in human lung adenocarcinoma A549 cells. J. Agric. Food Chem. 2007, 55, 11035–11043. [Google Scholar] [CrossRef]
- Gupta, A.K.; Soto, D.E.; Feldman, M.D.; Goldsmith, J.D.; Mick, R.; Hahn, S.M.; Machtay, M.; Muschel, R.J.; McKenna, W.G. Signaling pathways in NSCLC as a predictor of outcome and response to therapy. Lung 2004, 182, 151–162. [Google Scholar] [CrossRef]
- Osaki, M.; Oshimura, M.; Ito, H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis 2004, 9, 667–676. [Google Scholar] [CrossRef]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scialpi, R.; Arrè, V.; Giannelli, G.; Dituri, F. Laminin-332 γ2 Monomeric Chain Promotes Adhesion and Migration of Hepatocellular Carcinoma Cells. Cancers 2023, 15, 373. https://doi.org/10.3390/cancers15020373
Scialpi R, Arrè V, Giannelli G, Dituri F. Laminin-332 γ2 Monomeric Chain Promotes Adhesion and Migration of Hepatocellular Carcinoma Cells. Cancers. 2023; 15(2):373. https://doi.org/10.3390/cancers15020373
Chicago/Turabian StyleScialpi, Rosanna, Valentina Arrè, Gianluigi Giannelli, and Francesco Dituri. 2023. "Laminin-332 γ2 Monomeric Chain Promotes Adhesion and Migration of Hepatocellular Carcinoma Cells" Cancers 15, no. 2: 373. https://doi.org/10.3390/cancers15020373
APA StyleScialpi, R., Arrè, V., Giannelli, G., & Dituri, F. (2023). Laminin-332 γ2 Monomeric Chain Promotes Adhesion and Migration of Hepatocellular Carcinoma Cells. Cancers, 15(2), 373. https://doi.org/10.3390/cancers15020373





