Deep Learning for Fully Automatic Tumor Segmentation on Serially Acquired Dynamic Contrast-Enhanced MRI Images of Triple-Negative Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Image Acquisition
2.3. Data Curation
2.4. Automatic Segmentation Framework
2.5. Statistical Analysis
3. Results
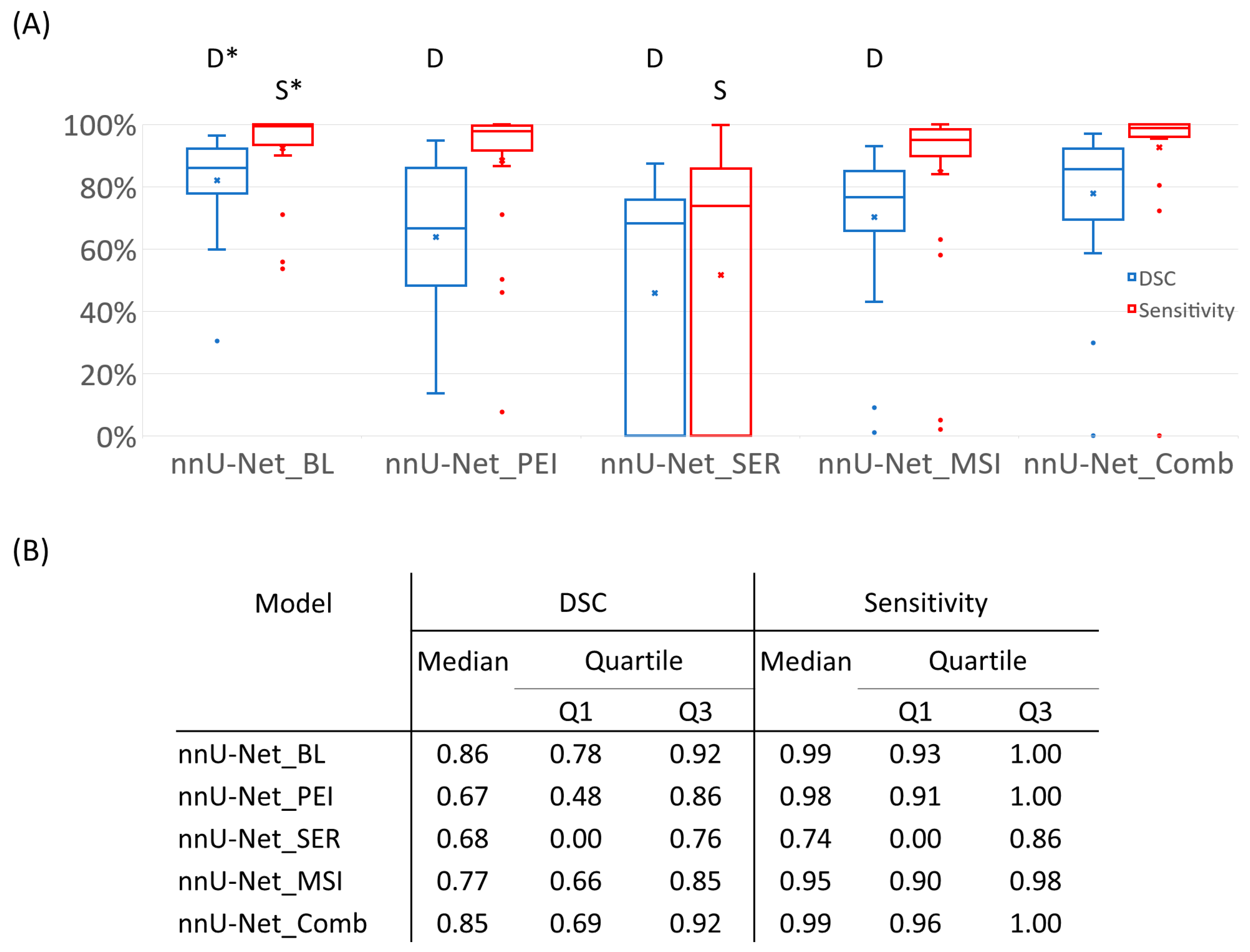
3.1. Segmentation Performance of Semiquantitative Parametric Maps
3.2. Mask Type Comparison
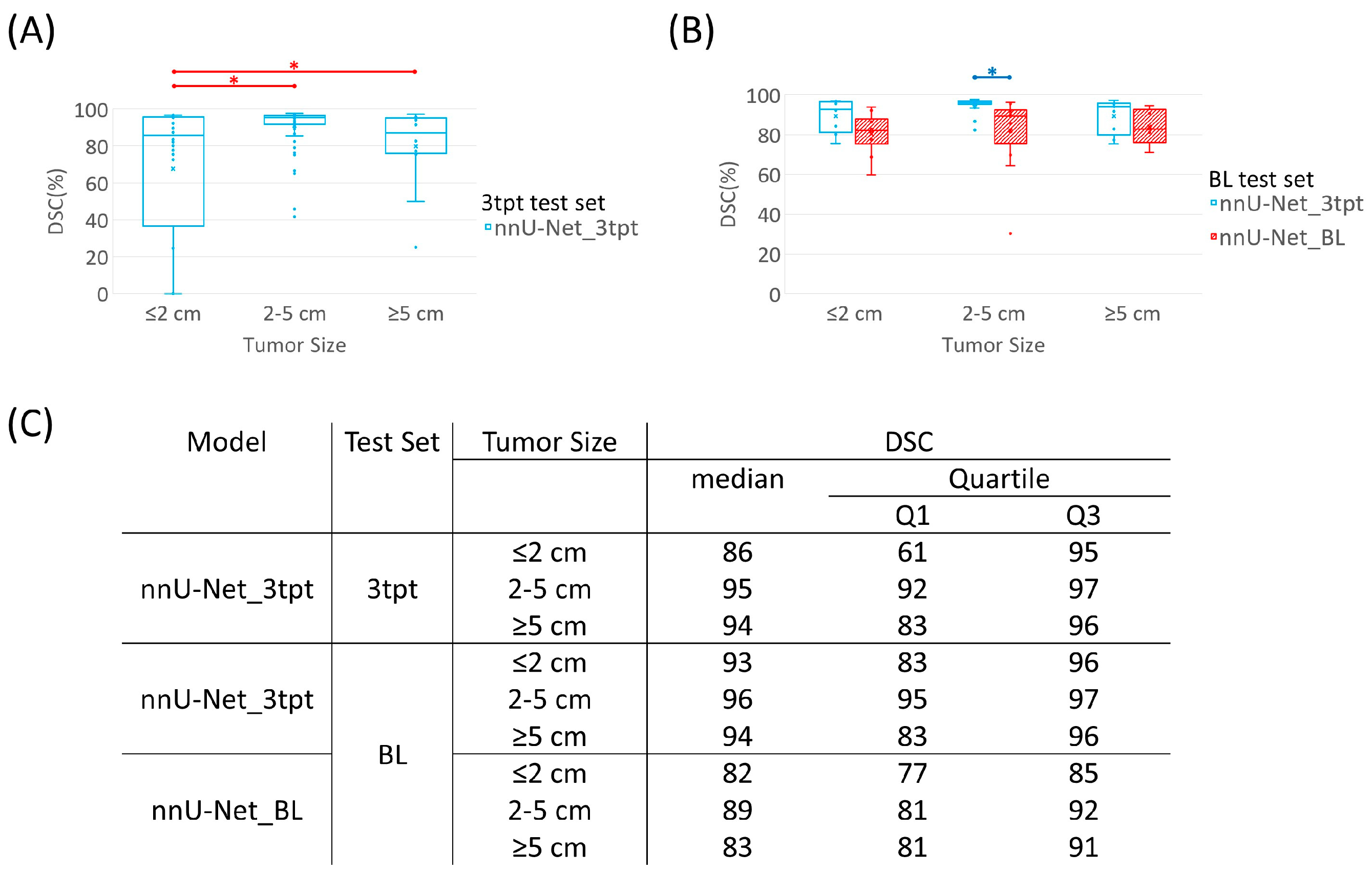
3.3. Segmentation Performance Using Datasets of Different Time Points
3.4. Tumor Size Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
- K.K.H. serves on the Medical Advisory Board for ArmadaHealth, AstraZeneca, and receives research funding from Cairn Surgical, Eli Lilly&Co., and Lumicell.
- K.H. is currently receiving research funding from Siemens Healthineers and has received research funding from GE.
- J.K.L. received grant or research support from Novartis, Medivation/Pfizer, Genentech, GSK, EMD-Serono, AstraZeneca, Medimmune, Zenith, Merck; participated in Speaker’s Bureau for MedLearning, Physician’s Education Resource, Prime Oncology, Medscape, Clinical Care Options, Medpage; and receives royalty from UpToDate.
- Spouse of A.T works for Eli Lilly.
- D.T. declares research contracts with Pfizer, Novartis, and Ployphor and is a consultant of AstraZeneca, GlaxoSmithKline, OncoPep, Gilead, Novartis, Pfizer, Personalis, and Sermonix.
- W.Y. receives royalties from Elsevier.
- J.M. is a consultant of C4 Imaging, L.L.C., and an inventor of United States patents licensed to Siemens Healthineers and GE Healthcare.
- For the remaining authors, none were declared.
References
- Wu, Q.; Siddharth, S.; Sharma, D. Triple Negative Breast Cancer: A Mountain Yet to Be Scaled Despite the Triumphs. Cancers 2021, 13, 3697. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-Negative Breast Cancer: Clinical Features and Patterns of Recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Zhang, Y.; Chan, S.; Park, V.Y.; Chang, K.-T.; Mehta, S.; Kim, M.J.; Combs, F.J.; Chang, P.; Chow, D.; Parajuli, R.; et al. Automatic Detection and Segmentation of Breast Cancer on MRI Using Mask R-CNN Trained on Non–Fat-Sat Images and Tested on Fat-Sat Images. Acad. Radiol. 2022, 29, S135–S144. [Google Scholar] [CrossRef]
- El Adoui, M.; Drisis, S.; Benjelloun, M. Multi-input deep learning architecture for predicting breast tumor response to chemotherapy using quantitative MR images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1491–1500. [Google Scholar] [CrossRef]
- Ha, R.; Chin, C.; Karcich, J.; Liu, M.Z.; Chang, P.; Mutasa, S.; Van Sant, E.P.; Wynn, R.T.; Connolly, E.; Jambawalikar, S. Prior to Initiation of Chemotherapy, Can We Predict Breast Tumor Response? Deep Learning Convolutional Neural Networks Approach Using a Breast MRI Tumor Dataset. J. Digit. Imaging 2018, 32, 693–701. [Google Scholar] [CrossRef]
- Kohli, M.D.; Summers, R.M.; Geis, J.R. Medical Image Data and Datasets in the Era of Machine Learning—Whitepaper from the 2016 C-MIMI Meeting Dataset Session. J. Digit. Imaging 2017, 30, 392–399. [Google Scholar] [CrossRef]
- Kupinski, M.A.; Giger, M.L. Automated seeded lesion segmentation on digital mammograms. IEEE Trans. Med. Imaging 1998, 17, 510–517. [Google Scholar] [CrossRef]
- Yuan, Y.; Giger, M.L.; Li, H.; Suzuki, K.; Sennett, C. A dual-stage method for lesion segmentation on digital mammograms. Med. Phys. 2007, 34, 4180–4193. [Google Scholar] [CrossRef]
- Horsch, K.; Giger, M.L.; Venta, L.A.; Vyborny, C.J. Automatic segmentation of breast lesions on ultrasound. Med. Phys. 2001, 28, 1652–1659. [Google Scholar] [CrossRef]
- Rojas Domínguez, A.; Nandi, A.K. Detection of masses in mammograms via statistically based enhancement, multilevel-thresholding segmentation, and region selection. Comput. Med. Imaging Graph. 2008, 32, 304–315. [Google Scholar] [CrossRef]
- Pereira, D.C.; Ramos, R.P.; do Nascimento, M.Z. Segmentation and detection of breast cancer in mammograms combining wavelet analysis and genetic algorithm. Comput. Methods Programs Biomed. 2014, 114, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Timp, S.; Karssemeijer, N. A new 2D segmentation method based on dynamic programming applied to computer aided detection in mammography. Med. Phys. 2004, 31, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Petrick, N.; Chan, H.P.; Sahiner, B.; Helvie, M.A. Combined adaptive enhancement and region-growing segmentation of breast masses on digitized mammograms. Med. Phys. 1999, 26, 1642–1654. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Luo, Y.; Zhang, Q. Breast ultrasound image segmentation: A survey. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 493–507. [Google Scholar] [CrossRef]
- Hu, Y.; Guo, Y.; Wang, Y.; Yu, J.; Li, J.; Zhou, S.; Chang, C. Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med. Phys. 2019, 46, 215–228. [Google Scholar] [CrossRef]
- Al-Antari, M.A.; Al-Masni, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int. J. Med. Inform. 2018, 117, 44–54. [Google Scholar] [CrossRef]
- Kooi, T.; Litjens, G.; van Ginneken, B.; Gubern-Merida, A.; Sanchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Baccouche, A.; Garcia-Zapirain, B.; Castillo Olea, C.; Elmaghraby, A.S. Connected-UNets: A deep learning architecture for breast mass segmentation. NPJ Breast Cancer 2021, 7, 151. [Google Scholar] [CrossRef]
- Balkenende, L.; Teuwen, J.; Mann, R.M. Application of Deep Learning in Breast Cancer Imaging. Semin. Nucl. Med. 2022, 52, 584–596. [Google Scholar] [CrossRef]
- Kuhl, C.K.; Mielcareck, P.; Klaschik, S.; Leutner, C.; Wardelmann, E.; Gieseke, J.; Schild, H.H. Dynamic breast MR imaging: Are signal intensity time course data useful for differential diagnosis of enhancing lesions? Radiology 1999, 211, 101–110. [Google Scholar] [CrossRef]
- Agner, S.C.; Xu, J.; Fatakdawala, H.; Ganesan, S.; Madabhushi, A.; Englander, S.; Rosen, M.; Thomas, K.; Schnall, M.; Feldman, M.; et al. Segmentation and classification of triple negative breast cancers using DCE-MRI. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1227–1230. [Google Scholar]
- Woods, B.J.; Clymer, B.D.; Kurc, T.; Heverhagen, J.T.; Stevens, R.; Orsdemir, A.; Bulan, O.; Knopp, M.V. Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. J. Magn. Reson. Imaging 2007, 25, 495–501. [Google Scholar] [CrossRef]
- Aljabar, P.; Heckemann, R.A.; Hammers, A.; Hajnal, J.V.; Rueckert, D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage 2009, 46, 726–738. [Google Scholar] [CrossRef]
- Wang, H.; Yushkevich, P.A. Multi-atlas segmentation without registration: A supervoxel-based approach. In Proceedings of the Medical image computing and computer-assisted intervention: MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention 2013, Nagoya, Japan, 22–26 September 2013; Volume 16, pp. 535–542. [Google Scholar]
- Chen, W.; Giger, M.L.; Bick, U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad. Radiol. 2006, 13, 63–72. [Google Scholar] [CrossRef]
- Keller, B.M.; Nathan, D.L.; Wang, Y.; Zheng, Y.; Gee, J.C.; Conant, E.F.; Kontos, D. Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Med. Phys. 2012, 39, 4903–4917. [Google Scholar] [CrossRef]
- Zhang, J.; Saha, A.; Zhu, Z.; Mazurowski, M.A. Hierarchical Convolutional Neural Networks for Segmentation of Breast Tumors in MRI With Application to Radiogenomics. IEEE Trans. Med. Imaging 2019, 38, 435–447. [Google Scholar] [CrossRef]
- Dalmis, M.U.; Litjens, G.; Holland, K.; Setio, A.; Mann, R.; Karssemeijer, N.; Gubern-Merida, A. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med. Phys. 2017, 44, 533–546. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- El Adoui, M.; Mahmoudi, S.A.; Larhmam, M.A.; Benjelloun, M. MRI Breast Tumor Segmentation Using Different Encoder and Decoder CNN Architectures. Computers 2019, 8, 52. [Google Scholar] [CrossRef]
- Hirsch, L.; Huang, Y.; Luo, S.; Saccarelli, C.R.; Gullo, R.L.; Naranjo, I.D.; Bitencourt, A.G.V.; Onishi, N.; Ko, E.S.; Leithner, D.; et al. Radiologist-Level Performance by Using Deep Learning for Segmentation of Breast Cancers on MRI Scans. Radiol. Artif. Intell. 2021, 15, e200231. [Google Scholar] [CrossRef]
- Zhang, L.; Mohamed, A.A.; Chai, R.; Guo, Y.; Zheng, B.; Wu, S. Automated deep learning method for whole-breast segmentation in diffusion-weighted breast MRI. J. Magn. Reson. Imaging 2019, 51, 635–643. [Google Scholar] [CrossRef]
- Chen, X.; Men, K.; Chen, B.; Tang, Y.; Zhang, T.; Wang, S.; Li, Y.; Dai, J. CNN-Based Quality Assurance for Automatic Segmentation of Breast Cancer in Radiotherapy. Front. Oncol. 2020, 10, 524. [Google Scholar] [CrossRef]
- Gao, J.; Zhong, X.; Li, W.; Li, Q.; Shao, H.; Wang, Z.; Dai, Y.; Ma, H.; Shi, Y.; Zhang, H.; et al. Attention-based Deep Learning for the Preoperative Differentiation of Axillary Lymph Node Metastasis in Breast Cancer on DCE-MRI. J. Magn. Reson. Imaging 2023, 57, 1842–1853. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; et al. The Medical Segmentation Decathlon. Nat. Commun. 2022, 13, 4128. [Google Scholar] [CrossRef]
- Panthi, B.; Adrada, B.E.; Candelaria, R.P.; Guirguis, M.S.; Yam, C.; Boge, M.; Chen, H.; Hunt, K.K.; Huo, L.; Hwang, K.-P.; et al. Assessment of Response to Neoadjuvant Systemic Treatment in Triple-Negative Breast Cancer Using Functional Tumor Volumes from Longitudinal Dynamic Contrast-Enhanced MRI. Cancers 2023, 15, 1025. [Google Scholar] [CrossRef]
- Saranathan, M.; Rettmann, D.W.; Hargreaves, B.A.; Clarke, S.E.; Vasanawala, S.S. DIfferential subsampling with cartesian ordering (DISCO): A high spatio-temporal resolution dixon imaging sequence for multiphasic contrast enhanced abdominal imaging. J. Magn. Reson. Imaging 2012, 35, 1484–1492. [Google Scholar] [CrossRef]
- Khiat, A.; Gianfelice, D.; Amara, M.; Boulanger, Y. Influence of post-treatment delay on the evaluation of the response to focused ultrasound surgery of breast cancer by dynamic contrast enhanced MRI. Br. J. Radiol. 2006, 79, 308–314. [Google Scholar] [CrossRef]
- Yang, W.; Qiang, J.W.; Tian, H.P.; Chen, B.; Wang, A.J.; Zhao, J.G. Multi-parametric MRI in cervical cancer: Early prediction of response to concurrent chemoradiotherapy in combination with clinical prognostic factors. Eur. Radiol. 2017, 28, 437–445. [Google Scholar] [CrossRef]
- Zhou, Z.; Adrada, B.E.; Candelaria, R.P.; Elshafeey, N.A.; Boge, M.; Mohamed, R.M.; Pashapoor, S.; Sun, J.; Xu, Z.; Panthi, B.; et al. Prediction of pathologic complete response to neoadjuvant systemic therapy in triple negative breast cancer using deep learning on multiparametric MRI. Sci. Rep. 2023, 13, 1171. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Rahimpour, M.; Saint Martin, M.J.; Frouin, F.; Akl, P.; Orlhac, F.; Koole, M.; Malhaire, C. Visual ensemble selection of deep convolutional neural networks for 3D segmentation of breast tumors on dynamic contrast enhanced MRI. Eur. Radiol. 2023, 33, 959–969. [Google Scholar] [CrossRef]
- Yue, W.; Zhang, H.; Zhou, J.; Li, G.; Tang, Z.; Sun, Z.; Cai, J.; Tian, N.; Gao, S.; Dong, J.; et al. Deep learning-based automatic segmentation for size and volumetric measurement of breast cancer on magnetic resonance imaging. Front. Oncol. 2022, 12, 984626. [Google Scholar] [CrossRef]
- Dogan, B.E.; Turnbull, L.W. Imaging of triple-negative breast cancer. Ann. Oncol. 2012, 23, vi23–vi29. [Google Scholar] [CrossRef]
- Milon, A.; Vande Perre, S.; Poujol, J.; Trop, I.; Kermarrec, E.; Bekhouche, A.; Thomassin-Naggara, I. Abbreviated breast MRI combining FAST protocol and high temporal resolution (HTR) dynamic contrast enhanced (DCE) sequence. Eur. J. Radiol. 2019, 117, 199–208. [Google Scholar] [CrossRef]
- Onishi, N.; Sadinski, M.; Hughes, M.C.; Ko, E.S.; Gibbs, P.; Gallagher, K.M.; Fung, M.M.; Hunt, T.J.; Martinez, D.F.; Shukla-Dave, A.; et al. Ultrafast dynamic contrast-enhanced breast MRI may generate prognostic imaging markers of breast cancer. Breast Cancer Res. 2020, 22, 58. [Google Scholar] [CrossRef]
- Janse, M.H.A.; Janssen, L.M.; van der Velden, B.H.M.; Moman, M.R.; Wolters-van der Ben, E.J.M.; Kock, M.; Viergever, M.A.; van Diest, P.J.; Gilhuijs, K.G.A. Deep Learning-Based Segmentation of Locally Advanced Breast Cancer on MRI in Relation to Residual Cancer Burden: A Multi-Institutional Cohort Study. J. Magn. Reson. Imaging 2023. online ahead of print. [Google Scholar] [CrossRef]
- Wang, S.; Sun, K.; Wang, L.; Qu, L.; Yan, F.; Wang, Q.; Shen, D. Breast Tumor Segmentation in DCE-MRI With Tumor Sensitive Synthesis. IEEE Trans. Neural Networks Learn. Syst. 2021, 34, 4990–5001. [Google Scholar] [CrossRef]
- Zhou, Z.; Sanders, J.W.; Johnson, J.M.; Gule-Monroe, M.; Chen, M.; Briere, T.M.; Wang, Y.; Son, J.B.; Pagel, M.D.; Ma, J.; et al. MetNet: Computer-aided segmentation of brain metastases in post-contrast T1-weighted magnetic resonance imaging. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2020, 153, 189–196. [Google Scholar] [CrossRef]
- Zhao, R.; Qian, B.; Zhang, X.; Li, Y.; Wei, R.; Liu, Y.; Pan, Y. Rethinking Dice Loss for Medical Image Segmentation. In Proceedings of the 2020 IEEE International Conference on Data Mining (ICDM), Sorrento, Italy, 17–20 November 2020; pp. 851–860. [Google Scholar]
- Li, Y.; Han, G.; Liu, X. DCNet: Densely Connected Deep Convolutional Encoder-Decoder Network for Nasopharyngeal Carcinoma Segmentation. Sensors 2021, 21, 7877. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, Z.; Zhang, D.; Hau, W.K.; Zhang, H. Progressive Perception Learning for Main Coronary Segmentation in X-Ray Angiography. IEEE Trans. Med. Imaging 2023, 42, 864–879. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]



| Characteristic | All Datasets | BL | C2 | C4 |
|---|---|---|---|---|
| No. of datasets | 744 | 285 | 207 | 252 |
| Age, mean ± SD, years | 50 ± 11 | 50 ± 11 | 50 ± 11 | 50 ± 11 |
| Longest tumor diameter, mean ± SD, cm | 2.7 ± 1.6 | 3.4 ± 1.5 | 2.6 ± 1.4 | 2.1 ± 1.5 |
| Clinical stage, n (%) | ||||
| I | 96 (13) | 37 (13) | 29 (14) | 30 (12) |
| II | 542 (73) | 210 (74) | 148 (72) | 184 (73) |
| III | 106 (14) | 38 (13) | 30 (14) | 38 (15) |
| T category, n (%) | ||||
| T1 | 139 (19) | 54 (19) | 39 (19) | 46 (18) |
| T2 | 509 (68) | 195 (68) | 141 (68) | 173 (69) |
| T3 | 83 (11) | 31 (11) | 23 (11) | 29 (12) |
| T4 | 13 (2) | 5 (2) | 4 (2) | 4 (2) |
| N category, n (%) | ||||
| N0 | 490 (66) | 188 (66) | 139 (67) | 163 (65) |
| N1 | 171 (23) | 67 (24) | 44 (21) | 60 (24) |
| N2 | 26 (3) | 9 (3) | 8 (4) | 9 (4) |
| N3 | 57 (8) | 21 (7) | 16 (8) | 20 (8) |
| Model Name | Input Dataset | DCE Metrics |
|---|---|---|
| nnU-Net_BL | BL | Sub |
| nnU-Net_PEI | BL | PEI |
| nnU-Net_SER | BL | SER |
| nnU-Net_MSI | BL | MSI |
| nnU-Net_Comb | BL | Sub + PEI + MSI + SER |
| nnU-Net_C2 | C2 | Sub |
| nnU-Net_C4 | C4 | Sub |
| nnU-Net_3tpt | BL + C2 + C4 | Sub |
| nnU-Net_Excl | BL | Sub |
| nnU-Net_Incl | BL | Sub |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Rauch, D.E.; Mohamed, R.M.; Pashapoor, S.; Zhou, Z.; Panthi, B.; Son, J.B.; Hwang, K.-P.; Musall, B.C.; Adrada, B.E.; et al. Deep Learning for Fully Automatic Tumor Segmentation on Serially Acquired Dynamic Contrast-Enhanced MRI Images of Triple-Negative Breast Cancer. Cancers 2023, 15, 4829. https://doi.org/10.3390/cancers15194829
Xu Z, Rauch DE, Mohamed RM, Pashapoor S, Zhou Z, Panthi B, Son JB, Hwang K-P, Musall BC, Adrada BE, et al. Deep Learning for Fully Automatic Tumor Segmentation on Serially Acquired Dynamic Contrast-Enhanced MRI Images of Triple-Negative Breast Cancer. Cancers. 2023; 15(19):4829. https://doi.org/10.3390/cancers15194829
Chicago/Turabian StyleXu, Zhan, David E. Rauch, Rania M. Mohamed, Sanaz Pashapoor, Zijian Zhou, Bikash Panthi, Jong Bum Son, Ken-Pin Hwang, Benjamin C. Musall, Beatriz E. Adrada, and et al. 2023. "Deep Learning for Fully Automatic Tumor Segmentation on Serially Acquired Dynamic Contrast-Enhanced MRI Images of Triple-Negative Breast Cancer" Cancers 15, no. 19: 4829. https://doi.org/10.3390/cancers15194829
APA StyleXu, Z., Rauch, D. E., Mohamed, R. M., Pashapoor, S., Zhou, Z., Panthi, B., Son, J. B., Hwang, K.-P., Musall, B. C., Adrada, B. E., Candelaria, R. P., Leung, J. W. T., Le-Petross, H. T. C., Lane, D. L., Perez, F., White, J., Clayborn, A., Reed, B., Chen, H., ... Ma, J. (2023). Deep Learning for Fully Automatic Tumor Segmentation on Serially Acquired Dynamic Contrast-Enhanced MRI Images of Triple-Negative Breast Cancer. Cancers, 15(19), 4829. https://doi.org/10.3390/cancers15194829






