Differential Expression of NEK Kinase Family Members in Esophageal Adenocarcinoma and Barrett’s Esophagus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Analyses of NEKs Gene Expression from TCGA Database and GEO Databases
2.2. Genomic Alterations of NEK Family Members in Esophageal Adenocarcinoma
2.3. Analyses of Association of NEKs Gene Expression and Patients’ Survival
2.4. Quantitative Real Time RT-PCR
2.5. Western Blotting Analysis
2.6. Immunohistochemistry
2.7. Immunofluorescence Staining
2.8. Statistical Analysis
3. Results
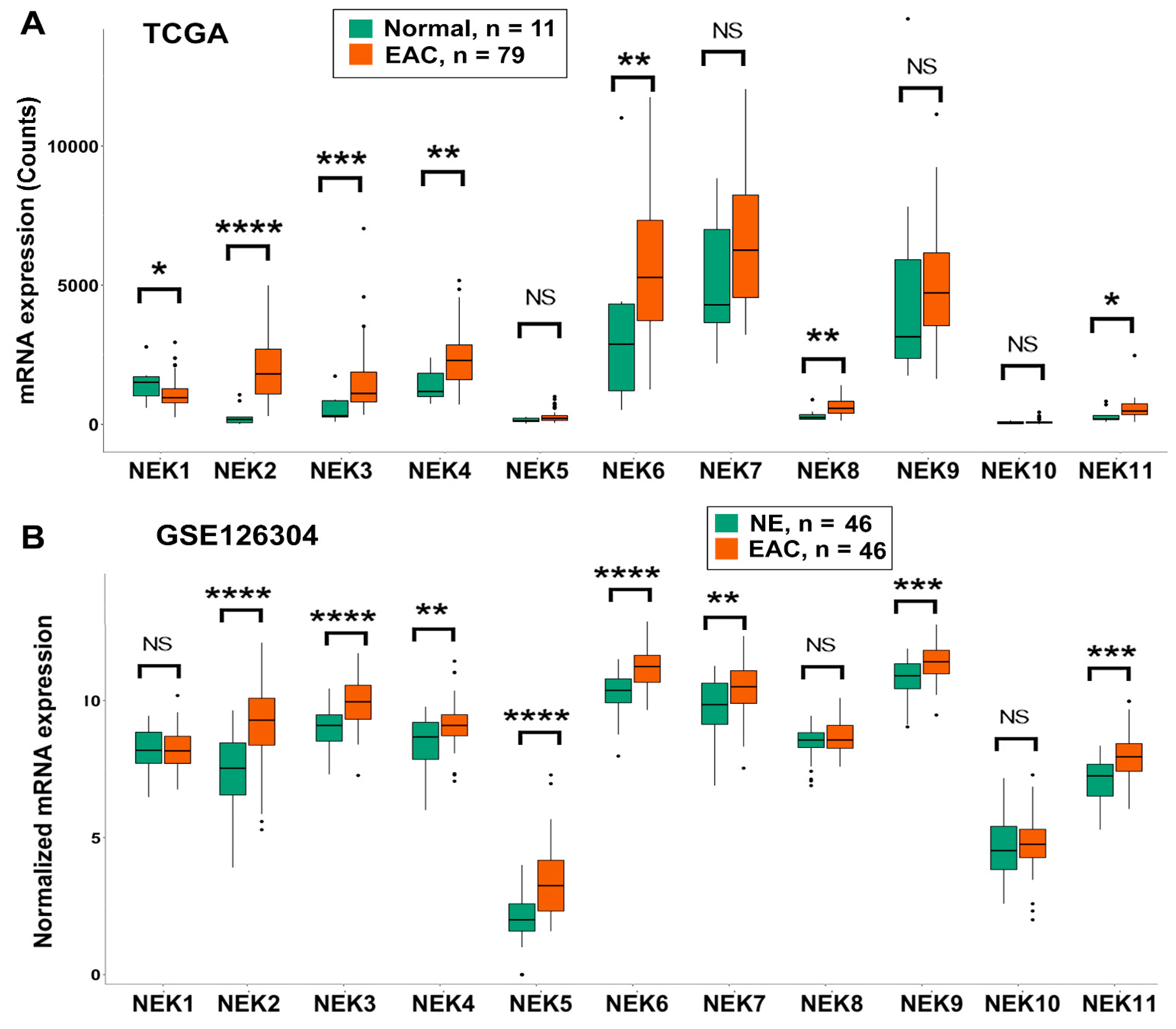
3.1. Differential Expression of NEK Family Members in Esophageal Adenocarcinoma from TCGA and GEO Databases
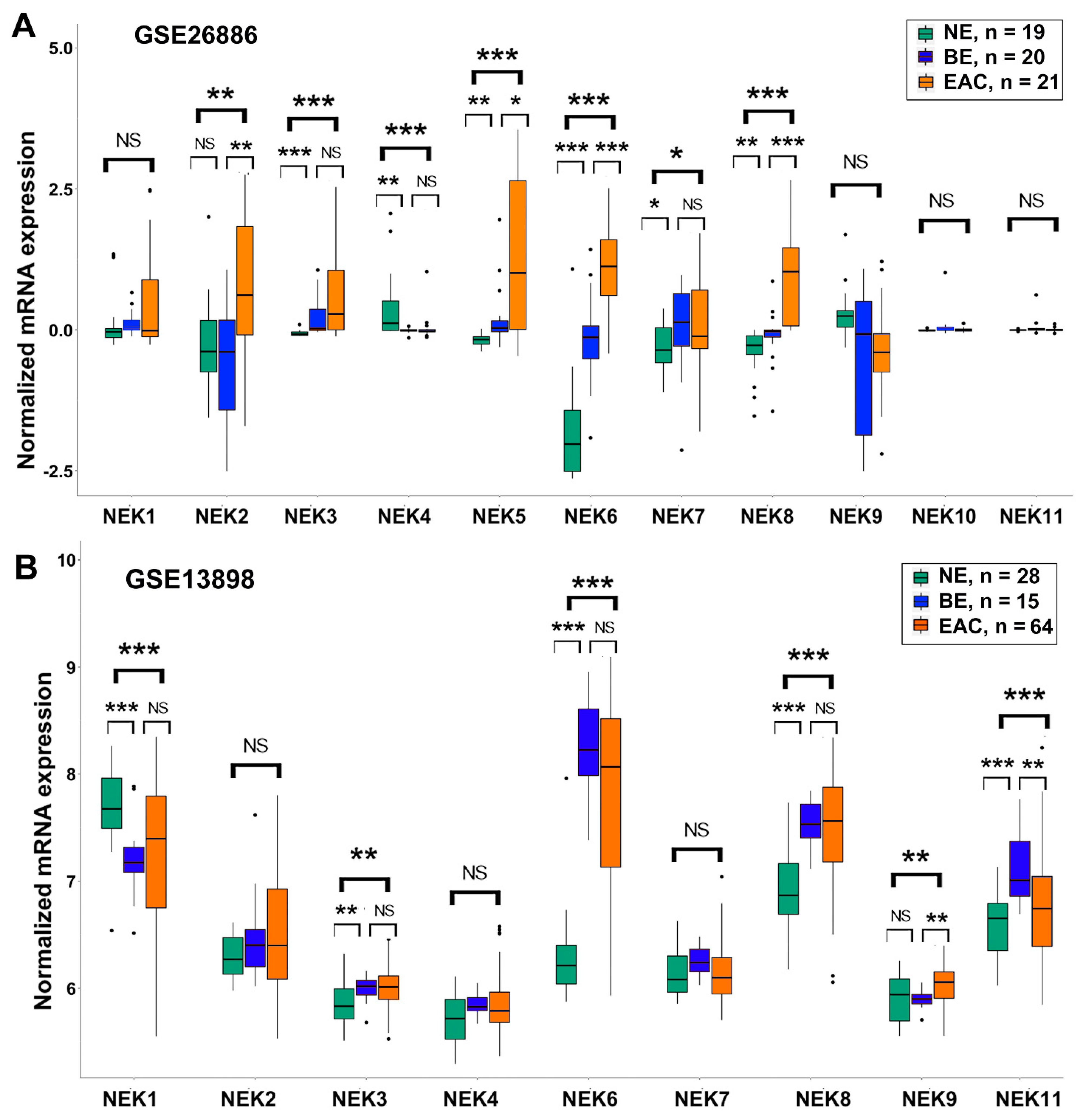
3.2. Differential Expression of NEK Family Members in Barrett’s Esophagus from GEO Databases
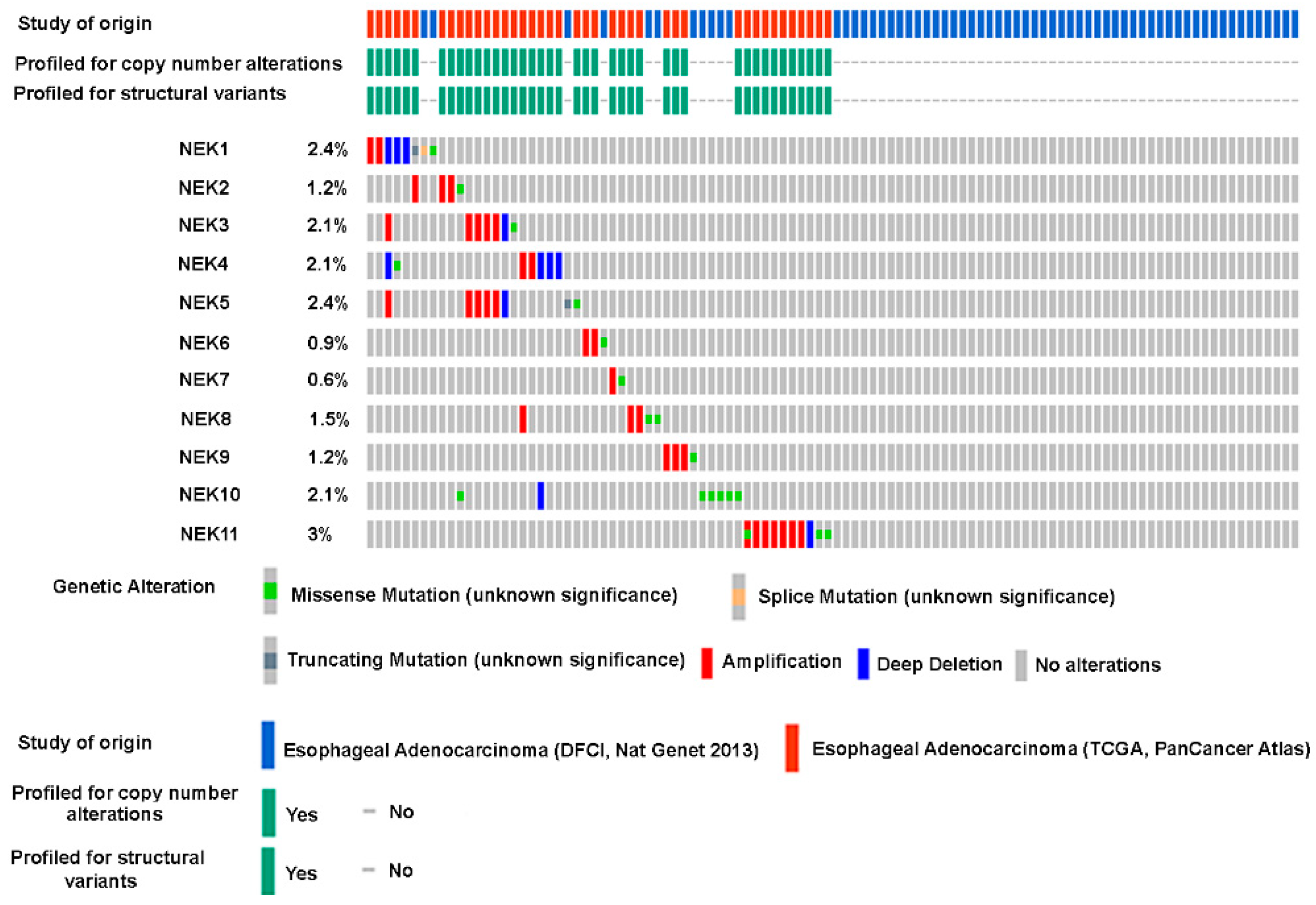
3.3. Genomic Alterations of NEK Family Members in Esophageal Adenocarcinoma
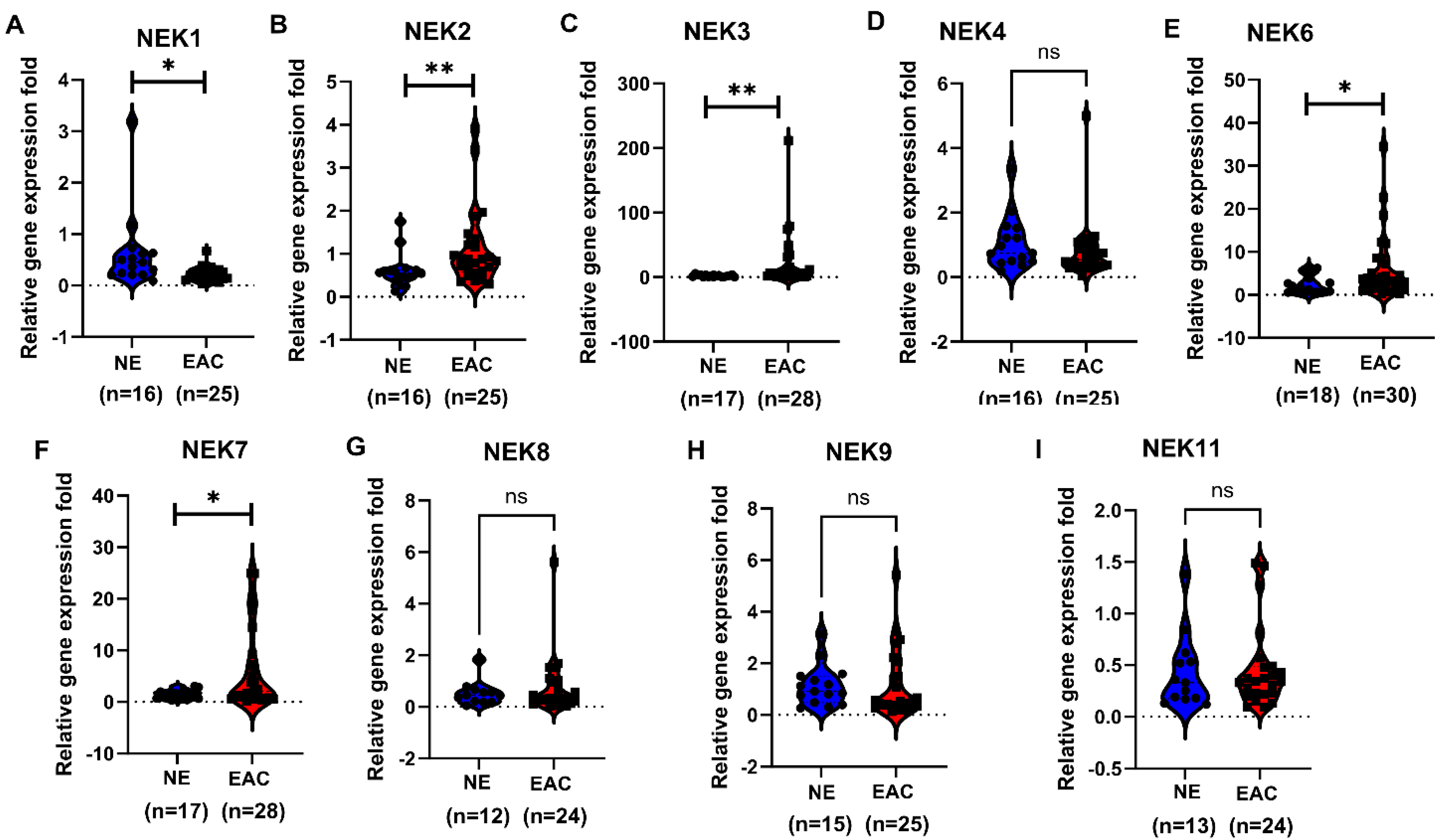
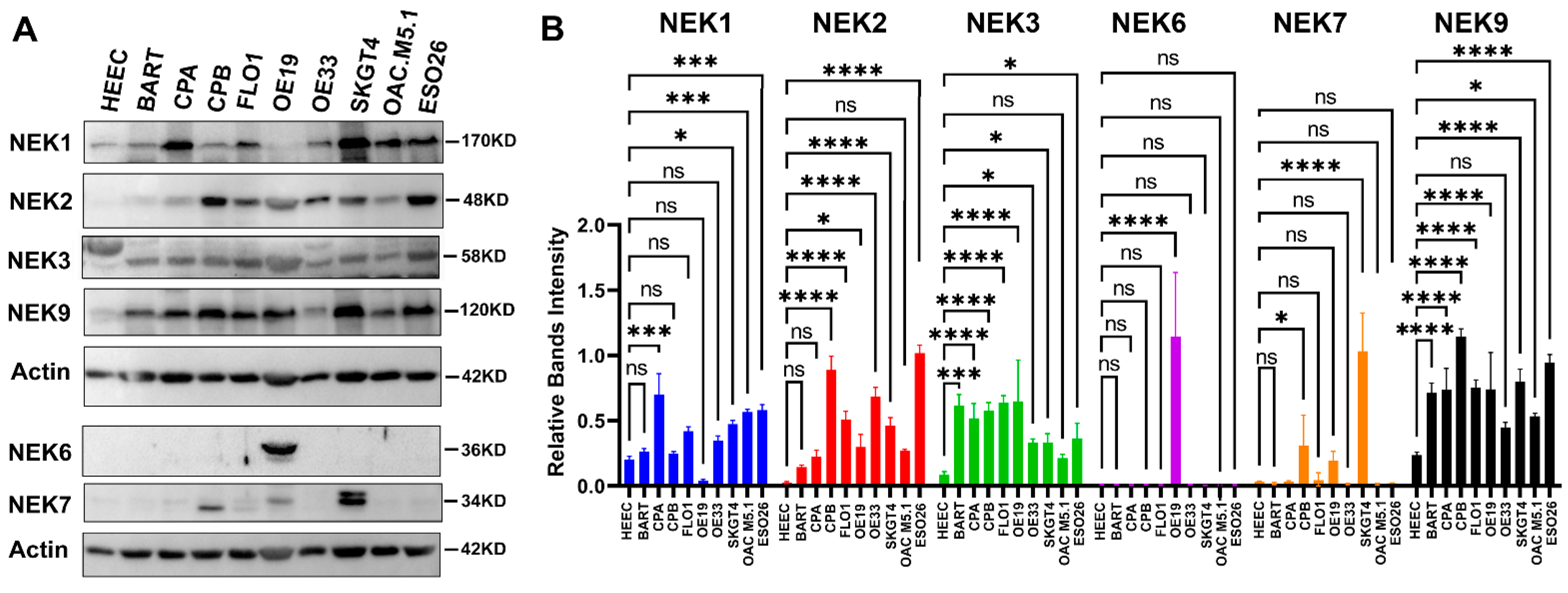
3.4. qRT-PCR Validation of NEKs Expression in Human Esophageal Cell Lines and Primary Tumor Tissues
3.5. NEKs Protein Expression in Human Esophageal Cell Lines and Primary Tumor Tissues
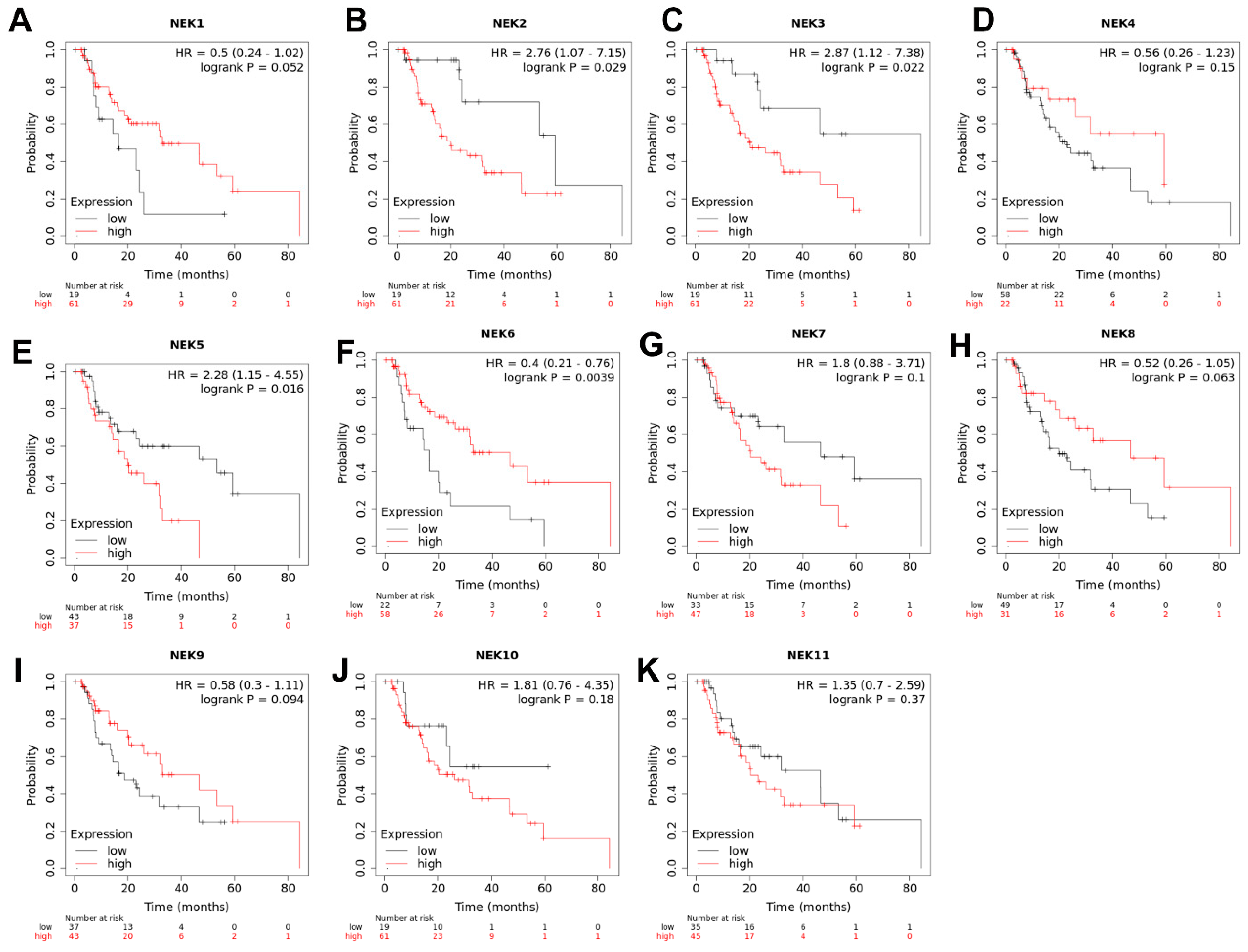
3.6. The Significance of NEKs Expression Associated with Patients’ Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Altorki, N.K.; Skinner, D.B. Adenocarcinoma in Barrett’s esophagus. Semin. Surg. Oncol. 1990, 6, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Sampliner, R.E. Adenocarcinoma of the esophagus and gastric cardia: Is there progress in the face of increasing cancer incidence? Ann. Intern. Med. 1999, 130, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Streitz, J.M., Jr. Barrett’s esophagus and esophageal cancer. Chest Surg. Clin. N. Am. 1994, 4, 227–240. [Google Scholar]
- van Soest, E.M.; Dieleman, J.P.; Siersema, P.D.; Sturkenboom, M.C.; Kuipers, E.J. Increasing incidence of Barrett’s oesophagus in the general population. Gut 2005, 54, 1062–1066. [Google Scholar] [CrossRef]
- Morgan, E.; Soerjomataram, I.; Rumgay, H.; Coleman, H.G.; Thrift, A.P.; Vignat, J.; Laversanne, M.; Ferlay, J.; Arnold, M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates From GLOBOCAN 2020. Gastroenterology 2022, 163, 649–658.e2. [Google Scholar] [CrossRef]
- Arnold, M.; Ferlay, J.; van Berge Henegouwen, M.I.; Soerjomataram, I. Global burden of oesophageal and gastric cancer by histology and subsite in 2018. Gut 2020, 69, 1564–1571. [Google Scholar] [CrossRef]
- Cook, M.B.; Thrift, A.P. Epidemiology of Barrett’s Esophagus and Esophageal Adenocarcinoma: Implications for Screening and Surveillance. Gastrointest. Endosc. Clin. N. Am. 2021, 31, 1–26. [Google Scholar] [CrossRef]
- Codipilly, D.C.; Sawas, T.; Dhaliwal, L.; Johnson, M.L.; Lansing, R.; Wang, K.K.; Leggett, C.L.; Katzka, D.A.; Iyer, P.G. Epidemiology and Outcomes of Young-Onset Esophageal Adenocarcinoma: An Analysis from a Population-Based Database. Cancer Epidemiol. Biomark. Prev. 2021, 30, 142–149. [Google Scholar] [CrossRef]
- van Nistelrooij, A.M.; van Marion, R.; Biermann, K.; Spaander, M.C.; van Lanschot, J.J.; Wijnhoven, B.P.; Dinjens, W.N. Early onset esophageal adenocarcinoma: A distinct molecular entity? Oncoscience 2016, 3, 42–48. [Google Scholar] [CrossRef]
- Spechler, S.J.; Souza, R.F. Barrett’s esophagus. N. Engl. J. Med. 2014, 371, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Burke, E.L.; Mullen, J.T. Barrett’s esophagus: A multifaceted condition. South. Med. J. 1976, 69, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Curtius, K.; Rubenstein, J.H.; Chak, A.; Inadomi, J.M. Computational modelling suggests that Barrett’s oesophagus may be the precursor of all oesophageal adenocarcinomas. Gut 2020, 70, 1435–1440. [Google Scholar] [CrossRef]
- Stawinski, P.M.; Dziadkowiec, K.N.; Kuo, L.A.; Echavarria, J.; Saligram, S. Barrett’s Esophagus: An Updated Review. Diagnostics 2023, 13, 321. [Google Scholar] [CrossRef]
- Brown, L.M.; Swanson, C.A.; Gridley, G.; Swanson, G.M.; Schoenberg, J.B.; Greenberg, R.S.; Silverman, D.T.; Pottern, L.M.; Hayes, R.B.; Schwartz, A.G.; et al. Adenocarcinoma of the esophagus: Role of obesity and diet. J. Natl. Cancer Inst. 1995, 87, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.M.; Silverman, D.T.; Pottern, L.M.; Schoenberg, J.B.; Greenberg, R.S.; Swanson, G.M.; Liff, J.M.; Schwartz, A.G.; Hayes, R.B.; Blot, W.J.; et al. Adenocarcinoma of the esophagus and esophagogastric junction in white men in the United States: Alcohol, tobacco, and socioeconomic factors. Cancer Causes Control 1994, 5, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Pera, M.; Manterola, C.; Vidal, O.; Grande, L. Epidemiology of esophageal adenocarcinoma. J. Surg. Oncol. 2005, 92, 151–159. [Google Scholar] [CrossRef]
- He, H.; Chen, N.; Hou, Y.; Wang, Z.; Zhang, Y.; Zhang, G.; Fu, J. Trends in the incidence and survival of patients with esophageal cancer: A SEER database analysis. Thorac. Cancer 2020, 11, 1121–1128. [Google Scholar] [CrossRef]
- Li, B.; Jin, J.; Guo, D.; Tao, Z.; Hu, X. Immune Checkpoint Inhibitors Combined with Targeted Therapy: The Recent Advances and Future Potentials. Cancers 2023, 15, 2858. [Google Scholar] [CrossRef]
- Ye, F.; Dewanjee, S.; Li, Y.; Jha, N.K.; Chen, Z.S.; Kumar, A.; Vishakha Behl, T.; Jha, S.K.; Tang, H. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol. Cancer 2023, 22, 105. [Google Scholar] [CrossRef]
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 2012, 125 Pt 19, 4423–4433. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.L.; O’Regan, L.; Fry, A.M. Novel insights into the mechanisms of mitotic spindle assembly by NEK kinases. Mol. Cell Oncol. 2016, 3, e1062952. [Google Scholar] [CrossRef]
- Quarmby, L.M.; Mahjoub, M.R. Caught Nek-ing: Cilia and centrioles. J. Cell Sci. 2005, 118 Pt 22, 5161–5169. [Google Scholar] [CrossRef] [PubMed]
- Moniz, L.; Dutt, P.; Haider, N.; Stambolic, V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Boehling, J.; Tran, M.N.; Cheng, T.; Rivera, A.; Collins-Burow, B.M.; Lee, S.B.; Drewry, D.H.; Burow, M.E. NEK Family Review and Correlations with Patient Survival Outcomes in Various Cancer Types. Cancers 2023, 15, 2067. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.L.; Niu, L.; Chen, W.L.; Zhang, Y.Q.; Huang, W.H. Integrative Analysis of the Expression Levels and Prognostic Values for NEK Family Members in Breast Cancer. Front. Genet. 2022, 13, 798170. [Google Scholar] [CrossRef]
- Kimchi, E.T.; Posner, M.C.; Park, J.O.; Darga, T.E.; Kocherginsky, M.; Karrison, T.; Hart, J.; Smith, K.D.; Mezhir, J.J.; Weichselbaum, R.R.; et al. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005, 65, 3146–3154. [Google Scholar] [CrossRef]
- Kim, S.M.; Park, Y.Y.; Park, E.S.; Cho, J.Y.; Izzo, J.G.; Zhang, D.; Kim, S.B.; Lee, J.H.; Bhutani, M.S.; Swisher, S.G.; et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS ONE 2010, 5, e15074. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, C.; Kemmner, W. Wdr66 is a novel marker for risk stratification and involved in epithelial-mesenchymal transition of esophageal squamous cell carcinoma. BMC Cancer 2013, 13, 137. [Google Scholar] [CrossRef]
- Nancarrow, D.J.; Clouston, A.D.; Smithers, B.M.; Gotley, D.C.; Drew, P.A.; Watson, D.I.; Tyagi, S.; Hayward, N.K.; Whiteman, D.C.; Australian Cancer Study and the Study of Digestive Health. Whole genome expression array profiling highlights differences in mucosal defense genes in Barrett’s esophagus and esophageal adenocarcinoma. PLoS ONE 2011, 6, e22513. [Google Scholar] [CrossRef]
- Ferrer-Torres, D.; Nancarrow, D.J.; Kuick, R.; Thomas, D.G.; Nadal, E.; Lin, J.; Chang, A.C.; Reddy, R.M.; Orringer, M.B.; Taylor, J.M.; et al. Genomic similarity between gastroesophageal junction and esophageal Barrett’s adenocarcinomas. Oncotarget 2016, 7, 54867–54882. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Guo, Y.; Chen, H.; Zhao, S.; Washington, K.; Hu, T.; Shyr, Y.; El-Rifai, W. Integrated molecular analysis reveals complex interactions between genomic and epigenomic alterations in esophageal adenocarcinomas. Sci. Rep. 2017, 7, 40729. [Google Scholar] [CrossRef]
- di Pietro, M.; Lao-Sirieix, P.; Boyle, S.; Cassidy, A.; Castillo, D.; Saadi, A.; Eskeland, R.; Fitzgerald, R.C. Evidence for a functional role of epigenetically regulated midcluster HOXB genes in the development of Barrett esophagus. Proc. Natl. Acad. Sci. USA 2012, 109, 9077–9082. [Google Scholar] [CrossRef] [PubMed]
- Hyland, P.L.; Hu, N.; Rotunno, M.; Su, H.; Wang, C.; Wang, L.; Pfeiffer, R.M.; Gherman, B.; Giffen, C.; Dykes, C.; et al. Global changes in gene expression of Barrett’s esophagus compared to normal squamous esophagus and gastric cardia tissues. PLoS ONE 2014, 9, e93219. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.S.; Na, D.; Lee, J.S.; Chae, J.; Kim, E.; Jang, G.; Lee, J.; Min, J.; Ock, C.Y.; Kong, S.H.; et al. Comprehensive Molecular Characterization of Adenocarcinoma of the Gastroesophageal Junction Between Esophageal and Gastric Adenocarcinomas. Ann. Surg. 2022, 275, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Peng, D.F.; Razvi, M.; Chen, H.; Washington, K.; Roessner, A.; Schneider-Stock, R.; El-Rifai, W. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut 2009, 58, 5–15. [Google Scholar] [CrossRef]
- Lee, O.J.; Hong, S.M.; Razvi, M.H.; Peng, D.; Powell, S.M.; Smoklin, M.; Moskaluk, C.A.; El-Rifai, W. Expression of calcium-binding proteins S100A2 and S100A4 in Barrett’s adenocarcinomas. Neoplasia 2006, 8, 843–850. [Google Scholar] [CrossRef][Green Version]
- Muir, C.S.; McKinney, P.A. Cancer of the oesophagus: A global overview. Eur. J. Cancer Prev. 1992, 1, 259–264. [Google Scholar] [CrossRef]
- Letwin, K.; Mizzen, L.; Motro, B.; Ben-David, Y.; Bernstein, A.; Pawson, T. A mammalian dual specificity protein kinase, Nek1, is related to the NIMA cell cycle regulator and highly expressed in meiotic germ cells. EMBO J. 1992, 11, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Brieno-Enriquez, M.A.; Moak, S.L.; Holloway, J.K.; Cohen, P.E. NIMA-related kinase 1 (NEK1) regulates meiosis I spindle assembly by altering the balance between alpha-Adducin and Myosin X. PLoS ONE 2017, 12, e0185780. [Google Scholar] [CrossRef] [PubMed]
- Pelegrini, A.L.; Moura, D.J.; Brenner, B.L.; Ledur, P.F.; Maques, G.P.; Henriques, J.A.; Saffi, J.; Lenz, G. Nek1 silencing slows down DNA repair and blocks DNA damage-induced cell cycle arrest. Mutagenesis 2010, 25, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, C.F.; Riley, D.J.; Chen, P.L. Nek1 kinase functions in DNA damage response and checkpoint control through a pathway independent of ATM and ATR. Cell Cycle 2011, 10, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Lin, H.; Wang, X.; Zuo, Q.; Qin, J.; Zhang, P. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim. Biophys. Sin. 2015, 47, 834–841. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, C.F.; Chiang, H.C.; Pena, M.; Polci, R.; Wei, R.L.; Edwards, R.A.; Hansel, D.E.; Chen, P.L.; Riley, D.J. Mutation of NIMA-related kinase 1 (NEK1) leads to chromosome instability. Mol. Cancer 2011, 10, 5. [Google Scholar] [CrossRef]
- Chen, Y.; Craigen, W.J.; Riley, D.J. Nek1 regulates cell death and mitochondrial membrane permeability through phosphorylation of VDAC1. Cell Cycle 2009, 8, 257–267. [Google Scholar] [CrossRef]
- Martins, M.B.; Perez, A.M.; Bohr, V.A.; Wilson, D.M.; 3rd Kobarg, J. NEK1 deficiency affects mitochondrial functions and the transcriptome of key DNA repair pathways. Mutagenesis 2021, 36, 223–236. [Google Scholar] [CrossRef]
- Patil, M.; Pabla, N.; Ding, H.F.; Dong, Z. Nek1 interacts with Ku80 to assist chromatin loading of replication factors and S-phase progression. Cell Cycle 2013, 12, 2608–2616. [Google Scholar] [CrossRef][Green Version]
- Patil, M.; Pabla, N.; Huang, S.; Dong, Z. Nek1 phosphorylates Von Hippel-Lindau tumor suppressor to promote its proteasomal degradation and ciliary destabilization. Cell Cycle 2013, 12, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Shalom, O.; Shalva, N.; Altschuler, Y.; Motro, B. The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 2008, 582, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, P.; Birkenmeier, E.H.; Birkenmeier, C.S.; Barker, J.E. Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. USA 2000, 97, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Naruse, H.; Ishiura, H.; Mitsui, J.; Takahashi, Y.; Matsukawa, T.; Yoshimura, J.; Doi, K.; Morishita, S.; Goto, J.; Toda, T.; et al. Loss-of-function variants in NEK1 are associated with an increased risk of sporadic ALS in the Japanese population. J. Hum. Genet. 2021, 66, 237–241. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Van Mossevelde, S.; Dillen, L.; De Bleecker, J.L.; Moisse, M.; Van Damme, P.; Van Broeckhoven, C.; van der Zee, J.; Consortium, B. NEK1 genetic variability in a Belgian cohort of ALS and ALS-FTD patients. Neurobiol. Aging 2018, 61, 255.e1–255.e7. [Google Scholar] [CrossRef]
- Kenna, K.P.; van Doormaal, P.T.; Dekker, A.M.; Ticozzi, N.; Kenna, B.J.; Diekstra, F.P.; van Rheenen, W.; van Eijk, K.R.; Jones, A.R.; Keagle, P.; et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 2016, 48, 1037–1042. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, C.F.; Polci, R.; Wei, R.; Riley, D.J.; Chen, P.L. Increased Nek1 expression in renal cell carcinoma cells is associated with decreased sensitivity to DNA-damaging treatment. Oncotarget 2014, 5, 4283–4294. [Google Scholar] [CrossRef][Green Version]
- Zhu, J.; Cai, Y.; Liu, P.; Zhao, W. Frequent Nek1 overexpression in human gliomas. Biochem. Biophys. Res. Commun. 2016, 476, 522–527. [Google Scholar] [CrossRef]
- Singh, V.; Jaiswal, P.K.; Ghosh, I.; Koul, H.K.; Yu, X.; De Benedetti, A. Targeting the TLK1/NEK1 DDR axis with Thioridazine suppresses outgrowth of androgen independent prostate tumors. Int. J. Cancer 2019, 145, 1055–1067. [Google Scholar] [CrossRef]
- Singh, V.; Jaiswal, P.K.; Ghosh, I.; Koul, H.K.; Yu, X.; De Benedetti, A. The TLK1-Nek1 axis promotes prostate cancer progression. Cancer Lett. 2019, 453, 31–141. [Google Scholar] [CrossRef]
- Ghosh, I.; Khalil, M.I.; Mirza, R.; King, J.; Olatunde, D.; De Benedetti, A. NEK1-Mediated Phosphorylation of YAP1 Is Key to Prostate Cancer Progression. Biomedicines 2023, 11, 734. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, X. LINC00883 Promotes Drug Resistance of Glioma Through a microRNA-136/NEK1-Dependent Mechanism. Front. Oncol. 2021, 11, 692265. [Google Scholar] [CrossRef] [PubMed]
- Schultz, S.J.; Fry, A.M.; Sutterlin, C.; Ried, T.; Nigg, E.A. Cell cycle-dependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 1994, 5, 625–635. [Google Scholar] [PubMed]
- Fry, A.M.; Schultz, S.J.; Bartek, J.; Nigg, E.A. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 1995, 270, 12899–12905. [Google Scholar] [CrossRef]
- Tanaka, K.; Parvinen, M.; Nigg, E.A. The in vivo expression pattern of mouse Nek2, a NIMA-related kinase, indicates a role in both mitosis and meiosis. Exp. Cell Res. 1997, 237, 264–274. [Google Scholar] [CrossRef]
- Rhee, K.; Wolgemuth, D.J. The NIMA-related kinase 2, Nek2, is expressed in specific stages of the meiotic cell cycle and associates with meiotic chromosomes. Development 1997, 124, 2167–2177. [Google Scholar] [CrossRef]
- Fry, A.M.; Meraldi, P.; Nigg, E.A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998, 17, 470–481. [Google Scholar] [CrossRef]
- Chen, Y.; Riley, D.J.; Zheng, L.; Chen, P.L.; Lee, W.H. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J. Biol. Chem. 2002, 277, 49408–49416. [Google Scholar] [CrossRef]
- Hayward, D.G.; Fry, A.M. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006, 237, 155–166. [Google Scholar] [CrossRef]
- Jeong, Y.; Lee, J.; Kim, K.; Yoo, J.C.; Rhee, K. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 2007, 120 Pt 12, 2106–2116. [Google Scholar] [CrossRef]
- Gu, Z.; Zhou, W.; Huang, J.; Yang, Y.; Wendlandt, E.; Xu, H.; He, X.; Tricot, G.; Zhan, F. Nek2 is a novel regulator of B cell development and immunological response. Biomed. Res. Int. 2014, 2014, 621082. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Barbagallo, F.; Chieffi, P.; Bourgeois, C.F.; Paronetto, M.P.; Sette, C. The centrosomal kinase NEK2 is a novel splicing factor kinase involved in cell survival. Nucleic Acids Res. 2014, 42, 3218–3227. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zheng, J.; Zhang, H.; Shen, N.; Luo, X.; Shen, C.; Song, P.; Zhang, Y.; Zhang, M.; Yang, S.; et al. Molecular characteristics, oncogenic roles, and relevant immune and pharmacogenomic features of NEK2 in gastric cancer. Int. Immunopharmacol. 2023, 116, 109737. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Yao, W.; Du, X.; Dong, J.; Zhang, X.; Shen, W.; Zhu, S. NEK2 promotes esophageal squamous cell carcinoma cell proliferation, migration and invasion through the Wnt/beta-catenin signaling pathway. Discov. Oncol. 2023, 14, 80. [Google Scholar] [CrossRef]
- Feng, X.; Jiang, Y.; Cui, Y.; Xu, Y.; Zhang, Q.; Xia, Q.; Chen, Y. NEK2 is associated with poor prognosis of clear cell renal cell carcinoma and promotes tumor cell growth and metastasis. Gene 2023, 851, 147040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.R.; Zheng, P.S. NEK2 inactivates the Hippo pathway to advance the proliferation of cervical cancer cells by cooperating with STRIPAK complexes. Cancer Lett. 2022, 549, 215917. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Barbagallo, F.; Caggiano, C.; De Musso, M.; Panzeri, V.; Di Agostino, S.; Paronetto, M.P.; Sette, C. Functional Interaction Between the Oncogenic Kinase NEK2 and Sam68 Promotes a Splicing Program Involved in Migration and Invasion in Triple-Negative Breast Cancer. Front. Oncol. 2022, 12, 880654. [Google Scholar] [CrossRef]
- Feng, X.; Guo, J.; An, G.; Wu, Y.; Liu, Z.; Meng, B.; He, N.; Zhao, X.; Chen, S.; Zhu, Y.; et al. Genetic Aberrations and Interaction of NEK2 and TP53 Accelerate Aggressiveness of Multiple Myeloma. Adv. Sci. 2022, 9, e2104491. [Google Scholar] [CrossRef]
- Bai, R.; Yuan, C.; Sun, W.; Zhang, J.; Luo, Y.; Gao, Y.; Li, Y.; Gong, Y.; Xie, C. NEK2 plays an active role in Tumorigenesis and Tumor Microenvironment in Non-Small Cell Lung Cancer: Retraction. Int. J. Biol. Sci. 2022, 18, 3943. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, X.; Xu, J.; Li, E.; Lao, M.; Tang, T.; Zhang, G.; Guo, C.; Zhang, X.; Chen, W.; et al. NEK2 inhibition triggers anti-pancreatic cancer immunity by targeting PD-L1. Nat. Commun. 2021, 12, 4536. [Google Scholar] [CrossRef]
- Wan, H.; Xu, L.; Zhang, H.; Wu, F.; Zeng, W.; Li, T. High expression of NEK2 promotes gastric cancer progression via activating AKT signaling. J. Physiol. Biochem. 2021, 77, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Hu, H.; Ding, Q.; Wang, M.; Zhu, Y.; Zhang, Z.; Geng, Z.; Lin, S.; Zhou, P. NEK2 promotes the migration and proliferation of ESCC via stabilization of YAP1 by phosphorylation at Thr-143. Cell Commun. Signal 2022, 20, 87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ding, L.; Gong, Y.; Zhao, J.; Zhang, J.; Mao, Z.; Wang, Z.; Zhang, W.; Zhou, R. NEK2 Promotes Cell Proliferation and Glycolysis by Regulating PKM2 Abundance via Phosphorylation in Diffuse Large B-Cell Lymphoma. Front. Oncol. 2021, 11, 677763. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zeng, Y.; Shi, L.; Yang, Q.; Chen, Y.; Wu, G.; Li, G.; Xu, S. Targeting NEK2 impairs oncogenesis and radioresistance via inhibiting the Wnt1/beta-catenin signaling pathway in cervical cancer. J. Exp. Clin. Cancer Res. 2020, 39, 183. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; He, Y.; Meng, B.; Chen, S.; Zhang, J.; Wu, X.; Zhu, Y.; Shen, Y.; Feng, X.; Guan, Y.; et al. NEK2 induces autophagy-mediated bortezomib resistance by stabilizing Beclin-1 in multiple myeloma. Mol. Oncol. 2020, 14, 763–778. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, G.; Tang, T.; Gao, X.; Liang, T. One shoot, three birds: Targeting NEK2 orchestrates chemoradiotherapy, targeted therapy, and immunotherapy in cancer treatment. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188696. [Google Scholar] [CrossRef]
- Melo-Hanchuk, T.D.; Martins, M.B.; Cunha, L.L.; Soares, F.A.; Ward, L.S.; Vassallo, J.; Kobarg, J. Expression of the NEK family in normal and cancer tissue: An immunohistochemical study. BMC Cancer 2020, 20, 23. [Google Scholar] [CrossRef]
- Cao, Y.; Song, J.; Chen, J.; Xiao, J.; Ni, J.; Wu, C. Overexpression of NEK3 is associated with poor prognosis in patients with gastric cancer. Medicine (Baltimore) 2018, 97, e9630. [Google Scholar] [CrossRef]
- Basei, F.L.; de Castro Ferezin, C.; Rodrigues de Oliveira, A.L.; Munoz, J.P.; Zorzano, A.; Kobarg, J. Nek4 regulates mitochondrial respiration and morphology. FEBS J. 2022, 289, 3262–3279. [Google Scholar] [CrossRef]
- Basei, F.L.; Meirelles, G.V.; Righetto, G.L.; Dos Santos Migueleti, D.L.; Smetana, J.H.; Kobarg, J. New interaction partners for Nek4.1 and Nek4.2 isoforms: From the DNA damage response to RNA splicing. Proteome Sci. 2015, 13, 11. [Google Scholar] [CrossRef]
- Nguyen, C.L.; Possemato, R.; Bauerlein, E.L.; Xie, A.; Scully, R.; Hahn, W.C. Nek4 regulates entry into replicative senescence and the response to DNA damage in human fibroblasts. Mol. Cell Biol. 2012, 32, 3963–3977. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.H.; Zhang, L.; Xiao, Z.; Rong, Z.X.; Li, Z.; He, J.; Chen, L.; Ou, D.M.; Liao, W.H.; Sun, L.Q. NEK4 kinase regulates EMT to promote lung cancer metastasis. J. Cell Mol. Med. 2018, 22, 5877–5887. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.L.; Sahota, N.K.; Pelletier, L.; Morrison, C.G.; Fry, A.M. Nek5 promotes centrosome integrity in interphase and loss of centrosome cohesion in mitosis. J. Cell Biol. 2015, 209, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Melo Hanchuk, T.D.; Papa, P.F.; La Guardia, P.G.; Vercesi, A.E.; Kobarg, J. Nek5 interacts with mitochondrial proteins and interferes negatively in mitochondrial mediated cell death and respiration. Cell Signal 2015, 27, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Melo-Hanchuk, T.D.; Slepicka, P.F.; Pelegrini, A.L.; Menck, C.F.M.; Kobarg, J. NEK5 interacts with topoisomerase IIbeta and is involved in the DNA damage response induced by etoposide. J. Cell Biochem. 2019, 120, 16853–16866. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Zhang, J.; Yang, X.; Wu, Z.; Sun, C.; Wang, Z.; Wang, B. NEK5 promotes breast cancer cell proliferation through up-regulation of Cyclin A2. Mol. Carcinog. 2019, 58, 933–943. [Google Scholar] [CrossRef]
- Matossian, M.D.; Elliott, S.; Van Hoang, T.; Burks, H.E.; Wright, M.K.; Alzoubi, M.S.; Yan, T.; Chang, T.; Wathieu, H.; Windsor, G.O.; et al. NEK5 activity regulates the mesenchymal and migratory phenotype in breast cancer cells. Breast Cancer Res. Treat. 2021, 189, 49–61. [Google Scholar] [CrossRef]
- Kandli, M.; Feige, E.; Chen, A.; Kilfin, G.; Motro, B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and nek7) related to the fungal mitotic regulator, NIMA. Genomics 2000, 68, 187–196. [Google Scholar] [CrossRef]
- Belham, C.; Roig, J.; Caldwell, J.A.; Aoyama, Y.; Kemp, B.E.; Comb, M.; Avruch, J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 2003, 278, 34897–34909. [Google Scholar] [CrossRef]
- O’Regan, L.; Fry, A.M. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell Biol. 2009, 29, 3975–3990. [Google Scholar] [CrossRef]
- Feige, E.; Motro, B. The related murine kinases, Nek6 and Nek7, display distinct patterns of expression. Mech. Dev. 2002, 110, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Minoguchi, S.; Minoguchi, M.; Yoshimura, A. Differential control of the NIMA-related kinases, Nek6 and Nek7, by serum stimulation. Biochem. Biophys. Res. Commun. 2003, 301, 899–906. [Google Scholar] [CrossRef] [PubMed]
- de Souza, E.E.; Meirelles, G.V.; Godoy, B.B.; Perez, A.M.; Smetana, J.H.; Doxsey, S.J.; McComb, M.E.; Costello, C.E.; Whelan, S.A.; Kobarg, J. Characterization of the human NEK7 interactome suggests catalytic and regulatory properties distinct from those of NEK6. J. Proteome Res. 2014, 13, 4074–4090. [Google Scholar] [CrossRef] [PubMed]
- Peres de Oliveira, A.; Kazuo Issayama, L.; Betim Pavan, I.C.; Riback Silva, F.; Diniz Melo-Hanchuk, T.; Moreira Simabuco, F.; Kobarg, J. Checking NEKs: Overcoming a Bottleneck in Human Diseases. Molecules 2020, 25, 1778. [Google Scholar] [CrossRef]
- Zhu, M.; Sun, Y.; Xue, H.; Wu, G.; Wang, Z.; Shi, J.; Ma, J.; Gu, B.; Yan, X. NEK6 Promotes the Progression of Osteosarcoma Through Activating STAT3 Signaling Pathway by Down-Regulation of miR-26a-5p. Int. J. Gen. Med. 2023, 16, 2831–2848. [Google Scholar] [CrossRef]
- He, Z.; Ni, X.; Xia, L.; Shao, Z. Overexpression of NIMA-related kinase 6 (NEK6) contributes to malignant growth and dismal prognosis in Human Breast Cancer. Pathol. Res. Pract. 2018, 214, 1648–1654. [Google Scholar] [CrossRef]
- Choudhury, A.D.; Schinzel, A.C.; Cotter, M.B.; Lis, R.T.; Labella, K.; Lock, Y.J.; Izzo, F.; Guney, I.; Bowden, M.; Li, Y.Y.; et al. Castration Resistance in Prostate Cancer Is Mediated by the Kinase NEK6. Cancer Res. 2017, 77, 753–765. [Google Scholar] [CrossRef]
- Kasap, E.; Boyacioglu, S.O.; Korkmaz, M.; Yuksel, E.S.; Unsal, B.; Kahraman, E.; Ozutemiz, O.; Yuceyar, H. Aurora kinase A (AURKA) and never in mitosis gene A-related kinase 6 (NEK6) genes are upregulated in erosive esophagitis and esophageal adenocarcinoma. Exp. Ther. Med. 2012, 4, 33–42. [Google Scholar] [CrossRef]
- Liu, R.; Liu, Y.; Liu, C.; Gao, A.; Wang, L.; Tang, H.; Wu, Q.; Wang, X.; Tian, D.; Qi, Z.; et al. NEK7-Mediated Activation of NLRP3 Inflammasome Is Coordinated by Potassium Efflux/Syk/JNK Signaling During Staphylococcus aureus Infection. Front. Immunol. 2021, 12, 747370. [Google Scholar] [CrossRef]
- Liu, H.; Gu, C.; Liu, M.; Liu, G.; Wang, Y. NEK7 mediated assembly and activation of NLRP3 inflammasome downstream of potassium efflux in ventilator-induced lung injury. Biochem. Pharmacol. 2020, 177, 113998. [Google Scholar] [CrossRef]
- Yan, Z.; Qu, J.; Li, Z.; Yi, J.; Su, Y.; Lin, Q.; Yu, G.; Lin, Z.; Yin, W.; Lu, F.; et al. NEK7 Promotes Pancreatic Cancer Progression And Its Expression Is Correlated With Poor Prognosis. Front. Oncol. 2021, 11, 705797. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.K.; Zhu, X.R.; Zhan, Y.; Yuan, W.Z.; Jin, W.L. NEK7 promotes gastric cancer progression as a cell proliferation regulator. Cancer Cell Int. 2021, 21, 438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Z.; Xu, X.; Wan, Y.; Qu, K.; Fan, H.; Chen, Q.; Sun, X.; Liu, C. Nek7 is overexpressed in hepatocellular carcinoma and promotes hepatocellular carcinoma cell proliferation in vitro and in vivo. Oncotarget 2016, 7, 18620–18630. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, G.; Yuan, Y.; Wu, G.; Wang, S.; Yuan, L. NEK7 interacts with NLRP3 to modulate the pyroptosis in inflammatory bowel disease via NF-kappaB signaling. Cell Death Dis. 2019, 10, 906. [Google Scholar] [CrossRef]
- Poehlmann, A.; Kuester, D.; Malfertheiner, P.; Guenther, T.; Roessner, A. Inflammation and Barrett’s carcinogenesis. Pathol. Res. Pract. 2012, 208, 269–280. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Zhang, Q.; Zhang, X.; Yu, C.; Huo, X.; Cheng, E.; Wang, D.H.; Spechler, S.J.; Souza, R.F. Cancer-related inflammation and Barrett’s carcinogenesis: Interleukin-6 and STAT3 mediate apoptotic resistance in transformed Barrett’s cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G454–G460. [Google Scholar] [CrossRef]
- Otto, E.A.; Trapp, M.L.; Schultheiss, U.T.; Helou, J.; Quarmby, L.M.; Hildebrandt, F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J. Am. Soc. Nephrol. 2008, 19, 587–592. [Google Scholar] [CrossRef]
- McCooke, J.K.; Appels, R.; Barrero, R.A.; Ding, A.; Ozimek-Kulik, J.E.; Bellgard, M.I.; Morahan, G.; Phillips, J.K. A novel mutation causing nephronophthisis in the Lewis polycystic kidney rat localises to a conserved RCC1 domain in Nek8. BMC Genom. 2012, 13, 393. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhao, Y.; Sun, Y.W.; Chen, Y. Never-in-Mitosis A-Related Kinase 8 (NEK8) Regulates Adipogenesis, Glucose Homeostasis, and Obesity. Oxid. Med. Cell Longev. 2022, 2022, 1947067. [Google Scholar] [CrossRef]
- Bowers, A.J.; Boylan, J.F. Nek8, a NIMA family kinase member, is overexpressed in primary human breast tumors. Gene 2004, 328, 135–142. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Ballout, F.; Lu, H.; Hu, T.; Zhu, S.; Chen, Z.; Peng, D. Differential Expression of NEK Kinase Family Members in Esophageal Adenocarcinoma and Barrett’s Esophagus. Cancers 2023, 15, 4821. https://doi.org/10.3390/cancers15194821
Chen L, Ballout F, Lu H, Hu T, Zhu S, Chen Z, Peng D. Differential Expression of NEK Kinase Family Members in Esophageal Adenocarcinoma and Barrett’s Esophagus. Cancers. 2023; 15(19):4821. https://doi.org/10.3390/cancers15194821
Chicago/Turabian StyleChen, Lei, Farah Ballout, Heng Lu, Tianling Hu, Shoumin Zhu, Zheng Chen, and Dunfa Peng. 2023. "Differential Expression of NEK Kinase Family Members in Esophageal Adenocarcinoma and Barrett’s Esophagus" Cancers 15, no. 19: 4821. https://doi.org/10.3390/cancers15194821
APA StyleChen, L., Ballout, F., Lu, H., Hu, T., Zhu, S., Chen, Z., & Peng, D. (2023). Differential Expression of NEK Kinase Family Members in Esophageal Adenocarcinoma and Barrett’s Esophagus. Cancers, 15(19), 4821. https://doi.org/10.3390/cancers15194821





