The Tetraspanin Tspan8 Associates with Endothelin Converting Enzyme ECE1 and Regulates Its Activity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Antibodies
2.3. Production of Tspan8 Knockout Mice
2.4. Endothelin Dosage
2.5. Preparation of Intestinal Tissues and DNA Dosage
2.6. Immunofluorescence
2.7. Immunoprecipitation (IP) and Biotinylation of Cell Surface Proteins
2.8. Mass Spectrometry Protein Identifications [15]
3. Results
3.1. Tspan8 Molecular Partners
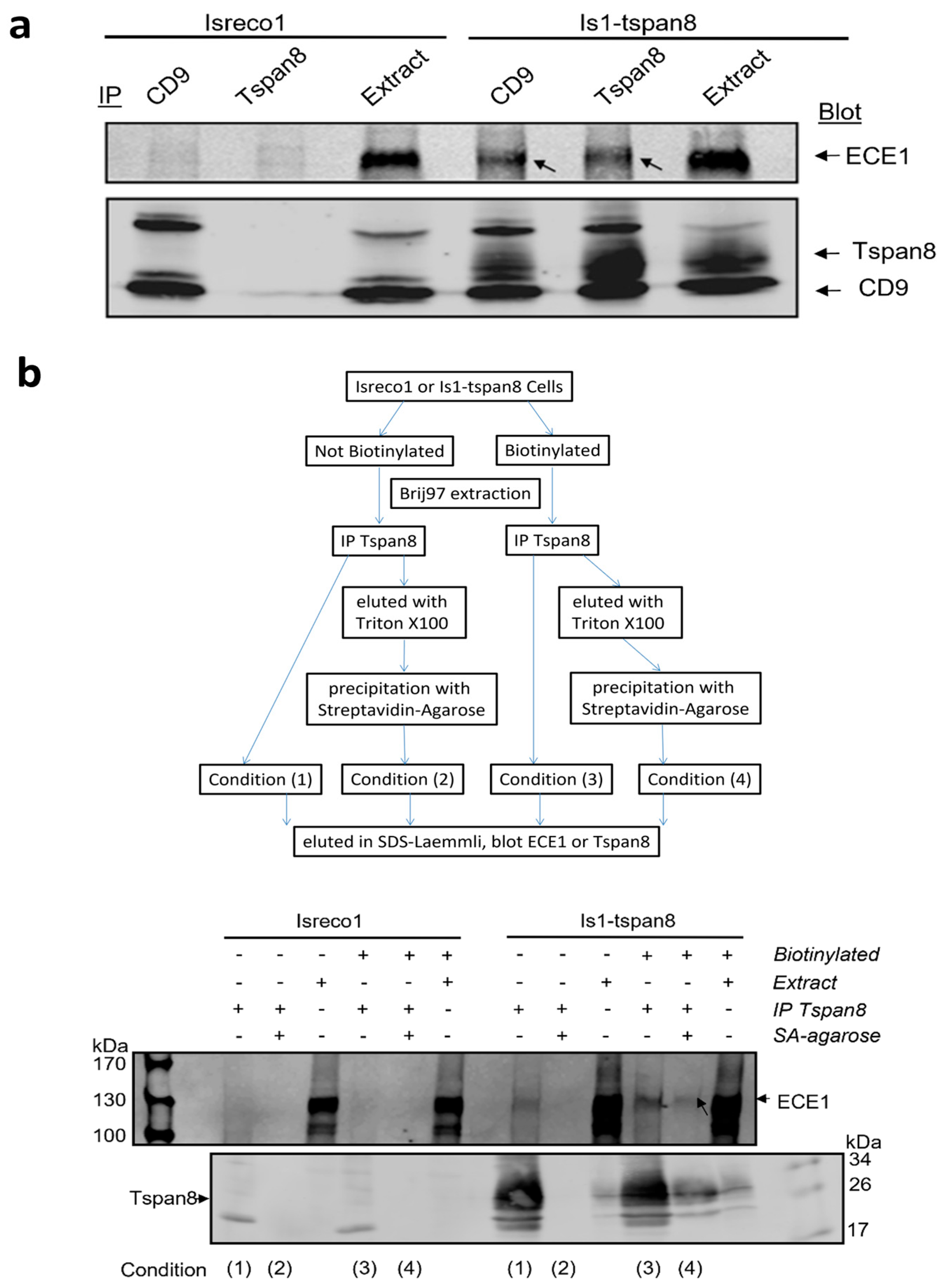
3.2. Association of ECE1 with Tspan8 Is Confirmed by Western Blot
3.3. ECE1 Is Associated with Tspan8 at the Plasma Membrane
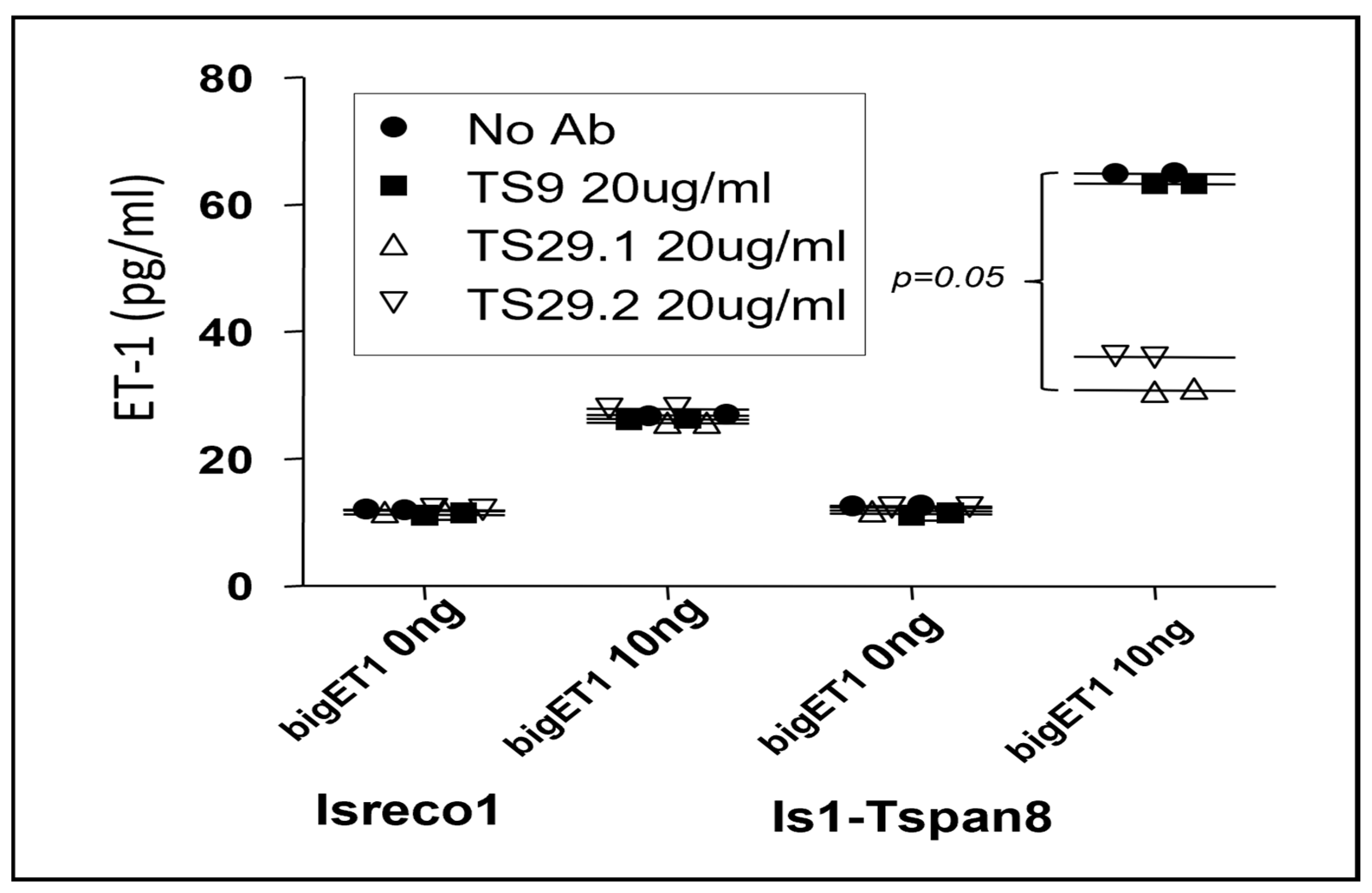
3.4. Modulation of bigET1 to Endothelin Conversion by Cellular ECE1 in the Presence of Tspan8
3.5. Tspan8 Regulation of BigET1 Conversion by Intestinal Tissue
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szala, S.; Kasai, Y.; Steplewski, Z.; Rodeck, U.; Koprowski, H.; Linnenbach, A.J. Molecular cloning of cDNA for the human tumor-associated antigen CO-029 and identification of related transmembrane antigens. Proc. Natl. Acad. Sci. USA 1990, 87, 6833–6837. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, E.; Le Naour, F.; Lagaudriere-Gesbert, C.; Billard, M.; Conjeaud, H.; Boucheix, C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol. 1996, 26, 2657–2665. [Google Scholar] [CrossRef] [PubMed]
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205. [Google Scholar] [CrossRef] [PubMed]
- Berditchevski, F. Complexes of tetraspanins with integrins: More than meets the eye. J. Cell Sci. 2001, 114, 4143–4151. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef]
- Le Naour, F.; Andre, M.; Greco, C.; Billard, M.; Sordat, B.; Emile, J.F.; Lanza, F.; Boucheix, C.; Rubinstein, E. Profiling of the tetraspanin web of human colon cancer cells. Mol. Cell. Proteom. 2006, 5, 845–857. [Google Scholar] [CrossRef]
- Charrin, S.; Le Naour, F.; Silvie, O.; Milhiet, P.E.; Boucheix, C.; Rubinstein, E. Lateral organization of membrane proteins: Tetraspanins spin their web. Biochem. J. 2009, 420, 133–154. [Google Scholar] [CrossRef]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef]
- Espenel, C.; Margeat, E.; Dosset, P.; Arduise, C.; Le Grimellec, C.; Royer, C.A.; Boucheix, C.; Rubinstein, E.; Milhiet, P.E. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 2008, 182, 765–776. [Google Scholar] [CrossRef]
- Peeters, R.; Cuenca-Escalona, J.; Zaal, E.A.; Hoekstra, A.T.; Balvert, A.C.G.; Vidal-Manrique, M.; Blomberg, N.; van Deventer, S.J.; Stienstra, R.; Jellusova, J.; et al. Fatty acid metabolism in aggressive B-cell lymphoma is inhibited by tetraspanin CD37. Nat. Commun. 2022, 13, 5371. [Google Scholar] [CrossRef]
- Jouannet, S.; Saint-Pol, J.; Fernandez, L.; Nguyen, V.; Charrin, S.; Boucheix, C.; Brou, C.; Milhiet, P.E.; Rubinstein, E. TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell. Mol. Life Sci. 2015, 73, 1895–1915. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.; Bralet, M.P.; Ailane, N.; Dubart-Kupperschmitt, A.; Rubinstein, E.; Le Naour, F.; Boucheix, C. E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell motility control in human colon carcinoma. Cancer Res. 2010, 70, 7674–7683. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Maisonial-Besset, A.; Zhu, Y.; Witkowski, T.; Roche, G.; Boucheix, C.; Greco, C.; Degoul, F. Targeting the Tetraspanins with Monoclonal Antibodies in Oncology: Focus on Tspan8/Co-029. Cancers 2019, 11, 179. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.; Lee, S. TSPAN8 as a Novel Emerging Therapeutic Target in Cancer for Monoclonal Antibody Therapy. Biomolecules 2020, 10, 388. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ailane, N.; Sala-Valdes, M.; Haghighi-Rad, F.; Billard, M.; Nguyen, V.; Saffroy, R.; Lemoine, A.; Rubinstein, E.; Boucheix, C.; et al. Multi-factorial modulation of colorectal carcinoma cells motility—Partial coordination by the tetraspanin Co-029/tspan8. Oncotarget 2017, 8, 27454–27470. [Google Scholar] [CrossRef]
- Inoue, A.; Yanagisawa, M.; Kimura, S.; Kasuya, Y.; Miyauchi, T.; Goto, K.; Masaki, T. The human endothelin family: Three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc. Natl. Acad. Sci. USA 1989, 86, 2863–2867. [Google Scholar] [CrossRef]
- Remuzzi, G.; Perico, N.; Benigni, A. New therapeutics that antagonize endothelin: Promises and frustrations. Nat. Rev. Drug Discov. 2002, 1, 986–1001. [Google Scholar] [CrossRef]
- Gray, G.A.; Webb, D.J. The endothelin system and its potential as a therapeutic target in cardiovascular disease. Pharmacol. Ther. 1996, 72, 109–148. [Google Scholar] [CrossRef]
- Gadea, A.; Schinelli, S.; Gallo, V. Endothelin-1 regulates astrocyte proliferation and reactive gliosis via a JNK/c-Jun signaling pathway. J. Neurosci. 2008, 28, 2394–2408. [Google Scholar] [CrossRef]
- Bagnato, A.; Rosano, L.; Spinella, F.; Di Castro, V.; Tecce, R.; Natali, P.G. Endothelin B receptor blockade inhibits dynamics of cell interactions and communications in melanoma cell progression. Cancer Res. 2004, 64, 1436–1443. [Google Scholar] [CrossRef]
- Okazawa, M.; Shiraki, T.; Ninomiya, H.; Kobayashi, S.; Masaki, T. Endothelin-induced apoptosis of A375 human melanoma cells. J. Biol. Chem. 1998, 273, 12584–12592. [Google Scholar] [CrossRef] [PubMed]
- Pflug, B.R.; Zheng, H.; Udan, M.S.; D’Antonio, J.M.; Marshall, F.F.; Brooks, J.D.; Nelson, J.B. Endothelin-1 promotes cell survival in renal cell carcinoma through the ET(A) receptor. Cancer Lett. 2007, 246, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.E.; Fozard, J.R. Regional vasodilation is a prominent feature of the haemodynamic response to endothelin in anaesthetized, spontaneously hypertensive rats. Eur. J. Pharmacol. 1988, 155, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Khodorova, A.; Montmayeur, J.P.; Strichartz, G. Endothelin receptors and pain. J. Pain 2009, 10, 4–28. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.P.; Haymond, T.; Smith, S.N.; Sweitzer, S.M. Evidence for the endothelin system as an emerging therapeutic target for the treatment of chronic pain. J. Pain Res. 2014, 7, 531–545. [Google Scholar] [CrossRef]
- Greco, C.; Basso, L.; Desormeaux, C.; Fournel, A.; Demuynck, B.; Lafendi, L.; Chapiro, S.; Lemoine, A.; Zhu, Y.Y.; Knauf, C.; et al. Endothelin-1 Exhibiting Pro-Nociceptive and Pro-Peristaltic Activities Is Increased in Peritoneal Carcinomatosis. Front. Pain Res. 2021, 2, 613187. [Google Scholar] [CrossRef]
- Irani, S.; Salajegheh, A.; Smith, R.A.; Lam, A.K. A review of the profile of endothelin axis in cancer and its management. Crit. Rev. Oncol. Hematol. 2014, 89, 314–321. [Google Scholar] [CrossRef]
- Enevoldsen, F.C.; Sahana, J.; Wehland, M.; Grimm, D.; Infanger, M.; Kruger, M. Endothelin Receptor Antagonists: Status Quo and Future Perspectives for Targeted Therapy. J. Clin. Med. 2020, 9, 824. [Google Scholar] [CrossRef]
- Cajot, J.F.; Sordat, I.; Silvestre, T.; Sordat, B. Differential display cloning identifies motility-related protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res. 1997, 57, 2593–2597. [Google Scholar]
- Zoller, M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef]
- Ashman, L.K.; Zoller, M. Tetraspanins in Cancer. In Tetraspanins; Berditchevski, F., Rubinstein, E., Eds.; Proteins and Cell Regulation; Springer: Dordrecht, The Netherlands, 2013; pp. 257–298. [Google Scholar]
- El Kharbili, M.; Cario, M.; Bechetoille, N.; Pain, C.; Boucheix, C.; Degoul, F.; Masse, I.; Berthier-Vergnes, O. Tspan8 Drives Melanoma Dermal Invasion by Promoting ProMMP-9 Activation and Basement Membrane Proteolysis in a Keratinocyte-Dependent Manner. Cancers 2020, 12, 1297. [Google Scholar] [CrossRef]
- Voglstaetter, M.; Thomsen, A.R.; Nouvel, J.; Koch, A.; Jank, P.; Navarro, E.G.; Gainey-Schleicher, T.; Khanduri, R.; Gross, A.; Rossner, F.; et al. Tspan8 is expressed in breast cancer and regulates E-cadherin/catenin signalling and metastasis accompanied by increased circulating extracellular vesicles. J. Pathol. 2019, 248, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Gires, O.; Zhu, L.; Liu, J.; Li, J.; Yang, H.; Ju, G.; Huang, J.; Ge, W.; Chen, Y.; et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat. Commun. 2019, 10, 2863. [Google Scholar] [CrossRef] [PubMed]
- Agaesse, G.; Barbollat-Boutrand, L.; Sulpice, E.; Bhajun, R.; El Kharbili, M.; Berthier-Vergnes, O.; Degoul, F.; de la Fouchardiere, A.; Berger, E.; Voeltzel, T.; et al. A large-scale RNAi screen identifies LCMR1 as a critical regulator of Tspan8-mediated melanoma invasion. Oncogene 2017, 36, 5084. [Google Scholar] [CrossRef] [PubMed]
- Agaesse, G.; Barbollat-Boutrand, L.; El Kharbili, M.; Berthier-Vergnes, O.; Masse, I. p53 targets TSPAN8 to prevent invasion in melanoma cells. Oncogenesis 2017, 6, e309. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, X.; Zhu, L.; Lao, Z.; Zhou, T.; Zang, L.; Ge, W.; Jiang, M.; Xu, J.; Cao, Y.; et al. SOX9 is a critical regulator of TSPAN8-mediated metastasis in pancreatic cancer. Oncogene 2021, 40, 4884–4893. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, H.; Wu, L.; Xu, L.; Li, J.; Wang, Q.; Pu, X. Silencing of long non-coding RNA SOX21-AS1 inhibits lung adenocarcinoma invasion and migration by impairing TSPAN8 via transcription factor GATA6. Int. J. Biol. Macromol. 2020, 164, 1294–1303. [Google Scholar] [CrossRef]
- Zhang, H.S.; Liu, H.Y.; Zhou, Z.; Sun, H.L.; Liu, M.Y. TSPAN8 promotes colorectal cancer cell growth and migration in LSD1-dependent manner. Life Sci. 2020, 241, 117114. [Google Scholar] [CrossRef]
- Jackson, C.D.; Turner, A.J. Detection and assessment of endothelin-converting enzyme activity. Methods Mol. Biol. 2002, 206, 107–124. [Google Scholar] [CrossRef]
- Xu, D.; Emoto, N.; Giaid, A.; Slaughter, C.; Kaw, S.; deWit, D.; Yanagisawa, M. ECE-1: A membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell 1994, 78, 473–485. [Google Scholar] [CrossRef]
- Schmidt, M.; Kroger, B.; Jacob, E.; Seulberger, H.; Subkowski, T.; Otter, R.; Meyer, T.; Schmalzing, G.; Hillen, H. Molecular characterization of human and bovine endothelin converting enzyme (ECE-1). FEBS Lett. 1994, 356, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Mzhavia, N.; Pan, H.; Che, F.Y.; Fricker, L.D.; Devi, L.A. Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of regulatory peptides. J. Biol. Chem. 2003, 278, 14704–14711. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Quinto, J.; Herdt, A.; Eckman, C.B.; Eckman, E.A. Endothelin-converting enzymes and related metalloproteases in Alzheimer’s disease. J. Alzheimers Dis. 2013, 33 (Suppl. S1), S101–S110. [Google Scholar] [CrossRef] [PubMed]
- Emoto, N.; Nurhantari, Y.; Alimsardjono, H.; Xie, J.; Yamada, T.; Yanagisawa, M.; Matsuo, M. Constitutive lysosomal targeting and degradation of bovine endothelin-converting enzyme-1a mediated by novel signals in its alternatively spliced cytoplasmic tail. J. Biol. Chem. 1999, 274, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Awano, S.; Dawson, L.A.; Hunter, A.R.; Turner, A.J.; Usmani, B.A. Endothelin system in oral squamous carcinoma cells: Specific siRNA targeting of ECE-1 blocks cell proliferation. Int. J. Cancer 2006, 118, 1645–1652. [Google Scholar] [CrossRef]
- Egidy, G.; Juillerat-Jeanneret, L.; Jeannin, J.F.; Korth, P.; Bosman, F.T.; Pinet, F. Modulation of human colon tumor-stromal interactions by the endothelin system. Am. J. Pathol. 2000, 157, 1863–1874. [Google Scholar] [CrossRef]
- Zhao, K.; Erb, U.; Hackert, T.; Zoller, M.; Yue, S. Corrigendum to “Distorted leukocyte migration, angiogenesis, wound repair and metastasis in Tspan8 and Tspan8/CD151 double knockout mice indicate complementary activities of Tspan8 and CD51” [Biochim. Biophys. Acta 1865(2) (2018) 379–391]. Biochim. Biophys. Acta. Mol. Cell Res. 2018, 1865, 674. [Google Scholar] [CrossRef]
- Lammerding, J.; Kazarov, A.R.; Huang, H.; Lee, R.T.; Hemler, M.E. Tetraspanin CD151 regulates α6β1 integrin adhesion strengthening. Proc. Natl. Acad. Sci. USA 2003, 100, 7616–7621. [Google Scholar] [CrossRef]
- Iwamoto, R.; Higashiyama, S.; Mitamura, T.; Taniguchi, N.; Klagsbrun, M.; Mekada, E. Heparin-binding EGF-like growth factor, which acts as the diphtheria toxin receptor, forms a complex with membrane protein DRAP27/CD9, which up-regulates functional receptors and diphtheria toxin sensitivity. EMBO J. 1994, 13, 2322–2330. [Google Scholar] [CrossRef]
- Feigelson, S.W.; Grabovsky, V.; Shamri, R.; Levy, S.; Alon, R. The CD81 tetraspanin facilitates instantaneous leukocyte VLA-4 adhesion strengthening to vascular cell adhesion molecule 1 (VCAM-1) under shear flow. J. Biol. Chem. 2003, 278, 51203–51212. [Google Scholar] [CrossRef]



| Proteins (Genes Names) | Isreco1—IP CD9 | Is1-tspan8—IP CD9 | Is1-tspan 8—IP Tspan8 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptides (n) | S-Area | Peptides (n) | S-Area | Peptides (n) | S-Area | ||||
| Total | Unique | Total | Unique | Total | Unique | ||||
| ECE1 | 0 | 0 | 0 | 10 | 3 | 2.62 × 107 | 15 | 7 | 1.13 × 108 |
| Tspan6 | 0 | 0 | 0 | 3 | 1 | 3.64 × 107 | 4 | 2 | 2.56 × 107 |
| ITGB1 | 29 | 18 | 2.72 × 109 | 28 | 17 | 3.03 × 109 | 27 | 16 | 2.43 × 109 |
| NOTCH2 | 5 | 1 | 7.98 × 106 | 3 | 1 | 1.30 × 107 | 5 | 3 | 5.07 × 106 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Saint-Pol, J.; Nguyen, V.; Rubinstein, E.; Boucheix, C.; Greco, C. The Tetraspanin Tspan8 Associates with Endothelin Converting Enzyme ECE1 and Regulates Its Activity. Cancers 2023, 15, 4751. https://doi.org/10.3390/cancers15194751
Zhu Y, Saint-Pol J, Nguyen V, Rubinstein E, Boucheix C, Greco C. The Tetraspanin Tspan8 Associates with Endothelin Converting Enzyme ECE1 and Regulates Its Activity. Cancers. 2023; 15(19):4751. https://doi.org/10.3390/cancers15194751
Chicago/Turabian StyleZhu, Yingying, Julien Saint-Pol, Viet Nguyen, Eric Rubinstein, Claude Boucheix, and Céline Greco. 2023. "The Tetraspanin Tspan8 Associates with Endothelin Converting Enzyme ECE1 and Regulates Its Activity" Cancers 15, no. 19: 4751. https://doi.org/10.3390/cancers15194751
APA StyleZhu, Y., Saint-Pol, J., Nguyen, V., Rubinstein, E., Boucheix, C., & Greco, C. (2023). The Tetraspanin Tspan8 Associates with Endothelin Converting Enzyme ECE1 and Regulates Its Activity. Cancers, 15(19), 4751. https://doi.org/10.3390/cancers15194751








