Genome-Wide Association Study Identifies Novel Candidate Variants Associated with Postoperative Nausea and Vomiting
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.1.1. Patients Who Underwent Elective Surgery under General Anesthesia with Propofol or Desflurane
2.1.2. Patients Who Underwent Laparoscopic Gynecological Surgery
2.2. Patient Characteristics and Clinical Data
2.2.1. Patient Characteristics and Clinical Data in Patients Who Underwent Elective Surgery under General Anesthesia with Propofol or Desflurane
2.2.2. Patient Characteristics and Clinical Data in Patients Who Underwent Laparoscopic Gynecological Surgery
2.3. Whole-Genome Genotyping and Quality Control
2.4. Statistical Analysis
2.5. Additional In Silico Analysis
2.5.1. Power Analysis
2.5.2. Reference of Databases
3. Results
3.1. Impact of Clinical Variables on the Incidence of PONV in Subjects Who Underwent Elective Surgery under General Anesthesia with Propofol or Desflurane
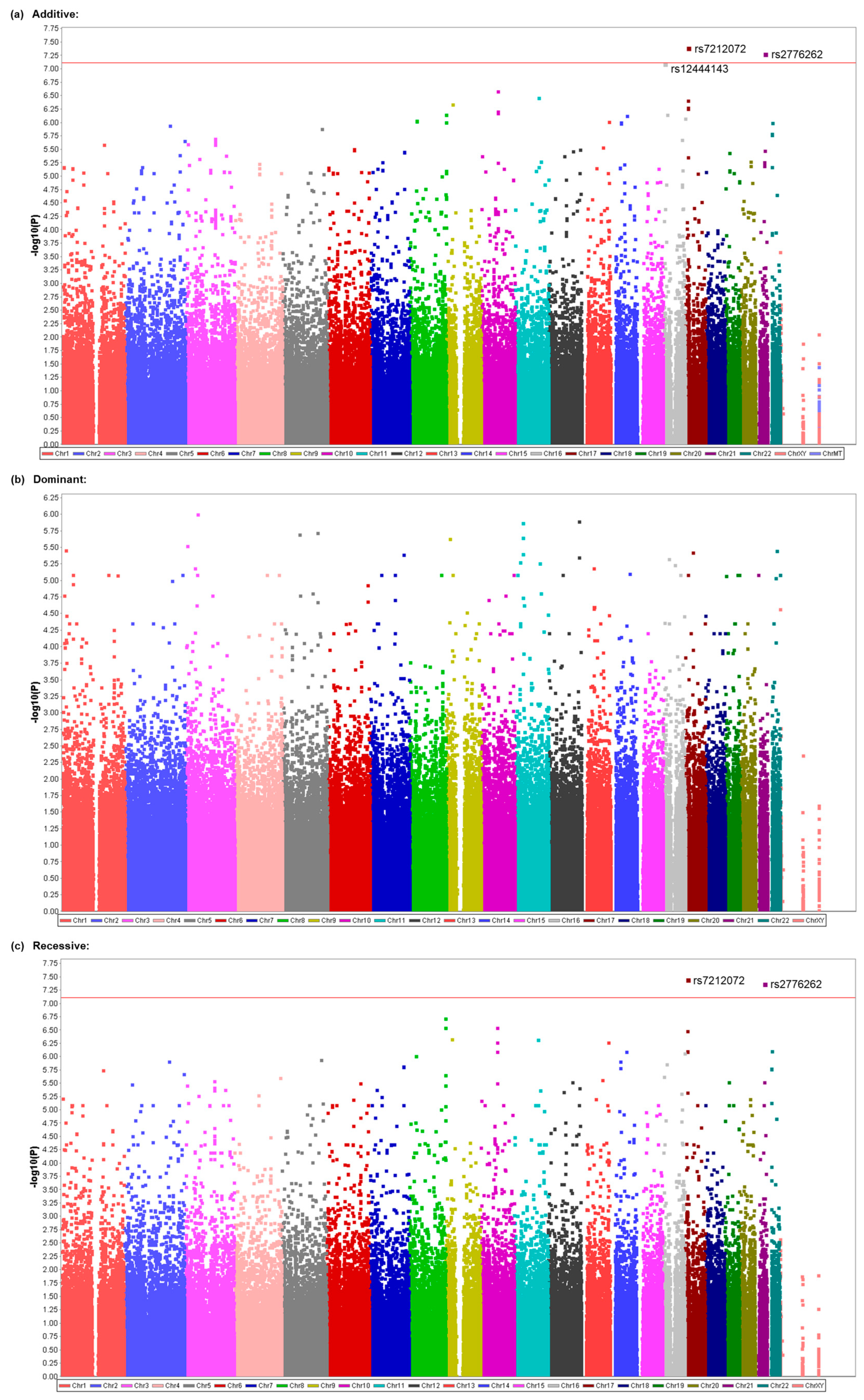
3.2. Identification of Genetic Polymorphisms Associated with PONV in All Patients Who Underwent Elective Surgery under General Anesthesia with Propofol or Desflurane
3.3. Identification of Genetic Polymorphisms Associated with PONV in Patients Who Underwent Elective Surgery under General Anesthesia with Propofol
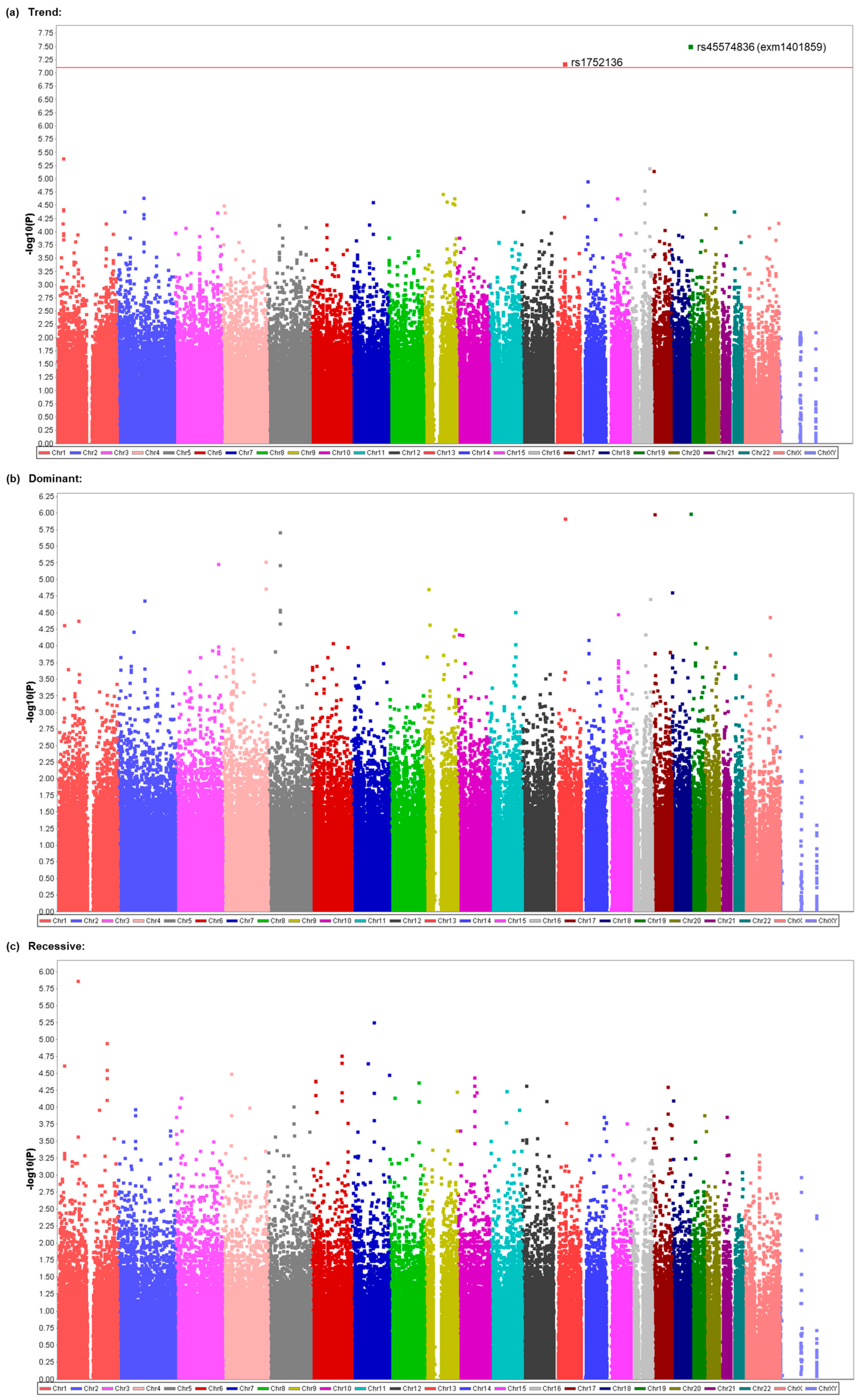
3.4. Replication of Possible Association between the rs45574836 (exm1401859) SNP and Vomiting in Patients Who Underwent Gynecological Laparoscopic Surgery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gan, T.J.; Belani, K.G.; Bergese, S.; Chung, F.; Diemunsch, P.; Habib, A.S.; Jin, Z.; Kovac, A.L.; Meyer, T.A.; Urman, R.D.; et al. Fourth consensus guidelines for the management of postoperative nausea and vomiting. Anesth. Analg. 2020, 131, 411–448. [Google Scholar] [CrossRef]
- Apfel, C.C.; Laara, E.; Koivuranta, M.; Greim, C.A.; Roewer, N. A simplified risk score for predicting postoperative nausea and vomiting: Conclusions from cross-validations between two centers. Anesthesiology 1999, 91, 693–700. [Google Scholar] [CrossRef]
- Gan, T.J.; Diemunsch, P.; Habib, A.S.; Kovac, A.; Kranke, P.; Meyer, T.A.; Watcha, M.; Chung, F.; Angus, S.; Apfel, C.C.; et al. Consensus guidelines for the management of postoperative nausea and vomiting. Anesth. Analg. 2014, 118, 85–113. [Google Scholar] [CrossRef]
- Parra-Sanchez, I.; Abdallah, R.; You, J.; Fu, A.Z.; Grady, M.; Cummings, K., 3rd; Apfel, C.; Sessler, D.I. A time-motion economic analysis of postoperative nausea and vomiting in ambulatory surgery. Can. J. Anaesth. 2012, 59, 366–375. [Google Scholar] [CrossRef]
- Watcha, M.F.; White, P.F. Postoperative nausea and vomiting: Its etiology, treatment, and prevention. Anesthesiology 1992, 77, 162–184. [Google Scholar] [CrossRef]
- Palazzo, M.G.; Strunin, L. Anaesthesia and emesis: I. Etiology. Can. Anaesth. Soc. J. 1984, 31, 178–187. [Google Scholar] [CrossRef]
- Camu, F.; Lauwers, M.H.; Verbessem, D. Incidence and aetiology of postoperative nausea and vomiting. Eur. J. Anaesthesiol. Suppl. 1992, 6, 25–31. [Google Scholar]
- Apfel, C.C.; Heidrich, F.M.; Jukar-Rao, S.; Jalota, L.; Hornuss, C.; Whelan, R.P.; Zhang, K.; Cakmakkaya, O.S. Evidence-based analysis of risk factors for postoperative nausea and vomiting. Br. J. Anaesth. 2012, 109, 742–753. [Google Scholar] [CrossRef]
- Gan, T.J.; Meyer, T.; Apfel, C.C.; Chung, F.; Davis, P.J.; Eubanks, S.; Kovac, A.; Philip, B.K.; Sessler, D.I.; Temo, J.; et al. Consensus guidelines for managing postoperative nausea and vomiting. Anesth. Analg. 2003, 97, 62–71. [Google Scholar] [CrossRef]
- Morino, R.; Ozaki, M.; Nagata, O.; Yokota, M. Incidence of and risk factors for postoperative nausea and vomiting at a Japanese Cancer Center: First large-scale study in Japan. J. Anesth. 2013, 27, 18–24. [Google Scholar] [CrossRef]
- Klenke, S.; Frey, U.H. Genetic variability in postoperative nausea and vomiting: A systematic review. Eur. J. Anaesthesiol. 2020, 37, 959–968. [Google Scholar] [CrossRef]
- Janicki, P.K.; Sugino, S. Genetic factors associated with pharmacotherapy and background sensitivity to postoperative and chemotherapy-induced nausea and vomiting. Exp. Brain Res. 2014, 232, 2613–2625. [Google Scholar] [CrossRef]
- Lopez-Morales, P.; Flores-Funes, D.; Sanchez-Migallon, E.G.; Liron-Ruiz, R.J.; Aguayo-Albasini, J.L. Genetic factors associated with postoperative nausea and vomiting: A systematic review. J. Gastrointest. Surg. 2018, 22, 1645–1651. [Google Scholar] [CrossRef]
- Klenke, S.; de Vries, G.J.; Schiefer, L.; Seyffert, N.; Bachmann, H.S.; Peters, J.; Frey, U.H. Genetic contribution to PONV risk. Anaesth. Crit. Care Pain Med. 2020, 39, 45–51. [Google Scholar] [CrossRef]
- Joy Lin, Y.M.; Hsu, C.D.; Hsieh, H.Y.; Tseng, C.C.; Sun, H.S. Sequence variants of the HTR3A gene contribute to the genetic prediction of postoperative nausea in Taiwan. J. Hum. Genet. 2014, 59, 655–660. [Google Scholar] [CrossRef]
- Rueffert, H.; Thieme, V.; Wallenborn, J.; Lemnitz, N.; Bergmann, A.; Rudlof, K.; Wehner, M.; Olthoff, D.; Kaisers, U.X. Do variations in the 5-HT3A and 5-HT3B serotonin receptor genes (HTR3A and HTR3B) influence the occurrence of postoperative vomiting? Anesth. Analg. 2009, 109, 1442–1447. [Google Scholar] [CrossRef]
- Ma, X.X.; Chen, Q.X.; Wu, S.J.; Hu, Y.; Fang, X.M. Polymorphisms of the HTR3B gene are associated with post-surgery emesis in a Chinese Han population. J. Clin. Pharm. Ther. 2013, 38, 150–155. [Google Scholar] [CrossRef]
- Kim, M.S.; Lee, J.R.; Choi, E.M.; Kim, E.H.; Choi, S.H. Association of 5-HT3B receptor gene polymorphisms with the efficacy of ondansetron for postoperative nausea and vomiting. Yonsei Med. J. 2015, 56, 1415–1420. [Google Scholar] [CrossRef]
- Frey, U.H.; Schnee, C.; Achilles, M.; Silvanus, M.T.; Esser, J.; Peters, J. Postoperative nausea and vomiting: The role of the dopamine D2 receptor TaqIA polymorphism. Eur. J. Anaesthesiol. 2016, 33, 84–89. [Google Scholar] [CrossRef]
- Nakagawa, M.; Kuri, M.; Kambara, N.; Tanigami, H.; Tanaka, H.; Kishi, Y.; Hamajima, N. Dopamine D2 receptor Taq IA polymorphism is associated with postoperative nausea and vomiting. J. Anesth. 2008, 22, 397–403. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, J.D.; Park, S.A.; Oh, C.S.; Kim, S.H. Effects of μ-opioid receptor gene polymorphism on postoperative nausea and vomiting in patients undergoing general anesthesia with remifentanil: Double blinded randomized trial. J. Korean Med. Sci. 2015, 30, 651–657. [Google Scholar] [CrossRef]
- Sugino, S.; Hayase, T.; Higuchi, M.; Saito, K.; Moriya, H.; Kumeta, Y.; Kurosawa, N.; Namiki, A.; Janicki, P.K. Association of μ-opioid receptor gene (OPRM1) haplotypes with postoperative nausea and vomiting. Exp. Brain Res. 2014, 232, 2627–2635. [Google Scholar] [CrossRef]
- Hayase, T.; Sugino, S.; Moriya, H.; Yamakage, M. TACR1 gene polymorphism and sex differences in postoperative nausea and vomiting. Anaesthesia 2015, 70, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Wesmiller, S.W.; Bender, C.M.; Sereika, S.M.; Ahrendt, G.; Bonaventura, M.; Bovbjerg, D.H.; Conley, Y. Association between serotonin transport polymorphisms and postdischarge nausea and vomiting in women following breast cancer surgery. Oncol. Nurs. Forum 2014, 41, 195–202. [Google Scholar] [CrossRef]
- Choi, E.M.; Lee, M.G.; Lee, S.H.; Choi, K.W.; Choi, S.H. Association of ABCB1 polymorphisms with the efficacy of ondansetron for postoperative nausea and vomiting. Anaesthesia 2010, 65, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Farhat, K.; Iqbal, J.; Waheed, A.; Mansoor, Q.; Ismail, M.; Pasha, A.K. Association of anti-emetic efficacy of ondansetron with G2677T polymorphism in a drug transporter gene ABCB1 in Pakistani population. J. Coll. Physicians Surg. Pak. 2015, 25, 486–490. [Google Scholar] [PubMed]
- Dzambazovska-Trajkovska, V.; Nojkov, J.; Kartalov, A.; Kuzmanovska, B.; Spiroska, T.; Seljmani, R.; Trajkovski, G.; Matevska-Geshkovska, N.; Dimovski, A. Association of single-nucleotide polymorhism C3435T in the ABCB1 gene with opioid sensitivity in treatment of postoperative pain. Pril 2016, 37, 73–80. [Google Scholar] [CrossRef]
- Balyan, R.; Zhang, X.; Chidambaran, V.; Martin, L.J.; Mizuno, T.; Fukuda, T.; Vinks, A.A.; Sadhasivam, S. OCT1 genetic variants are associated with postoperative morphine-related adverse effects in children. Pharmacogenomics 2017, 18, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Candiotti, K.A.; Birnbach, D.J.; Lubarsky, D.A.; Nhuch, F.; Kamat, A.; Koch, W.H.; Nikoloff, M.; Wu, L.; Andrews, D. The impact of pharmacogenomics on postoperative nausea and vomiting: Do CYP2D6 allele copy number and polymorphisms affect the success or failure of ondansetron prophylaxis? Anesthesiology 2005, 102, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Janicki, P.K.; Schuler, H.G.; Jarzembowski, T.M.; Rossi, M., 2nd. Prevention of postoperative nausea and vomiting with granisetron and dolasetron in relation to CYP2D6 genotype. Anesth. Analg. 2006, 102, 1127–1133. [Google Scholar] [CrossRef]
- Wesmiller, S.W.; Henker, R.A.; Sereika, S.M.; Donovan, H.S.; Meng, L.; Gruen, G.S.; Tarkin, I.S.; Conley, Y.P. The association of CYP2D6 genotype and postoperative nausea and vomiting in orthopedic trauma patients. Biol. Res. Nurs. 2013, 15, 382–389. [Google Scholar] [CrossRef]
- Janicki, P.K.; Vealey, R.; Liu, J.; Escajeda, J.; Postula, M.; Welker, K. Genome-wide association study using pooled DNA to identify candidate markers mediating susceptibility to postoperative nausea and vomiting. Anesthesiology 2011, 115, 54–64. [Google Scholar] [CrossRef]
- Klenke, S.; de Vries, G.J.; Schiefer, L.; Seyffert, N.; Bachmann, H.S.; Peters, J.; Frey, U.H. CHRM3 rs2165870 polymorphism is independently associated with postoperative nausea and vomiting, but combined prophylaxis is effective. Br. J. Anaesth. 2018, 121, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, S.E.; Bergman, P.H.; Jokela, R.M.; Olkkola, K.T.; Kaunisto, M.A.; Kalso, E.A. Clinical and genetic factors associated with post-operative nausea and vomiting after propofol anaesthesia. Acta Anaesthesiol. Scand. 2023, 67, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Sugino, S.; Konno, D.; Kawai, Y.; Nagasaki, M.; Endo, Y.; Hayase, T.; Yamazaki-Higuchi, M.; Kumeta, Y.; Tachibana, S.; Saito, K.; et al. Long non-coding RNA MIR4300HG polymorphisms are associated with postoperative nausea and vomiting: A genome-wide association study. Hum. Genom. 2020, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- Yeo, I.; Johnson, R. A new family of power transformations to improve normality or symmetry. Biometrika 2000, 87, 954–959. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.V.; Kontsevaya, G.V.; Feofanova, N.A.; Anisimova, M.V.; Serova, I.A.; Gerlinskaya, L.A.; Battulin, N.R.; Moshkin, M.P.; Serov, O.L. Unexpected phenotypic effects of a transgene integration causing a knockout of the endogenous Contactin-5 gene in mice. Transgenic Res. 2018, 27, 1–13. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.E. Postoperative nausea and vomiting. Korean J. Anesthesiol. 2014, 67, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Berisa, T.; Liu, J.Z.; Segurel, L.; Tung, J.Y.; Hinds, D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016, 48, 709–717. [Google Scholar] [CrossRef]
- Folmer, D.E.; Oude Elferink, R.P.J.; Paulusma, C.C. P4 ATPases: Lipid flippases and their role in disease. Biochim. Biophys. Acta 2009, 1791, 628–635. [Google Scholar] [CrossRef]
- Benyamin, R.; Trescot, A.M.; Datta, S.; Buenaventura, R.; Adlaka, R.; Sehgal, N.; Glaser, S.E.; Vallejo, R. Opioid complications and side effects. Pain Physician 2008, 11 (Suppl. 2), S105–S120. [Google Scholar] [CrossRef]

| Demographic Data: | n | Minimum | Maximum | Mean | SD | Median |
|---|---|---|---|---|---|---|
| Gender | ||||||
| male | 432 | |||||
| female | 374 | |||||
| Age [years] | 806 | 23 | 93 | 58.80 | 13.25 | 60.00 |
| Height [cm] | 806 | 140.8 | 184.9 | 162.39 | 8.16 | 161.90 |
| Weight [kg] | 806 | 31.5 | 109.1 | 59.01 | 11.15 | 57.65 |
| Body mass index (BMI) [kg/m2] | 806 | 14.46 | 39.05 | 22.29 | 3.39 | 22.02 |
| History of smoking | 806 | |||||
| absence | 453 | |||||
| presence | 353 | |||||
| Frequency of alcohol drinking per week | 806 | 0 | 7 | 2.65 | 7.00 | 1.00 |
| History of motion sickness | 806 | |||||
| absence | 495 | |||||
| presence | 311 | |||||
| History of PONV | 806 | |||||
| absence | 722 | |||||
| presence | 84 | |||||
| Surgery and clinical data for postoperative period: | n | Minimum | Maximum | Mean | SD | Median |
| Duration of anaesthesia [min] | 806 | 30 | 1050 | 258.62 | 1050.00 | 214.50 |
| Duration of surgery [min] | 806 | 4 | 977 | 203.34 | 977.00 | 163.00 |
| Type of anesthesia | 806 | |||||
| TIVA (propofol) | 442 | |||||
| inhalational anesthesia (desflurane) | 364 | |||||
| Total dose of remifentanil [μg] | 806 | 0 | 24,300 | 3135.61 | 24,300.00 | 2500.00 |
| Total dose of fentanyl [μg] | 806 | 0 | 800 | 198.26 | 800.00 | 200.00 |
| Postoperative administration of narcotic drugs | 806 | |||||
| absence | 427 | |||||
| presence | 379 | |||||
| Postoperative administration of opioids including pentazocine | 806 | |||||
| absence | 340 | |||||
| presence | 466 | |||||
| Experience of pain | 806 | |||||
| absence | 250 | |||||
| presence | 556 | |||||
| Frequency of pain | 806 | 0 | 15 | 2.90 | 15.00 | 2.00 |
| PONV: | n | Minimum | Maximum | Mean | SD | Median |
| Nausea | 806 | |||||
| absence | 541 | |||||
| presence | 265 | |||||
| Frequency of nausea | 806 | 0 | 10 | 0.83 | 10.00 | 0.00 |
| Vomiting | 806 | |||||
| absence | 657 | |||||
| presence | 149 | |||||
| PONV | 806 | |||||
| absence | 526 | |||||
| presence | 280 |
| Model | Rank | CHR | SNP | Position | p | Related Gene | Genotype (Patients) | Genotype (Mean) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/B | B/B | A/A | A/B | B/B | |||||||
| Additive | 1 | 21 | rs2776262 | 36940158 | 0.00000008533 | (LOC100506403) | 2 | 95 | 709 | 4.145 | 0.313 | 0.556 |
| Additive | 2 | 19 | rs12609817 | 53859182 | 0.0000001055 | 4 | 109 | 693 | 2.882 | 0.486 | 0.531 | |
| Additive | 3 | 7 | rs921634 | 47872845 | 0.0000001393 | PKD1L1 | 9 | 145 | 652 | 2.293 | 0.479 | 0.525 |
| Additive | 4 | 5 | rs1587176 | 174851190 | 0.0000002228 | 5 | 172 | 628 | 2.761 | 0.552 | 0.515 | |
| Additive | 5 | 5 | rs10061408 | 117292776 | 0.0000005159 | 6 | 109 | 691 | 2.446 | 0.61 | 0.508 | |
| Additive | 6 | 5 | rs10071777 | 117286831 | 0.000000524 | 6 | 109 | 690 | 2.446 | 0.61 | 0.509 | |
| Additive | 7 | 8 | rs6558049 | 28093041 | 0.0000006488 | 5 | 109 | 692 | 2.747 | 0.508 | 0.525 | |
| Additive | 8 | 16 | rs7192373 | 90032149 | 0.0000006522 | DEF8 | 2 | 58 | 746 | 3.612 | 0.588 | 0.524 |
| Additive | 9 | 7 | rs17173793 | 153523345 | 0.0000008597 | 2 | 111 | 691 | 3.766 | 0.707 | 0.499 | |
| Additive | 10 | 5 | rs13159091 | 148067337 | 0.0000008984 | 2 | 100 | 704 | 3.808 | 0.638 | 0.513 | |
| Additive | 11 | 5 | rs17414326 | 117297258 | 0.000001209 | 2 | 68 | 736 | 3.646 | 0.708 | 0.512 | |
| Additive | 12 | 5 | rs1157652 | 8565638 | 0.000001661 | 21 | 193 | 592 | 1.572 | 0.543 | 0.497 | |
| Additive | 13 | 2 | rs3112976 | 180440437 | 0.000001666 | ZNF385B | 2 | 118 | 686 | 3.449 | 0.595 | 0.518 |
| Additive | 14 | 4 | exm2265817 | 5023112 | 0.000001687 | 2 | 64 | 740 | 3.808 | 0.57 | 0.525 | |
| Additive | 14 | 4 | rs10937615 | 5023112 | 0.000001687 | 2 | 64 | 740 | 3.808 | 0.57 | 0.525 | |
| Additive | 16 | 12 | exm-rs11057830 | 125307053 | 0.000001762 | SCARB1 | 3 | 120 | 683 | 2.986 | 0.523 | 0.528 |
| Additive | 17 | 9 | exm-rs755109 | 100696203 | 0.000001821 | HEMGN | 13 | 176 | 617 | 1.713 | 0.542 | 0.51 |
| Additive | 18 | 9 | rs1475696 | 100691397 | 0.000001857 | HEMGN | 13 | 175 | 618 | 1.713 | 0.54 | 0.511 |
| Additive | 19 | 19 | rs11084950 | 2657397 | 0.000002102 | GNG7 | 2 | 104 | 700 | 3.766 | 0.503 | 0.532 |
| Additive | 20 | 12 | rs2137547 | 26261873 | 0.000002264 | 3 | 91 | 712 | 3.018 | 0.603 | 0.517 | |
| Dominant | 1 | 11 | exm2274524 | 100221580 | 0.00000005555 * | CNTN5 | 0 | 4 | 801 | NA | 2.913 | 0.52 |
| Dominant | 2 | 1 | exm1762808 | 220823972 | 0.0000005544 | MARK1 | 0 | 2 | 802 | NA | 3.766 | 0.53 |
| Dominant | 3 | 8 | rs10087234 | 68735313 | 0.000001101 | 0 | 7 | 799 | NA | 2.205 | 0.522 | |
| Dominant | 4 | 19 | exm1473939 | 42937953 | 0.000001221 | CXCL17 | 1 | 7 | 798 | 0 | 2.64 | 0.519 |
| Dominant | 5 | 3 | rs164464 | 8749012 | 0.000001845 | 0 | 2 | 803 | NA | 3.553 | 0.53 | |
| Dominant | 6 | 3 | rs241055 | 8738790 | 0.000001855 | 0 | 2 | 804 | NA | 3.553 | 0.529 | |
| Dominant | 6 | 3 | rs12497498 | 8751490 | 0.000001855 | 0 | 2 | 804 | NA | 3.553 | 0.529 | |
| Dominant | 6 | 3 | rs17049459 | 8751984 | 0.000001855 | 0 | 2 | 804 | NA | 3.553 | 0.529 | |
| Dominant | 9 | 15 | rs16948440 | 65255168 | 0.000002007 | 0 | 2 | 790 | NA | 3.612 | 0.524 | |
| Dominant | 9 | 17 | rs222843 | 7145981 | 0.000002007 | 121 | 373 | 311 | 0.438 | 0.384 | 0.759 | |
| Dominant | 11 | 2 | rs2075225 | 71063066 | 0.00000221 | 106 | 373 | 326 | 0.451 | 0.392 | 0.727 | |
| Dominant | 12 | 17 | exm1286317 | 7163739 | 0.000002647 | CLDN7 | 121 | 374 | 311 | 0.438 | 0.385 | 0.756 |
| Dominant | 13 | 15 | rs34636936 | 65297261 | 0.000002663 | MTFMT | 0 | 2 | 804 | NA | 3.612 | 0.529 |
| Dominant | 14 | 22 | rs1210829 | 20308800 | 0.000003367 | 21 | 211 | 569 | 0.523 | 0.818 | 0.437 | |
| Dominant | 15 | 4 | rs2204206 | 149713872 | 0.000003862 | 207 | 374 | 225 | 0.41 | 0.471 | 0.761 | |
| Dominant | 16 | 17 | rs222835 | 7134129 | 0.000004137 | DVL2 | 120 | 362 | 324 | 0.481 | 0.365 | 0.749 |
| Dominant | 17 | 17 | rs222837 | 7132556 | 0.000004902 | DVL2 | 118 | 363 | 325 | 0.477 | 0.368 | 0.746 |
| Dominant | 18 | 17 | rs739669 | 7122377 | 0.000005338 | DLG4 | 118 | 362 | 326 | 0.477 | 0.369 | 0.744 |
| Dominant | 19 | 22 | exm2010161 | 38474406 | 0.000006687 | SLC16A8 | 0 | 2 | 804 | NA | 3.278 | 0.53 |
| Dominant | 20 | 4 | rs11734518 | 152213113 | 0.000008271 | 107 | 377 | 322 | 0.705 | 0.642 | 0.356 | |
| Recessive | 1 | 21 | rs2776262 | 36940158 | 0.00000007573 * | (LOC100506403) | 2 | 95 | 709 | 4.145 | 0.313 | 0.556 |
| Recessive | 2 | 19 | rs12609817 | 53859182 | 0.00000009569 | 4 | 109 | 693 | 2.882 | 0.486 | 0.531 | |
| Recessive | 3 | 7 | rs921634 | 47872845 | 0.0000001274 | PKD1L1 | 9 | 145 | 652 | 2.293 | 0.479 | 0.525 |
| Recessive | 4 | 5 | rs1587176 | 174851190 | 0.0000002107 | 5 | 172 | 628 | 2.761 | 0.552 | 0.515 | |
| Recessive | 5 | 8 | rs6558049 | 28093041 | 0.0000006248 | 5 | 109 | 692 | 2.747 | 0.508 | 0.525 | |
| Recessive | 6 | 5 | rs10061408 | 117292776 | 0.0000006556 | 6 | 109 | 691 | 2.446 | 0.61 | 0.508 | |
| Recessive | 7 | 16 | rs7192373 | 90032149 | 0.0000006607 | DEF8 | 2 | 58 | 746 | 3.612 | 0.588 | 0.524 |
| Recessive | 8 | 5 | rs10071777 | 117286831 | 0.0000006649 | 6 | 109 | 690 | 2.446 | 0.61 | 0.509 | |
| Recessive | 9 | 5 | rs13159091 | 148067337 | 0.000001017 | 2 | 100 | 704 | 3.808 | 0.638 | 0.513 | |
| Recessive | 10 | 7 | rs17173793 | 153523345 | 0.00000114 | 2 | 111 | 691 | 3.766 | 0.707 | 0.499 | |
| Recessive | 11 | 5 | rs17414326 | 117297258 | 0.000001587 | 2 | 68 | 736 | 3.646 | 0.708 | 0.512 | |
| Recessive | 12 | 4 | exm2265817 | 5023112 | 0.000001705 | 2 | 64 | 740 | 3.808 | 0.57 | 0.525 | |
| Recessive | 12 | 4 | rs10937615 | 5023112 | 0.000001705 | 2 | 64 | 740 | 3.808 | 0.57 | 0.525 | |
| Recessive | 14 | 2 | rs3112976 | 180440437 | 0.00000171 | ZNF385B | 2 | 118 | 686 | 3.449 | 0.595 | 0.518 |
| Recessive | 15 | 12 | exm-rs11057830 | 125307053 | 0.00000185 | SCARB1 | 3 | 120 | 683 | 2.986 | 0.523 | 0.528 |
| Recessive | 16 | 10 | rs1342273 | 111973339 | 0.00000187 | MXI1 | 12 | 159 | 635 | 1.815 | 0.41 | 0.544 |
| Recessive | 17 | 5 | rs1157652 | 8565638 | 0.000001997 | 21 | 193 | 592 | 1.572 | 0.543 | 0.497 | |
| Recessive | 18 | 19 | rs11084950 | 2657397 | 0.00000201 | GNG7 | 2 | 104 | 700 | 3.766 | 0.503 | 0.532 |
| Recessive | 19 | 5 | rs2591580 | 165406607 | 0.000002074 | 12 | 165 | 629 | 1.801 | 0.438 | 0.538 | |
| Recessive | 20 | 9 | rs10115047 | 96631287 | 0.000002086 | 25 | 238 | 543 | 1.457 | 0.47 | 0.523 | |
| Model | Rank | CHR | SNP | Position | p | Related Gene | Genotype (Patients) | Genotype (Mean) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/B | B/B | A/A | A/B | B/B | |||||||
| Additive | 1 | 17 | rs7212072 | 11310500 | 0.00000003919 * | SHISA6 | 13 | 158 | 271 | 2.02 | 0.523 | 0.504 |
| Additive | 2 | 21 | rs2776262 | 36940158 | 0.00000005028 * | (LOC100506403) | 2 | 45 | 395 | 4.145 | 0.342 | 0.562 |
| Additive | 3 | 16 | rs12444143 | 6071910 | 0.0000000777 * | RBFOX1 | 84 | 197 | 161 | 0.981 | 0.572 | 0.312 |
| Additive | 4 | 10 | rs2499891 | 66680444 | 0.000000254 | 21 | 173 | 248 | 1.499 | 0.586 | 0.454 | |
| Additive | 5 | 11 | rs7110244 | 95910507 | 0.0000003371 | MAML2 | 3 | 61 | 378 | 3.262 | 0.71 | 0.509 |
| Additive | 6 | 17 | rs1513743 | 11306268 | 0.000000375 | SHISA6 | 14 | 159 | 269 | 1.875 | 0.52 | 0.508 |
| Additive | 7 | 9 | rs1937955 | 25721500 | 0.0000004509 | 2 | 60 | 380 | 3.808 | 0.559 | 0.538 | |
| Additive | 8 | 17 | rs4293419 | 11306043 | 0.0000005146 | SHISA6 | 3 | 96 | 343 | 3.413 | 0.646 | 0.505 |
| Additive | 9 | 17 | rs1034899 | 11306660 | 0.000000536 | SHISA6 | 3 | 96 | 342 | 3.413 | 0.646 | 0.507 |
| Additive | 10 | 10 | rs7911209 | 66726865 | 0.0000006068 | 20 | 176 | 245 | 1.501 | 0.573 | 0.467 | |
| Additive | 11 | 10 | rs10733810 | 66723680 | 0.0000006447 | 20 | 168 | 254 | 1.501 | 0.595 | 0.455 | |
| Additive | 12 | 8 | rs1735176 | 146074281 | 0.0000006883 | 18 | 164 | 259 | 1.733 | 0.443 | 0.547 | |
| Additive | 13 | 16 | rs11649132 | 17032787 | 0.0000006937 | 15 | 138 | 289 | 1.587 | 0.601 | 0.48 | |
| Additive | 14 | 14 | rs1256520 | 65737193 | 0.0000007361 | 2 | 81 | 359 | 4.162 | 0.606 | 0.524 | |
| Additive | 14 | 14 | rs1256526 | 65739905 | 0.0000007361 | 2 | 81 | 359 | 4.162 | 0.606 | 0.524 | |
| Additive | 16 | 16 | rs7192373 | 90032149 | 0.0000008101 | DEF8 | 2 | 29 | 411 | 3.612 | 0.677 | 0.532 |
| Additive | 17 | 8 | rs2322976 | 28106141 | 0.0000009019 | 2 | 57 | 383 | 3.808 | 0.601 | 0.532 | |
| Additive | 18 | 8 | rs6558049 | 28093041 | 0.0000009166 | 2 | 59 | 381 | 3.808 | 0.58 | 0.534 | |
| Additive | 19 | 13 | rs816958 | 108522895 | 0.0000009365 | 14 | 96 | 331 | 1.878 | 0.466 | 0.527 | |
| Additive | 20 | 8 | exm734131 | 146076708 | 0.0000009716 | COMMD5 | 18 | 164 | 260 | 1.687 | 0.448 | 0.545 |
| Additive | 20 | 8 | rs1209879 | 146076708 | 0.0000009716 | COMMD5 | 18 | 164 | 260 | 1.687 | 0.448 | 0.545 |
| Dominant | 1 | 3 | rs13100791 | 49641049 | 0.0000009768 | BSN | 0 | 4 | 438 | NA | 2.629 | 0.536 |
| Dominant | 1 | 3 | exm315855 | 49737954 | 0.0000009768 | RNF123 | 0 | 4 | 438 | NA | 2.629 | 0.536 |
| Dominant | 1 | 3 | exm316496 | 49869455 | 0.0000009768 | TRAIP | 0 | 4 | 438 | NA | 2.629 | 0.536 |
| Dominant | 4 | 12 | exm1046926 | 123340542 | 0.000001253 | HIP1R | 0 | 2 | 440 | NA | 3.808 | 0.541 |
| Dominant | 5 | 11 | rs7933966 | 32875597 | 0.000001333 | PRRG4 | 94 | 224 | 124 | 0.501 | 0.379 | 0.915 |
| Dominant | 5 | 11 | exm-rs10767971 | 32895664 | 0.000001333 | 94 | 224 | 124 | 0.501 | 0.379 | 0.915 | |
| Dominant | 5 | 11 | rs10767971 | 32895664 | 0.000001333 | 94 | 224 | 124 | 0.501 | 0.379 | 0.915 | |
| Dominant | 8 | 5 | rs252110 | 141339522 | 0.000001858 | 0 | 8 | 433 | NA | 2.209 | 0.523 | |
| Dominant | 9 | 5 | rs34221525 | 68798118 | 0.000001959 | OCLN | 0 | 7 | 435 | NA | 2.165 | 0.529 |
| Dominant | 10 | 11 | rs4755454 | 32903263 | 0.000002224 | 96 | 225 | 121 | 0.49 | 0.392 | 0.911 | |
| Dominant | 11 | 9 | rs17725257 | 16216102 | 0.000002292 | 0 | 6 | 436 | NA | 2.427 | 0.53 | |
| Dominant | 12 | 3 | rs241055 | 8738790 | 0.000002929 | 0 | 2 | 440 | NA | 3.553 | 0.542 | |
| Dominant | 12 | 3 | rs164464 | 8749012 | 0.000002929 | 0 | 2 | 440 | NA | 3.553 | 0.542 | |
| Dominant | 12 | 3 | rs12497498 | 8751490 | 0.000002929 | 0 | 2 | 440 | NA | 3.553 | 0.542 | |
| Dominant | 12 | 3 | rs17049459 | 8751984 | 0.000002929 | 0 | 2 | 440 | NA | 3.553 | 0.542 | |
| Dominant | 16 | 1 | rs198412 | 11900437 | 0.000003433 | CLCN6,NPPA-AS1 | 0 | 4 | 438 | NA | 2.964 | 0.533 |
| Dominant | 17 | 22 | exm2010161 | 38474406 | 0.000003506 | SLC16A8 | 0 | 2 | 440 | NA | 3.278 | 0.543 |
| Dominant | 18 | 17 | rs17780388 | 30606301 | 0.000003661 | RHBDL3 | 0 | 5 | 437 | NA | 2.671 | 0.531 |
| Dominant | 19 | 11 | rs197697 | 32834416 | 0.000003879 | 92 | 224 | 126 | 0.503 | 0.383 | 0.901 | |
| Dominant | 20 | 7 | rs1982436 | 134470632 | 0.000003965 | CALD1 | 10 | 128 | 304 | 0.414 | 0.918 | 0.407 |
| Recessive | 1 | 17 | rs7212072 | 11310500 | 0.00000003412 * | SHISA6 | 13 | 158 | 271 | 2.02 | 0.523 | 0.504 |
| Recessive | 2 | 21 | rs2776262 | 36940158 | 0.00000004121 * | (LOC100506403) | 2 | 45 | 395 | 4.145 | 0.342 | 0.562 |
| Recessive | 3 | 8 | rs1735176 | 146074281 | 0.0000001881 | 18 | 164 | 259 | 1.733 | 0.443 | 0.547 | |
| Recessive | 4 | 10 | rs2499891 | 66680444 | 0.0000002799 | 21 | 173 | 248 | 1.499 | 0.586 | 0.454 | |
| Recessive | 5 | 8 | exm734131 | 146076708 | 0.0000002805 | COMMD5 | 18 | 164 | 260 | 1.687 | 0.448 | 0.545 |
| Recessive | 5 | 8 | rs1209879 | 146076708 | 0.0000002805 | COMMD5 | 18 | 164 | 260 | 1.687 | 0.448 | 0.545 |
| Recessive | 7 | 17 | rs1513743 | 11306268 | 0.0000003181 | SHISA6 | 14 | 159 | 269 | 1.875 | 0.52 | 0.508 |
| Recessive | 8 | 9 | rs1937955 | 25721500 | 0.0000004615 | 2 | 60 | 380 | 3.808 | 0.559 | 0.538 | |
| Recessive | 9 | 11 | rs7110244 | 95910507 | 0.0000004699 | MAML2 | 3 | 61 | 378 | 3.262 | 0.71 | 0.509 |
| Recessive | 10 | 13 | rs816958 | 108522895 | 0.000000523 | 14 | 96 | 331 | 1.878 | 0.466 | 0.527 | |
| Recessive | 11 | 10 | rs7911209 | 66726865 | 0.0000005299 | 20 | 176 | 245 | 1.501 | 0.573 | 0.467 | |
| Recessive | 12 | 17 | rs4293419 | 11306043 | 0.0000007621 | SHISA6 | 3 | 96 | 343 | 3.413 | 0.646 | 0.505 |
| Recessive | 13 | 22 | rs8142156 | 19692751 | 0.0000007685 | 4 | 64 | 374 | 2.599 | 0.367 | 0.566 | |
| Recessive | 13 | 22 | rs9618670 | 19694333 | 0.0000007685 | 4 | 63 | 375 | 2.599 | 0.373 | 0.564 | |
| Recessive | 15 | 14 | rs1256520 | 65737193 | 0.0000007818 | 2 | 81 | 359 | 4.162 | 0.606 | 0.524 | |
| Recessive | 15 | 14 | rs1256526 | 65739905 | 0.0000007818 | 2 | 81 | 359 | 4.162 | 0.606 | 0.524 | |
| Recessive | 17 | 10 | rs10733810 | 66723680 | 0.0000007878 | 20 | 168 | 254 | 1.501 | 0.595 | 0.455 | |
| Recessive | 18 | 17 | rs1034899 | 11306660 | 0.0000007904 | SHISA6 | 3 | 96 | 342 | 3.413 | 0.646 | 0.507 |
| Recessive | 19 | 16 | rs7192373 | 90032149 | 0.0000008464 | DEF8 | 2 | 29 | 411 | 3.612 | 0.677 | 0.532 |
| Recessive | 20 | 8 | rs6558049 | 28093041 | 0.0000009534 | 2 | 59 | 381 | 3.808 | 0.58 | 0.534 | |
| Recessive | 20 | 8 | rs2322976 | 28106141 | 0.0000009534 | 2 | 57 | 383 | 3.808 | 0.601 | 0.532 | |
| Model | Rank | CHR | SNP | Position | p | Related Gene | Genotype (Vomiting +) | Genotype (Vomiting −) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/B | B/B | A/A | A/B | B/B | |||||||
| Trend | 1 | 19 | exm1401859 | 1806667 | 0.00000002972 * | ATP8B3 | 4 | 31 | 62 | 1 | 44 | 300 |
| Trend | 2 | 13 | rs1752136 | 48726219 | 0.00000006384 * | (LOC105370198) | 0 | 20 | 77 | 0 | 14 | 331 |
| Trend | 3 | 1 | rs6675501 | 24672641 | 0.000003994 | GRHL3 | 42 | 41 | 14 | 73 | 165 | 107 |
| Trend | 4 | 16 | rs16954219 | 80925781 | 0.000006147 | 4 | 43 | 50 | 4 | 83 | 258 | |
| Trend | 5 | 17 | rs16944600 | 11245099 | 0.000006816 | SHISA6 | 4 | 23 | 70 | 27 | 167 | 151 |
| Trend | 6 | 14 | rs10438059 | 33026271 | 0.00001081 | AKAP6 | 13 | 48 | 36 | 16 | 123 | 205 |
| Trend | 7 | 16 | rs12928123 | 61178279 | 0.00001632 | 5 | 33 | 59 | 5 | 60 | 280 | |
| Trend | 7 | 16 | rs9929283 | 61182342 | 0.00001632 | 5 | 33 | 59 | 5 | 60 | 280 | |
| Trend | 9 | 9 | rs10867734 | 83933198 | 0.00001865 | 2 | 14 | 81 | 0 | 16 | 329 | |
| Trend | 10 | 2 | exm-rs6726639 | 112753097 | 0.00002221 | MERTK | 31 | 46 | 15 | 65 | 132 | 131 |
| Trend | 11 | 15 | rs765 | 48035685 | 0.00002252 | SEMA6D | 7 | 49 | 41 | 11 | 106 | 228 |
| Trend | 12 | 9 | rs7850697 | 131095112 | 0.00002275 | COQ4 | 2 | 41 | 54 | 38 | 189 | 118 |
| Trend | 12 | 9 | rs7030121 | 131101919 | 0.00002275 | 2 | 41 | 54 | 38 | 189 | 118 | |
| Trend | 14 | 9 | rs12684445 | 98805260 | 0.00002621 | 11 | 47 | 39 | 14 | 117 | 214 | |
| Trend | 15 | 7 | rs12704714 | 93930713 | 0.00002643 | 32 | 39 | 26 | 42 | 164 | 138 | |
| Trend | 16 | 16 | rs12445491 | 61145947 | 0.00002755 | 8 | 30 | 59 | 6 | 66 | 273 | |
| Trend | 17 | 9 | rs306772 | 124092355 | 0.00002782 | GSN | 1 | 32 | 64 | 1 | 51 | 293 |
| Trend | 18 | 9 | rs2240960 | 131039250 | 0.00002911 | SWI5 | 4 | 37 | 56 | 39 | 186 | 118 |
| Trend | 19 | 4 | rs2086431 | 9509791 | 0.00003047 | 4 | 28 | 65 | 3 | 49 | 293 | |
| Trend | 20 | 14 | rs17098983 | 32868873 | 0.00003088 | AKAP6 | 0 | 20 | 77 | 19 | 128 | 198 |
| Dominant | 1 | 19 | exm1401859 | 1806667 | 0.0000009938 | ATP8B3 | 4 | 31 | 62 | 1 | 44 | 300 |
| Dominant | 2 | 17 | rs16944600 | 11245099 | 0.000001015 | SHISA6 | 4 | 23 | 70 | 27 | 167 | 151 |
| Dominant | 3 | 13 | rs1752136 | 48726219 | 0.000001177 | 0 | 20 | 77 | 0 | 14 | 331 | |
| Dominant | 4 | 5 | rs7715247 | 50318550 | 0.000001902 | 7 | 22 | 68 | 31 | 167 | 147 | |
| Dominant | 5 | 4 | rs11946898 | 181926518 | 0.000005267 | 23 | 62 | 12 | 65 | 157 | 123 | |
| Dominant | 6 | 3 | rs1524511 | 179642067 | 0.000005693 | PEX5L | 10 | 53 | 34 | 23 | 111 | 211 |
| Dominant | 7 | 5 | rs622304 | 50279857 | 0.000005922 | 7 | 20 | 70 | 27 | 159 | 159 | |
| Dominant | 8 | 4 | rs12642493 | 181933056 | 0.00001346 | 23 | 60 | 14 | 64 | 153 | 128 | |
| Dominant | 9 | 9 | rs3808657 | 19127877 | 0.00001358 | 11 | 24 | 61 | 47 | 167 | 131 | |
| Dominant | 10 | 18 | rs8093227 | 2078441 | 0.00001524 | 11 | 62 | 24 | 34 | 140 | 171 | |
| Dominant | 11 | 16 | rs16954219 | 80925781 | 0.00001919 | 4 | 43 | 50 | 4 | 83 | 258 | |
| Dominant | 12 | 2 | exm-rs6726639 | 112753097 | 0.00002021 | MERTK | 31 | 46 | 15 | 65 | 132 | 131 |
| Dominant | 13 | 5 | rs250216 | 50281358 | 0.00002777 | 4 | 21 | 72 | 19 | 152 | 174 | |
| Dominant | 14 | 5 | rs13175573 | 50297517 | 0.00002949 | 5 | 23 | 69 | 22 | 161 | 162 | |
| Dominant | 15 | 11 | rs624584 | 107344724 | 0.00003038 | 4 | 12 | 81 | 12 | 122 | 211 | |
| Dominant | 16 | 15 | rs765 | 48035685 | 0.00003277 | SEMA6D | 7 | 49 | 41 | 11 | 106 | 228 |
| Dominant | 17 | 23 | rs12688309 | 113796171 | 0.00003574 | 0 | 8 | 81 | 5 | 79 | 199 | |
| Dominant | 18 | 1 | rs12027987 | 85206774 | 0.00004063 | 8 | 31 | 58 | 53 | 167 | 125 | |
| Dominant | 19 | 5 | rs12659587 | 50269410 | 0.00004483 | 4 | 21 | 72 | 19 | 150 | 176 | |
| Dominant | 20 | 9 | rs9792672 | 23087926 | 0.00004684 | 1 | 4 | 92 | 4 | 73 | 268 | |
| Recessive | 1 | 1 | rs1195866 | 81678254 | 0.00000133 | 14 | 28 | 55 | 5 | 117 | 219 | |
| Recessive | 2 | 7 | rs12704714 | 93930713 | 0.000005402 | 32 | 39 | 26 | 42 | 164 | 138 | |
| Recessive | 3 | 1 | rs734591 | 203449615 | 0.00001109 | PRELP | 43 | 29 | 25 | 72 | 174 | 99 |
| Recessive | 4 | 6 | rs4897427 | 130810954 | 0.00001688 | 29 | 34 | 33 | 38 | 177 | 130 | |
| Recessive | 5 | 6 | rs9492605 | 130796396 | 0.00002139 | 24 | 31 | 42 | 27 | 154 | 162 | |
| Recessive | 6 | 7 | rs2904188 | 68504050 | 0.00002176 | 11 | 21 | 65 | 4 | 96 | 245 | |
| Recessive | 6 | 7 | rs2869745 | 68539977 | 0.00002176 | 11 | 22 | 64 | 4 | 98 | 243 | |
| Recessive | 8 | 1 | rs6675501 | 24672641 | 0.00002366 | GRHL3 | 42 | 41 | 14 | 73 | 165 | 107 |
| Recessive | 9 | 1 | rs880878 | 203435718 | 0.00002728 | 43 | 30 | 24 | 76 | 172 | 97 | |
| Recessive | 10 | 4 | rs16994732 | 38717953 | 0.00003138 | 8 | 27 | 62 | 1 | 89 | 255 | |
| Recessive | 11 | 7 | rs12698219 | 158145606 | 0.0000321 | PTPRN2 | 9 | 18 | 70 | 2 | 65 | 278 |
| Recessive | 12 | 10 | rs7895191 | 70221267 | 0.00003529 | DNA2 | 0 | 46 | 51 | 41 | 143 | 161 |
| Recessive | 12 | 10 | rs12220316 | 70230840 | 0.00003529 | DNA2 | 0 | 46 | 51 | 41 | 143 | 161 |
| Recessive | 14 | 1 | rs3766902 | 203478370 | 0.00003617 | 39 | 36 | 22 | 65 | 158 | 122 | |
| Recessive | 15 | 6 | rs4395717 | 20826281 | 0.00003964 | CDKAL1 | 7 | 59 | 30 | 89 | 174 | 82 |
| Recessive | 16 | 6 | rs4077405 | 20876683 | 0.00004018 | CDKAL1 | 7 | 60 | 30 | 88 | 174 | 83 |
| Recessive | 17 | 8 | exm719103 | 124440174 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | rs6999234 | 124440174 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | exm719120 | 124448736 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | rs7014678 | 124448736 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | exm719122 | 124448804 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | exm719125 | 124449466 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | rs3824250 | 124449466 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | rs13269287 | 124453662 | 0.00004181 | WDYHV1 | 28 | 44 | 25 | 38 | 181 | 126 |
| Recessive | 17 | 8 | rs7822061 | 124456727 | 0.00004181 | 28 | 44 | 25 | 38 | 181 | 126 | |
| Phenotype (+/−) | GWAS | Replication Study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | p | p | p | Genotype | p | p | p | |||||
| G/G | G/A | A/A | (Trend) | (Dominant) | (Recessive) | G/G | G/A | A/A | (Trend) | (Dominant) | (Recessive) | |
| Nausea (−) | 252 | 39 | 2 | 0.001456 | 0.002497 | 0.3409 | 236 | 54 | 3 | 0.1010 | 0.06452 | 1 |
| Nausea (+) | 110 | 36 | 3 | 27 | 13 | 0 | ||||||
| Vomiting (−) | 300 | 44 | 1 | 2.972 × 10−8 † | 9.938 × 10−7 | 0.009167 | 253 | 59 | 3 | 0.02389 * | 0.03131 * | 1 |
| Vomiting (+) | 62 | 31 | 4 | 10 | 8 | 0 | ||||||
| PONV (−) | 244 | 37 | 1 | 0.0003522 | 0.001215 | 0.05993 | 234 | 52 | 3 | 0.04636 * | 0.02885 * | 1 |
| PONV (+) | 118 | 38 | 4 | 29 | 15 | 0 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishizawa, D.; Morino, R.; Inoue, R.; Ohka, S.; Kasai, S.; Hasegawa, J.; Ebata, Y.; Nakayama, K.; Sumikura, H.; Hayashida, M.; et al. Genome-Wide Association Study Identifies Novel Candidate Variants Associated with Postoperative Nausea and Vomiting. Cancers 2023, 15, 4729. https://doi.org/10.3390/cancers15194729
Nishizawa D, Morino R, Inoue R, Ohka S, Kasai S, Hasegawa J, Ebata Y, Nakayama K, Sumikura H, Hayashida M, et al. Genome-Wide Association Study Identifies Novel Candidate Variants Associated with Postoperative Nausea and Vomiting. Cancers. 2023; 15(19):4729. https://doi.org/10.3390/cancers15194729
Chicago/Turabian StyleNishizawa, Daisuke, Ryozo Morino, Rie Inoue, Seii Ohka, Shinya Kasai, Junko Hasegawa, Yuko Ebata, Kyoko Nakayama, Hiroyuki Sumikura, Masakazu Hayashida, and et al. 2023. "Genome-Wide Association Study Identifies Novel Candidate Variants Associated with Postoperative Nausea and Vomiting" Cancers 15, no. 19: 4729. https://doi.org/10.3390/cancers15194729
APA StyleNishizawa, D., Morino, R., Inoue, R., Ohka, S., Kasai, S., Hasegawa, J., Ebata, Y., Nakayama, K., Sumikura, H., Hayashida, M., Yokota, M., & Ikeda, K. (2023). Genome-Wide Association Study Identifies Novel Candidate Variants Associated with Postoperative Nausea and Vomiting. Cancers, 15(19), 4729. https://doi.org/10.3390/cancers15194729





