Dysregulation of DNAM-1-Mediated NK Cell Anti-Cancer Responses in the Tumor Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
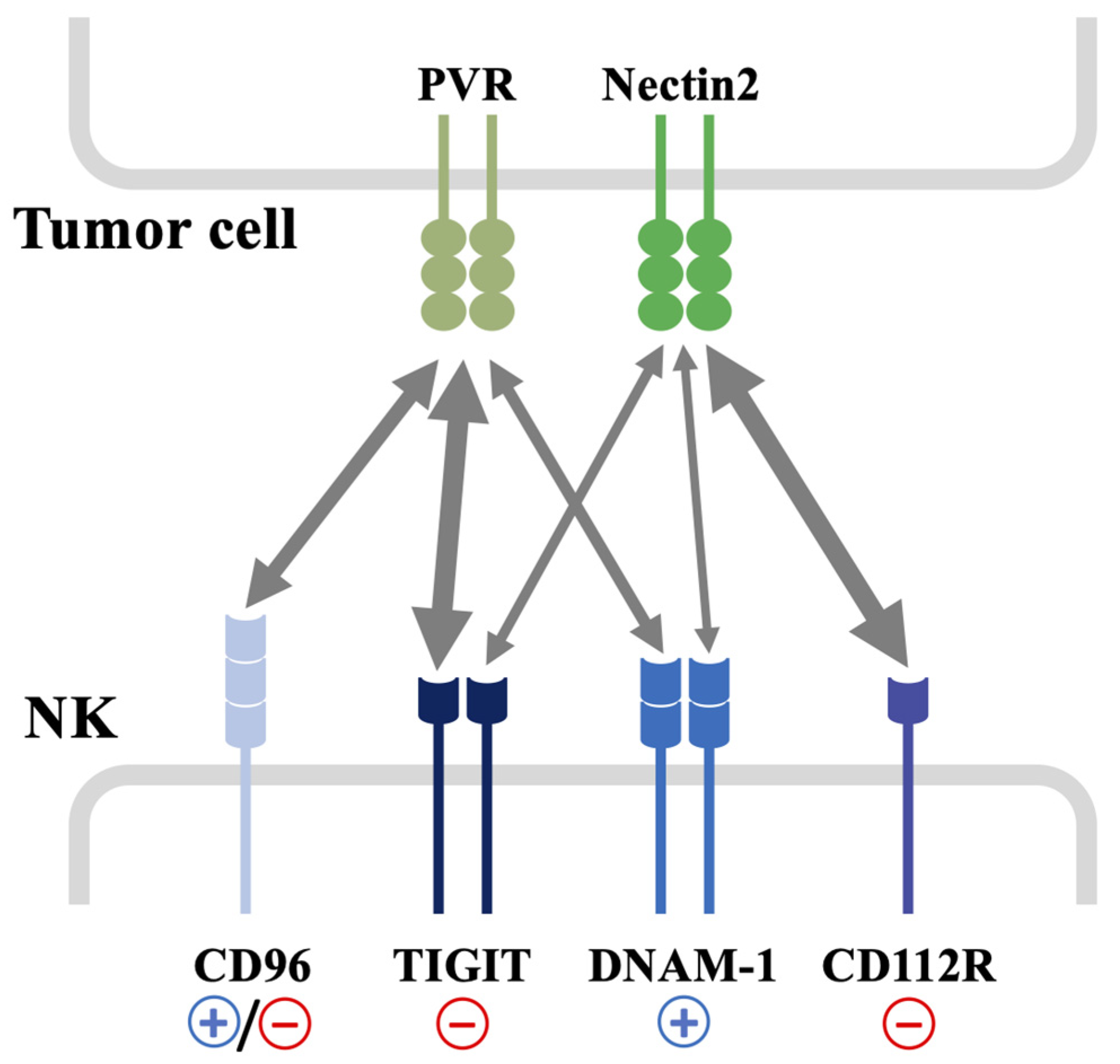
2. DNAM-1 Role in NK Cell Biology
3. DNAM-1 and Its Ligands: Regulation and Function in the Tumor Microenvironment
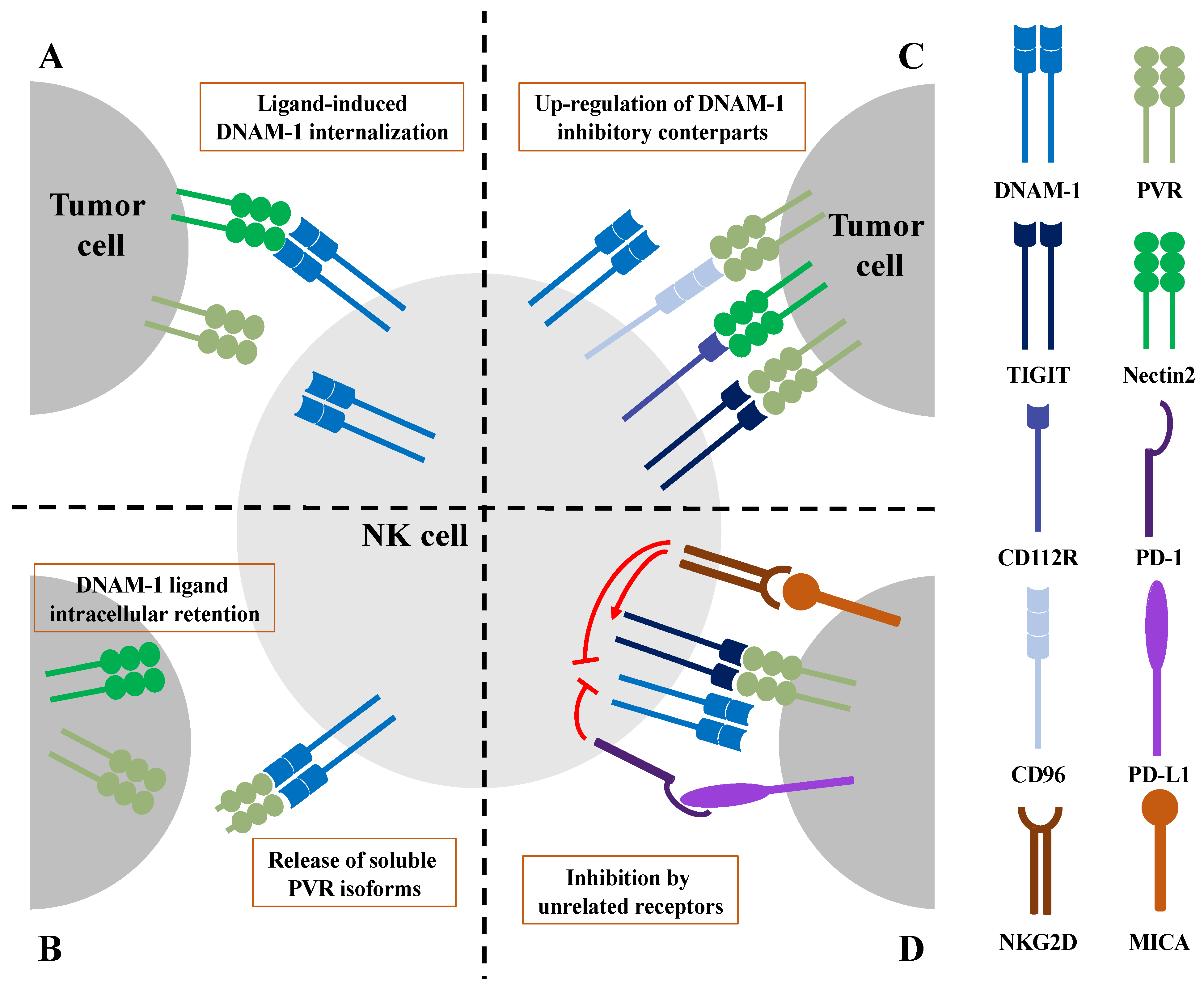
3.1. Ligand-Induced DNAM-1 Internalization Results in Impaired NK Cell Effector Functions
3.2. Impairment of DNAM-1 Functionality by Altered Ligand Expression on Tumor Cells
4. DNAM-1 Dysfunction Caused by Inhibitory Checkpoints and Other Unrelated Receptors
4.1. DNAM-1 Inhibition by Checkpoint Inhibitory Receptors
4.2. DNAM-1 Inhibition upon Chronic Stimulation of NKG2D
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caligiuri, M.A. Human natural killer cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef]
- Morvan, M.G.; Lanier, L.L. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 2016, 16, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Molgora, M.; Cortez, V.S.; Colonna, M. Killing the Invaders: NK Cell Impact in Tumors and Anti-Tumor Therapy. Cancers 2021, 13, 595. [Google Scholar] [CrossRef]
- Lopes, N.; Vivier, E.; Narni-Mancinelli, E. Natural killer cells and type 1 innate lymphoid cells in cancer. Semin. Immunol. 2023, 66, 101709. [Google Scholar] [CrossRef]
- Raulet, D.H.; Gasser, S.; Gowen, B.G.; Deng, W.; Jung, H. Regulation of Ligands for the NKG2D Activating Receptor. Annu. Rev. Immunol. 2013, 31, 413–441. [Google Scholar] [CrossRef]
- Lanier, L.L. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 2015, 3, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Bottino, C.; Castriconi, R.; Pende, D.; Rivera, P.; Nanni, M.; Carnemolla, B.; Cantoni, C.; Grassi, J.; Marcenaro, S.; Reymond, N.; et al. Identification of PVR (CD155) and Nectin-2 (CD112) as Cell Surface Ligands for the Human DNAM-1 (CD226) Activating Molecule. J. Exp. Med. 2003, 198, 557–567. [Google Scholar] [CrossRef]
- Tahara-Hanaoka, S.; Shibuya, K.; Onoda, Y.; Zhang, H.; Yamazaki, S.; Miyamoto, A.; Honda, S.; Lanier, L.L.; Shibuya, A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 2004, 16, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Barrow, A.D.; Martin, C.J.; Colonna, M. The Natural Cytotoxicity Receptors in Health and Disease. Front. Immunol. 2019, 10, 909. [Google Scholar] [CrossRef]
- Ljunggren, H.G.; Kärre, K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 1990, 11, 237–244. [Google Scholar] [CrossRef]
- Guia, S.; Fenis, A.; Vivier, E.; Narni-Mancinelli, E. Activating and inhibitory receptors expressed on innate lymphoid cells. Semin. Immunopathol. 2018, 40, 331–341. [Google Scholar] [CrossRef]
- Corvino, D.; Kumar, A.; Bald, T. Plasticity of NK cells in Cancer. Front. Immunol. 2022, 13, 888313. [Google Scholar] [CrossRef]
- Judge, S.J.; Murphy, W.J.; Canter, R.J. Characterizing the Dysfunctional NK Cell: Assessing the Clinical Relevance of Exhaustion, Anergy, and Senescence. Front. Cell. Infect. Microbiol. 2020, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Pesce, S.; Greppi, M.; Tabellini, G.; Rampinelli, F.; Parolini, S.; Olive, D.; Moretta, L.; Moretta, A.; Marcenaro, E. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J. Allergy Clin. Immunol. 2017, 139, 335–346.e3. [Google Scholar] [CrossRef]
- Bi, J.; Tian, Z. NK Cell Dysfunction and Checkpoint Immunotherapy. Front. Immunol. 2019, 10, 1999. [Google Scholar] [CrossRef] [PubMed]
- Mamessier, E.; Sylvain, A.; Thibult, M.-L.; Houvenaeghel, G.; Jacquemier, J.; Castellano, R.; Gonçalves, A.; André, P.; Romagné, F.; Thibault, G.; et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J. Clin. Investig. 2011, 121, 3609–3622. [Google Scholar] [CrossRef] [PubMed]
- Platonova, S.; Cherfils-Vicini, J.; Damotte, D.; Crozet, L.; Vieillard, V.; Validire, P.; André, P.; Dieu-Nosjean, M.-C.; Alifano, M.; Régnard, J.-F.; et al. Profound Coordinated Alterations of Intratumoral NK Cell Phenotype and Function in Lung Carcinoma. Cancer Res. 2011, 71, 5412–5422. [Google Scholar] [CrossRef]
- Peng, Y.-P.; Zhu, Y.; Zhang, J.-J.; Xu, Z.-K.; Qian, Z.-Y.; Dai, C.-C.; Jiang, K.-R.; Wu, J.-L.; Gao, W.-T.; Li, Q.; et al. Comprehensive analysis of the percentage of surface receptors and cytotoxic granules positive natural killer cells in patients with pancreatic cancer, gastric cancer, and colorectal cancer. J. Transl. Med. 2013, 11, 262. [Google Scholar] [CrossRef]
- Paul, S.; Kulkarni, N.; Shilpi; Lal, G. Intratumoral natural killer cells show reduced effector and cytolytic properties and control the differentiation of effector Th1 cells. OncoImmunology 2016, 5, e1235106. [Google Scholar] [CrossRef]
- Gill, S.; Vasey, A.E.; De Souza, A.; Baker, J.; Smith, A.T.; Kohrt, H.E.; Florek, M.; Gibbs, K.D.; Tate, K.; Ritchie, D.S.; et al. Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood 2012, 119, 5758–5768. [Google Scholar] [CrossRef] [PubMed]
- Conner, M.; Hance, K.W.; Yadavilli, S.; Smothers, J.; Waight, J.D. Emergence of the CD226 Axis in Cancer Immunotherapy. Front. Immunol. 2022, 13, 914406. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Smyth, M.J. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 2015, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Stanietsky, N.; Rovis, T.L.; Glasner, A.; Seidel, E.; Tsukerman, P.; Yamin, R.; Enk, J.; Jonjic, S.; Mandelboim, O. Mouse TIGIT inhibits NK -cell cytotoxicity upon interaction with CD155. Eur. J. Immunol. 2013, 43, 2138–2150. [Google Scholar] [CrossRef]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef]
- Zhu, Y.; Paniccia, A.; Schulick, A.C.; Chen, W.; Koenig, M.R.; Byers, J.T.; Yao, S.; Bevers, S.; Edil, B.H. Identification of CD112R as a novel checkpoint for human T cells. J. Exp. Med. 2016, 213, 167–176. [Google Scholar] [CrossRef]
- Fuchs, A.; Colonna, M. The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance. Semin. Cancer Biol. 2006, 16, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Andrews, D.M.; McLaughlin, N.M.; Yagita, H.; Gilfillan, S.; Colonna, M.; Smyth, M.J. DNAM-1/CD155 Interactions promote cytokine and NK cell-mediated suppression of poorly immunogenic melanoma metastases. J. Immunol. 2010, 184, 902–911. [Google Scholar] [CrossRef]
- Weulersse, M.; Asrir, A.; Pichler, A.C.; Lemaitre, L.; Braun, M.; Carrié, N.; Joubert, M.-V.; Le Moine, M.; Souto, L.D.; Gaud, G.; et al. Eomes-Dependent Loss of the Co-activating Receptor CD226 Restrains CD8+ T Cell Anti-tumor Functions and Limits the Efficacy of Cancer Immunotherapy. Immunity 2020, 53, 824–839.e10. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.-S.; Ko, M.; Choi, D.-S.; Kim, J.H.; Lee, D.-H.; Kang, S.-H.; Kim, I.; Lee, H.J.; Choi, E.K.; Kim, K.-P.; et al. CD226hiCD8+ T Cells Are a Prerequisite for Anti-TIGIT Immunotherapy. Cancer Immunol. Res. 2020, 8, 912–925. [Google Scholar] [CrossRef]
- Burns, G.F.; Triglia, T.; Werkmeister, J.A.; Begley, C.G.; Boyd, A.W. TLiSA1, a human T lineage-specific activation antigen involved in the differentiation of cytotoxic T lymphocytes and anomalous killer cells from their precursors. J. Exp. Med. 1985, 161, 1063–1078. [Google Scholar] [CrossRef]
- Shibuya, A.; Campbell, D.; Hannum, C.; Yssel, H.; Franz-Bacon, K.; McClanahan, T.; Kitamura, T.; Nicholl, J.; Sutherland, G.R.; Lanier, L.L.; et al. DNAM-1, A Novel Adhesion Molecule Involved in the Cytolytic Function of T Lymphocytes. Immunity 1996, 4, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, K.; Lanier, L.L.; Phillips, J.H.; Ochs, H.D.; Shimizu, K.; Nakayama, E.; Nakauchi, H.; Shibuya, A. Physical and Functional Association of LFA-1 with DNAM-1 Adhesion Molecule. Immunity 1999, 11, 615–623. [Google Scholar] [CrossRef]
- Shibuya, A.; Lanier, L.L.; Phillips, J.H. Protein Kinase C Is Involved in the Regulation of Both Signaling and Adhesion Mediated by DNAX Accessory Molecule-1 Receptor. J. Immunol. 1998, 161, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, K.; Shirakawa, J.; Kameyama, T.; Honda, S.-I.; Tahara-Hanaoka, S.; Miyamoto, A.; Onodera, M.; Sumida, T.; Nakauchi, H.; Miyoshi, H.; et al. CD226 (DNAM-1) Is Involved in Lymphocyte Function–associated Antigen 1 Costimulatory Signal for Naive T Cell Differentiation and Proliferation. J. Exp. Med. 2003, 198, 1829–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, N.; Lu, Y.; Davidson, D.; Colonna, M.; Veillette, A. DNAM-1 controls NK cell activation via an ITT-like motif. J. Exp. Med. 2015, 212, 2165–2182. [Google Scholar] [CrossRef] [PubMed]
- Bryceson, Y.T.; March, M.E.; Ljunggren, H.-G.; Long, E.O. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood 2006, 107, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Long, E.O. Complementary Phosphorylation Sites in the Adaptor Protein SLP-76 Promote Synergistic Activation of Natural Killer Cells. Sci. Signal. 2012, 5, ra49. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, F.; Grzywacz, B.; Miller, J.S. Human NK Cell Development: One Road or Many? Front. Immunol. 2019, 10, 2078. [Google Scholar] [CrossRef]
- Bi, J.; Wang, X. Molecular Regulation of NK Cell Maturation. Front. Immunol. 2020, 11, 1945. [Google Scholar] [CrossRef]
- Anfossi, N.; André, P.; Guia, S.; Falk, C.S.; Roetynck, S.; Stewart, C.A.; Breso, V.; Frassati, C.; Reviron, D.; Middleton, D.; et al. Human NK Cell Education by Inhibitory Receptors for MHC Class I. Immunity 2006, 25, 331–342. [Google Scholar] [CrossRef]
- Wagner, A.K.; Kadri, N.; Snäll, J.; Brodin, P.; Gilfillan, S.; Colonna, M.; Bernhardt, G.; Höglund, P.; Kärre, K.; Chambers, B.J. Expression of CD226 is associated to but not required for NK cell education. Nat. Commun. 2017, 8, 15627. [Google Scholar] [CrossRef]
- Martinet, L.; De Andrade, L.F.; Guillerey, C.; Lee, J.S.; Liu, J.; Souza-Fonseca-Guimaraes, F.; Hutchinson, D.S.; Kolesnik, T.B.; Nicholson, S.E.; Huntington, N.D.; et al. DNAM-1 Expression Marks an Alternative Program of NK Cell Maturation. Cell Rep. 2015, 11, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Enqvist, M.; Ask, E.H.; Forslund, E.; Carlsten, M.; Abrahamsen, G.; Béziat, V.; Andersson, S.; Schaffer, M.; Spurkland, A.; Bryceson, Y.; et al. Coordinated Expression of DNAM-1 and LFA-1 in Educated NK Cells. J. Immunol. 2015, 194, 4518–4527. [Google Scholar] [CrossRef] [PubMed]
- López-Botet, M.; De Maria, A.; Muntasell, A.; Della Chiesa, M.; Vilches, C. Adaptive NK cell response to human cytomegalovirus: Facts and open issues. Semin. Immunol. 2023, 65, 101706. [Google Scholar] [CrossRef]
- Nabekura, T.; Kanaya, M.; Shibuya, A.; Fu, G.; Gascoigne, N.R.; Lanier, L.L. Costimulatory Molecule DNAM-1 Is Essential for Optimal Differentiation of Memory Natural Killer Cells during Mouse Cytomegalovirus Infection. Immunity 2014, 40, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Rikitake, Y.; Mandai, K.; Takai, Y. The role of nectins in different types of cell–cell adhesion. J. Cell Sci. 2012, 125 Pt 16, 3713–3722. [Google Scholar] [CrossRef]
- Iwasaki, A.; Welker, R.; Mueller, S.; Linehan, M.; Nomoto, A.; Wimmer, E. Immunofluorescence Analysis of Poliovirus Receptor Expression in Peyer’s Patches of Humans, Primates, and CD155 Transgenic Mice: Implications for Poliovirus Infection. J. Infect. Dis. 2002, 186, 585–592. [Google Scholar] [CrossRef]
- Escalante, N.K.; von Rossum, A.; Lee, M.; Choy, J.C.; Manes, T.D.; Pober, J.S.; Clement, M.; Guedj, K.; Andreata, F.; Morvan, M.; et al. CD155 on Human Vascular Endothelial Cells Attenuates the Acquisition of Effector Functions in CD8 T Cells. Arter. Thromb. Vasc. Biol. 2011, 31, 1177–1184. [Google Scholar] [CrossRef]
- Pende, D.; Castriconi, R.; Romagnani, P.; Spaggiari, G.M.; Marcenaro, S.; Dondero, A.; Lazzeri, E.; Lasagni, L.; Martini, S.; Rivera, P.; et al. Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: Relevance for natural killer-dendritic cell interaction. Blood 2006, 107, 2030–2036. [Google Scholar] [CrossRef]
- Kamran, N.; Takai, Y.; Miyoshi, J.; Biswas, S.K.; Wong, J.S.B.; Gasser, S. Toll-Like Receptor Ligands Induce Expression of the Costimulatory Molecule CD155 on Antigen-Presenting Cells. PLoS ONE 2013, 8, e54406. [Google Scholar] [CrossRef]
- Bellora, F.; Castriconi, R.; Dondero, A.; Reggiardo, G.; Moretta, L.; Mantovani, A.; Moretta, A.; Bottino, C. The interaction of human natural killer cells with either unpolarized or polarized macrophages results in different functional outcomes. Proc. Natl. Acad. Sci. USA 2010, 107, 21659–21664. [Google Scholar] [CrossRef] [PubMed]
- Solecki, D.J.; Gromeier, M.; Mueller, S.; Bernhardt, G.; Wimmer, E. Expression of the Human Poliovirus Receptor/CD155 Gene Is Activated by Sonic Hedgehog. J. Biol. Chem. 2002, 277, 25697–25702. [Google Scholar] [CrossRef] [PubMed]
- Hirota, T.; Irie, K.; Okamoto, R.; Ikeda, W.; Takai, Y. Transcriptional activation of the mouse Necl-5/Tage4/PVR/CD155 gene by fibroblast growth factor or oncogenic Ras through the Raf–MEK–ERK–AP-1 pathway. Oncogene 2005, 24, 2229–2235. [Google Scholar] [CrossRef]
- Soriani, A.; Zingoni, A.; Cerboni, C.; Iannitto, M.L.; Ricciardi, M.R.; Di Gialleonardo, V.; Cippitelli, M.; Fionda, C.; Petrucci, M.T.; Guarini, A.; et al. ATM-ATR–dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood 2009, 113, 3503–3511. [Google Scholar] [CrossRef]
- Fionda, C.; Abruzzese, M.P.; Zingoni, A.; Soriani, A.; Ricci, B.; Molfetta, R.; Paolini, R.; Santoni, A.; Cippitelli, M. Nitric oxide donors increase PVR/CD155 DNAM-1 ligand expression in multiple myeloma cells: Role of DNA damage response activation. BMC Cancer 2015, 15, 17. [Google Scholar] [CrossRef]
- Mekhloufi, A.; Kosta, A.; Stabile, H.; Molfetta, R.; Zingoni, A.; Soriani, A.; Cippitelli, M.; Paolini, R.; Gismondi, A.; Ricciardi, M.R.; et al. Bone Marrow Stromal Cell-Derived IL-8 Upregulates PVR Expression on Multiple Myeloma Cells via NF-kB Transcription Factor. Cancers 2020, 12, 440. [Google Scholar] [CrossRef]
- Briukhovetska, D.; Suarez-Gosalvez, J.; Voigt, C.; Markota, A.; Giannou, A.D.; Schübel, M.; Jobst, J.; Zhang, T.; Dörr, J.; Märkl, F.; et al. T cell-derived interleukin-22 drives the expression of CD155 by cancer cells to suppress NK cell function and promote metastasis. Immunity 2023, 56, 143–161.e11. [Google Scholar] [CrossRef]
- Koike, S.; Horie, H.; Ise, I.; Okitsu, A.; Yoshida, M.; Iizuka, N.; Takeuchi, K.; Takegami, T.; Nomoto, A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990, 9, 3217–3224. [Google Scholar] [CrossRef] [PubMed]
- Baury, B.; Masson, D.; McDermott, B.M., Jr.; Jarry, A.; Blottiere, H.M.; Blanchardie, P.; Laboisse, C.L.; Lustenberger, P.; Racaniello, V.R.; Denis, M.G. Identification of secreted CD155 isoforms. Biochem. Biophys. Res. Commun. 2003, 309, 175–182. [Google Scholar] [CrossRef]
- Kucan Brlić, P.; Lenac Roviš, T.; Cinamon, G.; Tsukerman, P.; Mandelboim, O.; Jonjić, S. Targeting PVR (CD155) and its receptors in anti-tumor therapy. Cell. Mol. Immunol. 2019, 16, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Molfetta, R.; Zitti, B.; Lecce, M.; Milito, N.D.; Stabile, H.; Fionda, C.; Cippitelli, M.; Gismondi, A.; Santoni, A.; Paolini, R. CD155: A Multi-Functional Molecule in Tumor Progression. Int. J. Mol. Sci. 2020, 21, 922. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Madore, J.; Li, X.-Y.; Smyth, M.J. Tumor intrinsic and extrinsic immune functions of CD155. Semin. Cancer Biol. 2020, 65, 189–196. [Google Scholar] [CrossRef]
- Castriconi, R.; Dondero, A.; Corrias, M.V.; Lanino, E.; Pende, D.; Moretta, L.; Bottino, C.; Moretta, A. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: Critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res. 2004, 64, 9180–9184. [Google Scholar] [CrossRef]
- Pende, D.; Spaggiari, G.M.; Marcenaro, S.; Martini, S.; Rivera, P.; Capobianco, A.; Falco, M.; Lanino, E.; Pierri, I.; Zambello, R.; et al. Analysis of the receptor-ligand interactions in the natural killer–mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: Evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112). Blood 2005, 105, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Carlsten, M.; Björkström, N.K.; Norell, H.; Bryceson, Y.; van Hall, T.; Baumann, B.C.; Hanson, M.; Schedvins, K.; Kiessling, R.; Ljunggren, H.-G.; et al. DNAX Accessory Molecule-1 Mediated Recognition of Freshly Isolated Ovarian Carcinoma by Resting Natural Killer Cells. Cancer Res 2007, 67, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- El-Sherbiny, Y.M.; Meade, J.L.; Holmes, T.D.; McGonagle, D.; Mackie, S.L.; Morgan, A.W.; Cook, G.; Feyler, S.; Richards, S.J.; Davies, F.E.; et al. The Requirement for DNAM-1, NKG2D, and NKp46 in the Natural Killer Cell-Mediated Killing of Myeloma Cells. Cancer Res 2007, 67, 8444–8449. [Google Scholar] [CrossRef] [PubMed]
- Lakshmikanth, T.; Burke, S.; Ali, T.H.; Kimpfler, S.; Ursini, F.; Ruggeri, L.; Capanni, M.; Umansky, V.; Paschen, A.; Sucker, A.; et al. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J. Clin. Investig. 2009, 119, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Correa, B.; Morgado, S.; Gayoso, I.; Bergua, J.M.; Casado, J.G.; Arcos, M.J.; Bengochea, M.L.; Duran, E.; Solana, R.; Tarazona, R. Human NK cells in acute myeloid leukaemia patients: Analysis of NK cell-activating receptors and their ligands. Cancer Immunol. Immunother. 2011, 60, 1195–1205. [Google Scholar] [CrossRef]
- Torelli, G.F.; Peragine, N.; Raponi, S.; Pagliara, D.; De Propris, M.S.; Vitale, A.; Bertaina, A.; Barberi, W.; Moretta, L.; Basso, G.; et al. Recognition of adult and pediatric acute lymphoblastic leukemia blasts by natural killer cells. Haematologica 2014, 99, 1248–1254. [Google Scholar] [CrossRef][Green Version]
- Stamm, H.; Klingler, F.; Grossjohann, E.-M.; Muschhammer, J.; Vettorazzi, E.; Heuser, M.; Mock, U.; Thol, F.; Vohwinkel, G.; Latuske, E.; et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene 2018, 37, 5269–5280. [Google Scholar] [CrossRef]
- Han, Y.; Zou, C.; Zhu, C.; Liu, T.; Shen, S.; Cheng, P.; Cheng, W.; Wu, A. The Systematic Landscape of Nectin Family and Nectin-Like Molecules: Functions and Prognostic Value in Low Grade Glioma. Front. Genet. 2021, 12, 718717. [Google Scholar] [CrossRef]
- Martínez-Sánchez, M.V.; Fuster, J.L.; Campillo, J.A.; Galera, A.M.; Bermúdez-Cortés, M.; Llinares, M.E.; Ramos-Elbal, E.; Pascual-Gázquez, J.F.; Fita, A.M.; Martínez-Banaclocha, H.; et al. Expression of NK Cell Receptor Ligands on Leukemic Cells Is Associated with the Outcome of Childhood Acute Leukemia. Cancers 2021, 13, 2294. [Google Scholar] [CrossRef] [PubMed]
- Tahara-Hanaoka, S.; Shibuya, K.; Kai, H.; Miyamoto, A.; Morikawa, Y.; Ohkochi, N.; Honda, S.-I.; Shibuya, A. Tumor rejection by the poliovirus receptor family ligands of the DNAM-1 (CD226) receptor. Blood 2006, 107, 1491–1496. [Google Scholar] [CrossRef] [PubMed]
- Croxford, J.L.; Tang, M.L.F.; Pan, M.F.; Huang, C.W.; Kamran, N.; Phua, C.M.L.; Chng, W.J.; Ng, S.B.; Raulet, D.H.; Gasser, S. ATM-dependent spontaneous regression of early Eμ-myc–induced murine B-cell leukemia depends on natural killer and T cells. Blood 2013, 121, 2512–2521. [Google Scholar] [CrossRef] [PubMed]
- Iguchi-Manaka, A.; Kai, H.; Yamashita, Y.; Shibata, K.; Tahara-Hanaoka, S.; Honda, S.-I.; Yasui, T.; Kikutani, H.; Shibuya, K.; Shibuya, A. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J. Exp. Med. 2008, 205, 2959–2964. [Google Scholar] [CrossRef]
- Gilfillan, S.; Chan, C.J.; Cella, M.; Haynes, N.M.; Rapaport, A.S.; Boles, K.S.; Andrews, D.M.; Smyth, M.J.; Colonna, M. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 2008, 205, 2965–2973. [Google Scholar] [CrossRef]
- Guillerey, C.; De Andrade, L.F.; Vuckovic, S.; Miles, K.; Ngiow, S.F.; Yong, M.C.; Teng, M.W.; Colonna, M.; Ritchie, D.S.; Chesi, M.; et al. Immunosurveillance and therapy of multiple myeloma are CD226 dependent. J. Clin. Investig. 2015, 125, 2077–2089. [Google Scholar] [CrossRef]
- Molfetta, R.; Quatrini, L.; Zitti, B.; Capuano, C.; Galandrini, R.; Santoni, A.; Paolini, R. Regulation of NKG2D Expression and Signaling by Endocytosis. Trends Immunol. 2016, 37, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Costello, R.T.; Sivori, S.; Marcenaro, E.; Lafage-Pochitaloff, M.; Mozziconacci, M.-J.; Reviron, D.; Gastaut, J.-A.; Pende, D.; Olive, D.; Moretta, A. Defective expression and function of natural killer cell–triggering receptors in patients with acute myeloid leukemia. Blood 2002, 99, 3661–3667. [Google Scholar] [CrossRef]
- Fauriat, C.; Just-Landi, S.; Mallet, F.; Arnoulet, C.; Sainty, D.; Olive, D.; Costello, R.T. Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood 2007, 109, 323–330. [Google Scholar] [CrossRef]
- Garcia-Iglesias, T.; Del Toro-Arreola, A.; Albarran-Somoza, B.; Del Toro-Arreola, S.; Sanchez-Hernandez, P.E.; Ramirez-Dueñas, M.G.; Balderas-Peña, L.-M.-A.; Bravo-Cuellar, A.; Ortiz-Lazareno, P.C.; Daneri-Navarro, A. Low NKp30, NKp46 and NKG2D expression and reduced cytotoxic activity on NK cells in cervical cancer and precursor lesions. BMC Cancer 2009, 9, 186. [Google Scholar] [CrossRef]
- Semeraro, M.; Rusakiewicz, S.; Zitvogel, L.; Kroemer, G. Natural killer cell mediated immunosurveillance of pediatric neuroblastoma. OncoImmunology 2015, 4, e1042202. [Google Scholar] [CrossRef] [PubMed]
- Pesce, S.; Tabellini, G.; Cantoni, C.; Patrizi, O.; Coltrini, D.; Rampinelli, F.; Matta, J.; Vivier, E.; Moretta, A.; Parolini, S.; et al. B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. OncoImmunology 2015, 4, e1001224. [Google Scholar] [CrossRef] [PubMed]
- Carlsten, M.; Norell, H.; Bryceson, Y.T.; Poschke, I.; Schedvins, K.; Ljunggren, H.-G.; Kiessling, R.; Malmberg, K.-J. Primary Human Tumor Cells Expressing CD155 Impair Tumor Targeting by Down-Regulating DNAM-1 on NK Cells. J. Immunol. 2009, 183, 4921–4930. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, C.F.; Martínez-Sánchez, M.V.; Gimeno, L.; Mrowiec, A.; Martínez-García, J.; Server-Pastor, G.; Martínez-Escribano, J.A.; Torroba, A.; Ferri, B.; Abellán, D.J.; et al. NK Cell Education in Tumor Immune Surveillance: DNAM-1/KIR Receptor Ratios as Predictive Biomarkers for Solid Tumor Outcome. Cancer Immunol. Res. 2018, 6, 1537–1547. [Google Scholar] [CrossRef]
- Guillamón, C.F.; Martínez-Sánchez, M.V.; Gimeno, L.; Campillo, J.A.; Server-Pastor, G.; Martínez-García, J.; Martínez-Escribano, J.; Torroba, A.; Ferri, B.; Abellán, D.J.; et al. Activating KIRs on Educated NK Cells Support Downregulation of CD226 and Inefficient Tumor Immunosurveillance. Cancer Immunol. Res. 2019, 7, 1307–1317. [Google Scholar] [CrossRef]
- Carlsten, M.; Baumann, B.C.; Simonsson, M.; Jädersten, M.; Forsblom, A.-M.; Hammarstedt, C.; Bryceson, Y.T.; Ljunggren, H.-G.; Hellström-Lindberg, E.; Malmberg, K.-J. Reduced DNAM-1 expression on bone marrow NK cells associated with impaired killing of CD34+ blasts in myelodysplastic syndrome. Leukemia 2010, 24, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Correa, B.; Gayoso, I.; Bergua, J.M.; Casado, J.G.; Morgado, S.; Solana, R.; Tarazona, R. Decreased expression of DNAM-1 on NK cells from acute myeloid leukemia patients. Immunol. Cell Biol. 2012, 90, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Aguilera, A.R.; Sundarrajan, A.; Corvino, D.; Stannard, K.; Krumeich, S.; Das, I.; Lima, L.G.; Guzman, L.G.M.; Li, K.; et al. CD155 on Tumor Cells Drives Resistance to Immunotherapy by Inducing the Degradation of the Activating Receptor CD226 in CD8+ T Cells. Immunity 2020, 53, 805–823.e15. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Jantrapirom, S.; Piccolo, L.; Pruksakorn, D.; Potikanond, S.; Nimlamool, W. Ubiquilin Networking in Cancers. Cancers 2020, 12, 1586. [Google Scholar] [CrossRef] [PubMed]
- Molfetta, R.; Petillo, S.; Cippitelli, M.; Paolini, R. SUMOylation and related post-translational modifications in natural killer cell anti-cancer responses. Front. Cell Dev. Biol. 2023, 11, 1213114. [Google Scholar] [CrossRef]
- Gong, J.; Fang, L.; Liu, R.; Wang, Y.; Xing, J.; Chen, Y.; Zhuang, R.; Zhang, Y.; Zhang, C.; Yang, A.; et al. UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. Eur. J. Immunol. 2014, 44, 3758–3767. [Google Scholar] [CrossRef] [PubMed]
- Zitti, B.; Molfetta, R.; Fionda, C.; Quatrini, L.; Stabile, H.; Lecce, M.; de Turris, V.; Ricciardi, M.R.; Petrucci, M.T.; Cippitelli, M.; et al. Innate immune activating ligand SUMOylation affects tumor cell recognition by NK cells. Sci. Rep. 2017, 7, 10445. [Google Scholar] [CrossRef] [PubMed]
- Molfetta, R.; Milito, N.D.; Zitti, B.; Lecce, M.; Fionda, C.; Cippitelli, M.; Santoni, A.; Paolini, R. The Ubiquitin-proteasome pathway regulates Nectin2/CD112 expression and impairs NK cell recognition and killing. Eur. J. Immunol. 2019, 49, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Fionda, C.; Stabile, H.; Molfetta, R.; Soriani, A.; Bernardini, G.; Zingoni, A.; Gismondi, A.; Paolini, R.; Cippitelli, M.; Santoni, A. Translating the anti-myeloma activity of Natural Killer cells into clinical application. Cancer Treat. Rev. 2018, 70, 255–264. [Google Scholar] [CrossRef]
- Okumura, G.; Iguchi-Manaka, A.; Murata, R.; Yamashita-Kanemaru, Y.; Shibuya, A.; Shibuya, K. Tumor-derived soluble CD155 inhibits DNAM-1–mediated antitumor activity of natural killer cells. J. Exp. Med. 2020, 217, e20191290. [Google Scholar] [CrossRef]
- Iguchi-Manaka, A.; Okumura, G.; Kojima, H.; Cho, Y.; Hirochika, R.; Bando, H.; Sato, T.; Yoshikawa, H.; Hara, H.; Shibuya, A.; et al. Increased Soluble CD155 in the Serum of Cancer Patients. PLoS ONE 2016, 11, e0152982. [Google Scholar] [CrossRef]
- Iguchi-Manaka, A.; Okumura, G.; Ichioka, E.; Kiyomatsu, H.; Ikeda, T.; Bando, H.; Shibuya, A.; Shibuya, K. High expression of soluble CD155 in estrogen receptor-negative breast cancer. Breast Cancer 2020, 27, 92–99. [Google Scholar] [CrossRef]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Yu, X.; Harden, K.; Gonzalez, L.C.; Francesco, M.; Chiang, E.; Irving, B.; Tom, I.; Ivelja, S.; Refino, C.J.; Clark, H.; et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2008, 10, 48–57. [Google Scholar] [CrossRef]
- Joller, N.; Hafler, J.P.; Brynedal, B.; Kassam, N.; Spoerl, S.; Levin, S.D.; Sharpe, A.H.; Kuchroo, V.K. Cutting Edge: TIGIT Has T Cell-Intrinsic Inhibitory Functions. J. Immunol. 2011, 186, 1338–1342. [Google Scholar] [CrossRef]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H.; et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with CD155 and CD155L2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed]
- Lozano, E.; Dominguez-Villar, M.; Kuchroo, V.; Hafler, D.A. The TIGIT/CD226 Axis Regulates Human T Cell Function. J. Immunol. 2012, 188, 3869–3875. [Google Scholar] [CrossRef] [PubMed]
- Stengel, K.F.; Harden-Bowles, K.; Yu, X.; Rouge, L.; Yin, J.; Comps-Agrar, L.; Wiesmann, C.; Bazan, J.F.; Eaton, D.L.; Grogan, J.L. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell–cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc. Natl. Acad. Sci. USA 2012, 109, 5399–5404. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Li, M.; Hu, D.; Li, C.; Ge, B.; Jin, B.; Fan, Z. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 2013, 20, 456–464. [Google Scholar] [CrossRef]
- Li, M.; Xia, P.; Du, Y.; Liu, S.; Huang, G.; Chen, J.; Zhang, H.; Hou, N.; Cheng, X.; Zhou, L.; et al. T-cell Immunoglobulin and ITIM Domain (TIGIT) Receptor/Poliovirus Receptor (CD155) Ligand Engagement Suppresses Interferon-γ Production of Natural Killer Cells via β-Arrestin 2-mediated Negative Signaling. J. Biol. Chem. 2014, 289, 17647–17657. [Google Scholar] [CrossRef]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8+ T Cell Effector Function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef]
- Chauvin, J.-M.; Pagliano, O.; Fourcade, J.; Sun, Z.; Wang, H.; Sander, C.; Kirkwood, J.M.; Chen, T.-H.T.; Maurer, M.; Korman, A.J.; et al. TIGIT and PD-1 impair tumor antigen–specific CD8+ T cells in melanoma patients. J. Clin. Investig. 2015, 125, 2046–2058. [Google Scholar] [CrossRef]
- Thibaudin, M.; Limagne, E.; Hampe, L.; Ballot, E.; Truntzer, C.; Ghiringhelli, F. Targeting PD-L1 and TIGIT could restore intratumoral CD8 T cell function in human colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 2549–2563. [Google Scholar] [CrossRef]
- Harjunpää, H.; Guillerey, C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020, 200, 108–119. [Google Scholar] [CrossRef]
- Florou, V.; Garrido-Laguna, I. Clinical Development of Anti-TIGIT Antibodies for Immunotherapy of Cancer. Curr. Oncol. Rep. 2022, 24, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Seth, S.; Maier, M.; Qiu, Q.; Ravens, I.; Kremmer, E.; Förster, R.; Bernhardt, G. The murine pan T cell marker CD96 is an adhesion receptor for CD155 and nectin-1. Biochem. Biophys. Res. Commun. 2007, 364, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Cella, M.; Giurisato, E.; Shaw, A.S.; Colonna, M. Cutting Edge: CD96 (Tactile) Promotes NK Cell-Target Cell Adhesion by Interacting with the Poliovirus Receptor (CD155). J. Immunol. 2004, 172, 3994–3998. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Huang, Q.; Huang, M.; Wen, H.; Lin, R.; Zheng, M.; Qu, K.; Li, K.; Wei, H.; Xiao, W.; et al. Human CD96 Correlates to Natural Killer Cell Exhaustion and Predicts the Prognosis of Human Hepatocellular Carcinoma. Hepatology 2019, 70, 168–183. [Google Scholar] [CrossRef]
- Georgiev, H.; Ravens, I.; Papadogianni, G.; Bernhardt, G. Coming of Age: CD96 Emerges as Modulator of Immune Responses. Front. Immunol. 2018, 9, 1072. [Google Scholar] [CrossRef]
- Meyer, D.; Seth, S.; Albrecht, J.; Maier, M.K.; du Pasquier, L.; Ravens, I.; Dreyer, L.; Burger, R.; Gramatzki, M.; Schwinzer, R.; et al. CD96 Interaction with CD155 via Its First Ig-like Domain Is Modulated by Alternative Splicing or Mutations in Distal Ig-like Domains. J. Biol. Chem. 2009, 284, 2235–2244. [Google Scholar] [CrossRef]
- Blake, S.J.; Stannard, K.; Liu, J.; Allen, S.; Yong, M.C.R.; Mittal, D.; Aguilera, A.R.; Miles, J.J.; Lutzky, V.P.; De Andrade, L.F.; et al. Suppression of Metastases Using a New Lymphocyte Checkpoint Target for Cancer Immunotherapy. Cancer Discov. 2016, 6, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.R.; Lutzky, V.P.; Mittal, D.; Li, X.-Y.; Stannard, K.; Takeda, K.; Bernhardt, G.; Teng, M.W.L.; Dougall, W.C.; Smyth, M.J. CD96 targeted antibodies need not block CD96-CD155 interactions to promote NK cell anti-metastatic activity. OncoImmunology 2018, 7, e1424677. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.; Fleischmann-Mundt, B.; Woller, N.; Niemann, J.; Ribback, S.; Peters, K.; Demir, I.E.; Armbrecht, N.; Ceyhan, G.O.; Manns, M.P.; et al. Perioperative, Spatiotemporally Coordinated Activation of T and NK Cells Prevents Recurrence of Pancreatic Cancer. Cancer Res. 2018, 78, 475–488. [Google Scholar] [CrossRef]
- Husain, B.; Ramani, S.R.; Chiang, E.; Lehoux, I.; Paduchuri, S.; Arena, T.A.; Patel, A.; Wilson, B.; Chan, P.; Franke, Y.; et al. A Platform for Extracellular Interactome Discovery Identifies Novel Functional Binding Partners for the Immune Receptors B7-H3/CD276 and PVR/CD155. Mol. Cell. Proteom. 2019, 18, 2310–2323. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, C.F.; Gimeno, L.; Server, G.; Martínez-Sánchez, M.V.; Escudero, J.F.; López-Cubillana, P.; Cabezas-Herrera, J.; Campillo, J.A.; Abellan, D.J.; Martínez-García, J.; et al. Immunological Risk Stratification of Bladder Cancer Based on Peripheral Blood Natural Killer Cell Biomarkers. Eur. Urol. Oncol. 2021, 4, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Peng, M.; Xing, P.; Wei, Y.; Galbo, P.M.; Corrigan, D.; Wang, H.; Su, Y.; Dong, X.; Sun, Q.; et al. Blockade of the immunosuppressive KIR2DL5/PVR pathway elicits potent human NK cell–mediated antitumor immunity. J. Clin. Investig. 2022, 132, e163620. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Ophir, E.; Kotturi, M.F.; Levy, O.; Ganguly, S.; Leung, L.; Vaknin, I.; Kumar, S.; Dassa, L.; Hansen, K.; et al. PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8+ T-cell Function. Cancer Immunol. Res. 2019, 7, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Sunderland, A.; Zhou, Y.; Schulick, R.D.; Edil, B.H.; Zhu, Y. Blockade of CD112R and TIGIT signaling sensitizes human natural killer cell functions. Cancer Immunol. Immunother. 2017, 66, 1367–1375. [Google Scholar] [CrossRef]
- Murter, B.; Pan, X.; Ophir, E.; Alteber, Z.; Azulay, M.; Sen, R.; Levy, O.; Dassa, L.; Vaknin, I.; Fridman-Kfir, T.; et al. Mouse PVRIG Has CD8+ T Cell–Specific Coinhibitory Functions and Dampens Antitumor Immunity. Cancer Immunol. Res. 2019, 7, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Cao, G.; Zheng, X.; Sun, C.; Wei, H.; Tian, Z.; Xiao, W.; Sun, R.; Sun, H. Blockade of checkpoint receptor PVRIG unleashes anti-tumor immunity of NK cells in murine and human solid tumors. J. Hematol. Oncol. 2021, 14, 100. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, W.; Jankovic, V.; Golubov, J.; Poon, P.; Oswald, E.M.; Gurer, C.; Wei, J.; Ramos, I.; Wu, Q.; et al. Combination cancer immunotherapy targeting PD-1 and GITR can rescue CD8+ T cell dysfunction and maintain memory phenotype. Sci. Immunol. 2018, 3, eaat7061. [Google Scholar] [CrossRef]
- Banta, K.L.; Xu, X.; Chitre, A.S.; Au-Yeung, A.; Takahashi, C.; O’Gorman, W.E.; Wu, T.D.; Mittman, S.; Cubas, R.; Comps-Agrar, L.; et al. Mechanistic conver-gence of the TIGIT and PD- 1 inhibitory pathways necessitates co-blockade to optimize anti-tumor CD8+ T cell responses. Immunity 2022, 55, 512. [Google Scholar] [CrossRef] [PubMed]
- Milito, N.D.; Zingoni, A.; Stabile, H.; Soriani, A.; Capuano, C.; Cippitelli, M.; Gismondi, A.; Santoni, A.; Paolini, R.; Molfetta, R. NKG2D engagement on human NK cells leads to DNAM-1 hypo-responsiveness through different converging mechanisms. Eur. J. Immunol. 2023, 53, e2250198. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Simonetta, F.; Baker, J.; Pierini, A.; Wenokur, A.S.; Morrison, A.R.; Murphy, W.J.; Negrin, R.S. Regulation of murine NK cell exhaustion through the activation of the DNA damage repair pathway. J. Clin. Investig. 2019, 4, e127729. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.A.; Schirm, D.; Bendzick, L.; Hopps, R.; Selleck, C.; Hinderlie, P.; Felices, M.; Miller, J.S. Balanced engagement of activating and inhibitory receptors mitigates human NK cell exhaustion. J. Clin. Investig. 2022, 7, e150079. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paolini, R.; Molfetta, R. Dysregulation of DNAM-1-Mediated NK Cell Anti-Cancer Responses in the Tumor Microenvironment. Cancers 2023, 15, 4616. https://doi.org/10.3390/cancers15184616
Paolini R, Molfetta R. Dysregulation of DNAM-1-Mediated NK Cell Anti-Cancer Responses in the Tumor Microenvironment. Cancers. 2023; 15(18):4616. https://doi.org/10.3390/cancers15184616
Chicago/Turabian StylePaolini, Rossella, and Rosa Molfetta. 2023. "Dysregulation of DNAM-1-Mediated NK Cell Anti-Cancer Responses in the Tumor Microenvironment" Cancers 15, no. 18: 4616. https://doi.org/10.3390/cancers15184616
APA StylePaolini, R., & Molfetta, R. (2023). Dysregulation of DNAM-1-Mediated NK Cell Anti-Cancer Responses in the Tumor Microenvironment. Cancers, 15(18), 4616. https://doi.org/10.3390/cancers15184616





