Machine Learning for Digital Scoring of PRMT6 in Immunohistochemical Labeled Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. Immunohistochemical Staining
2.3. Evaluation of Staining
2.4. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Subjects
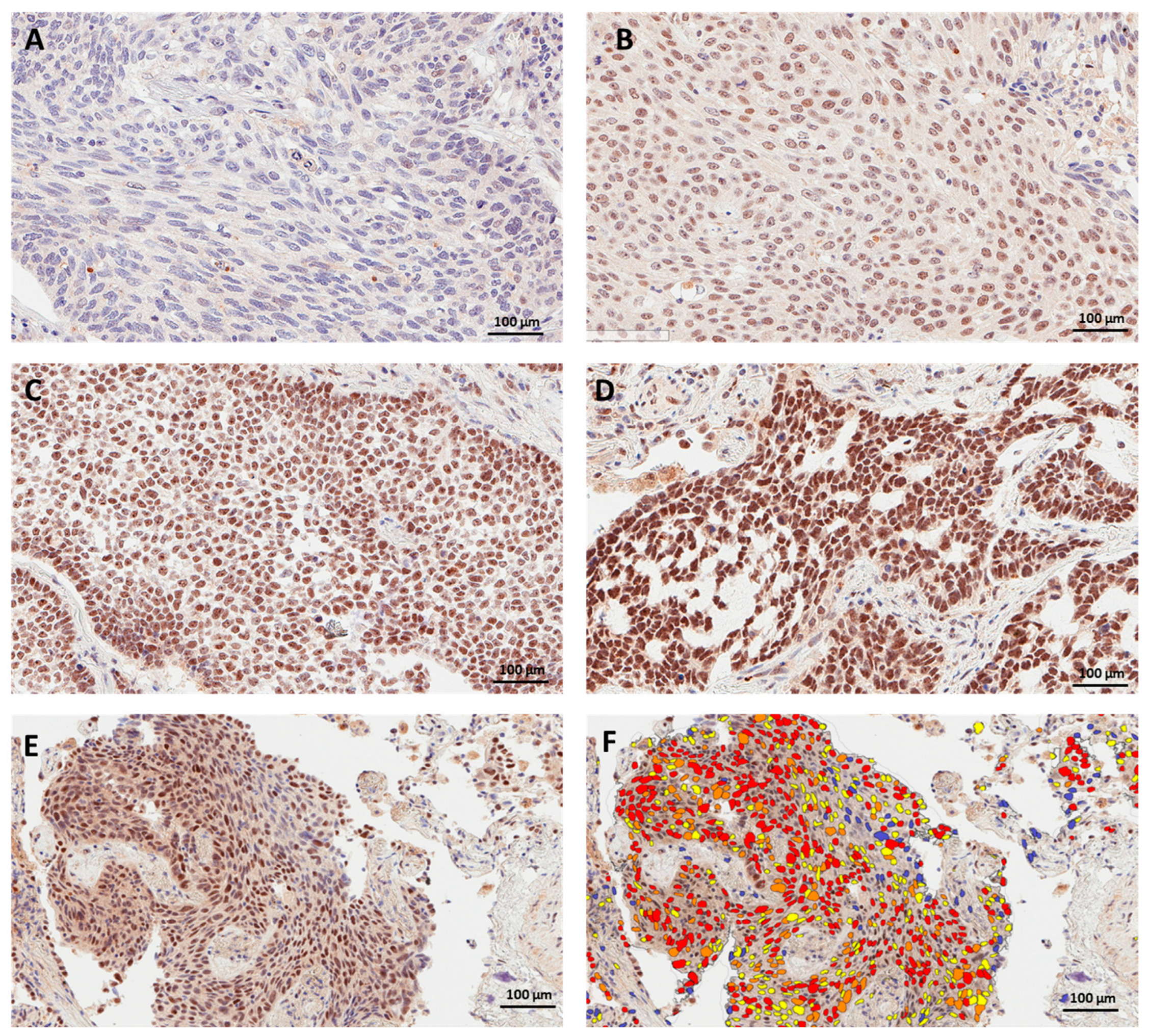
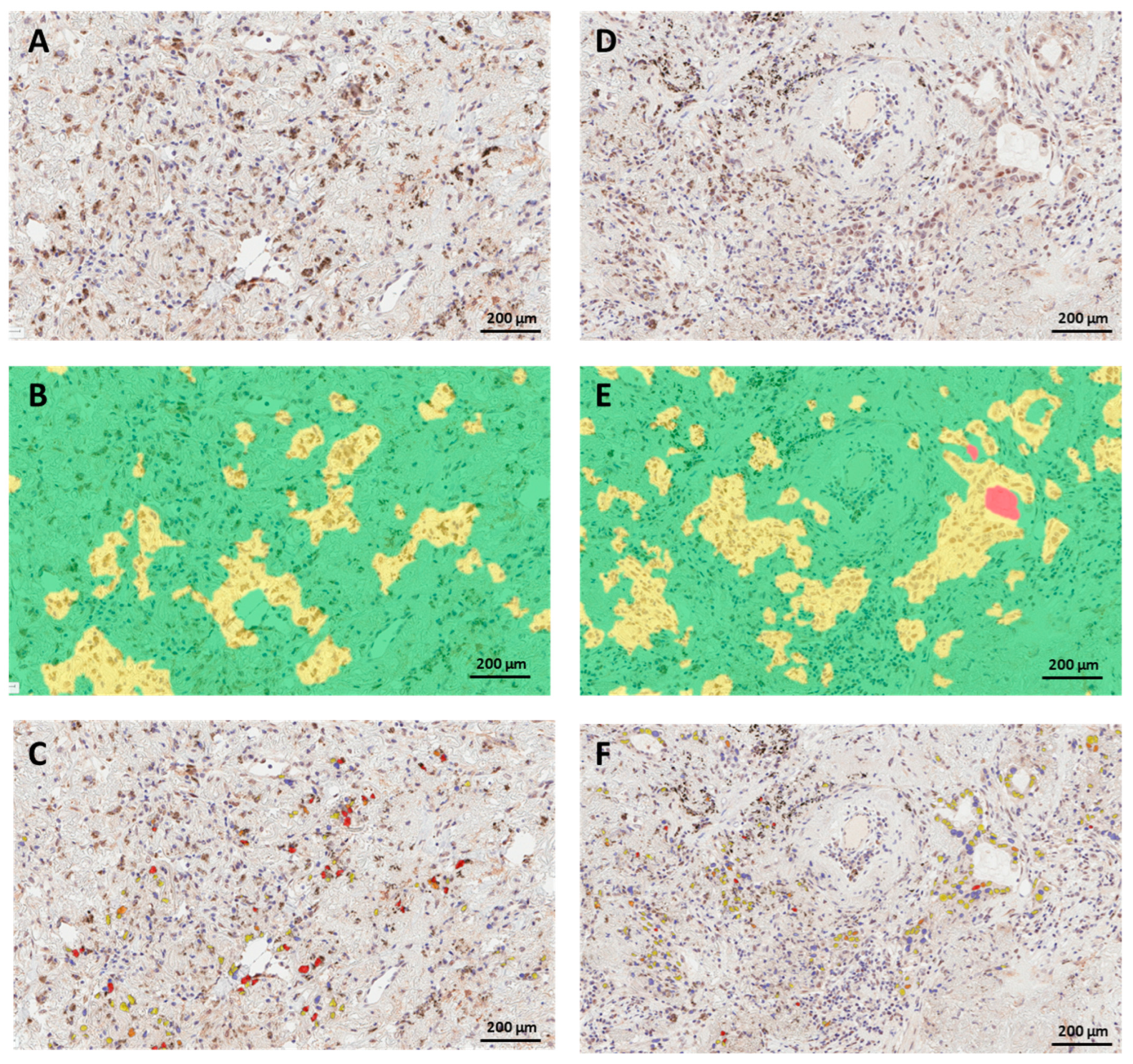
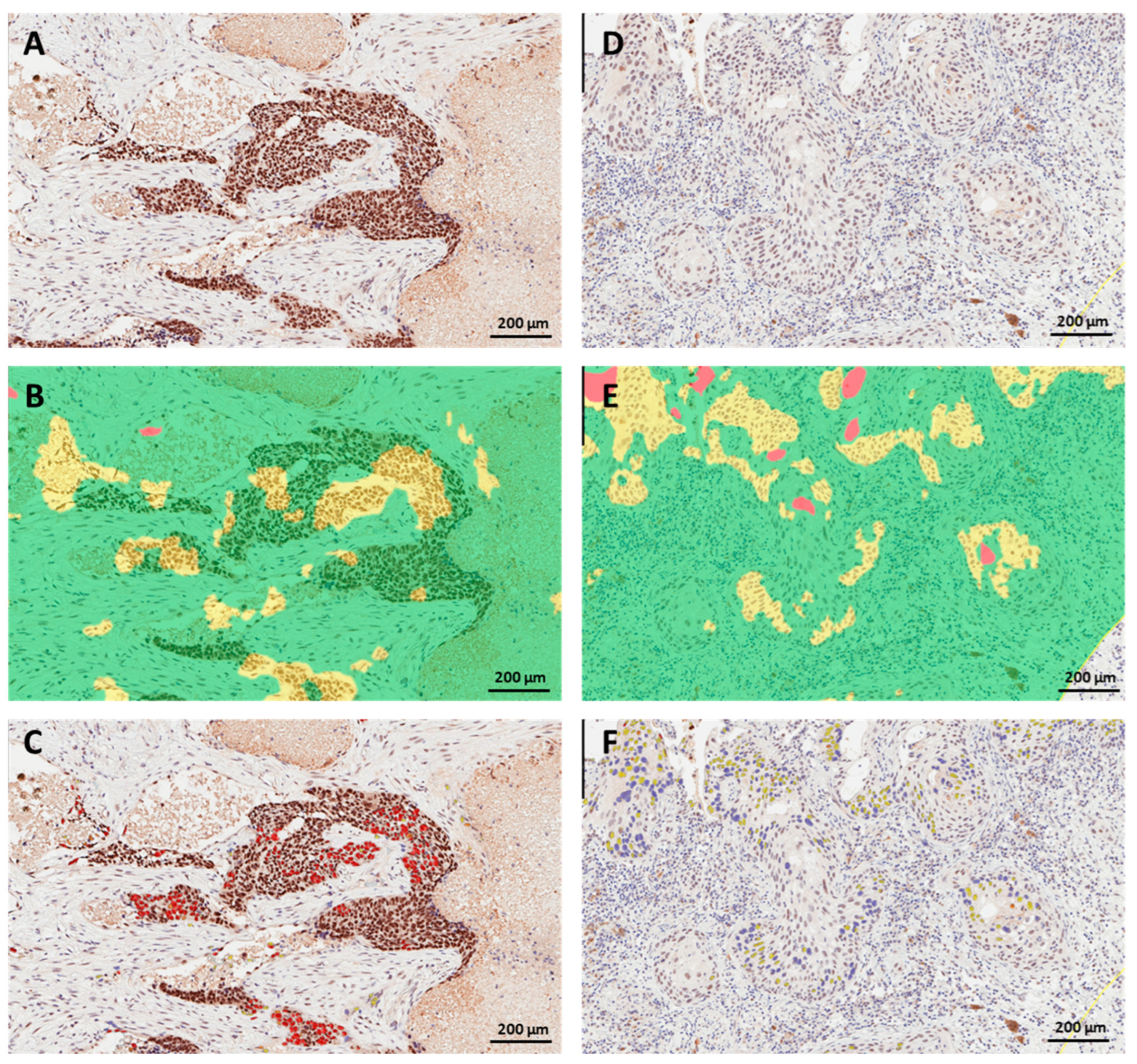
3.2. PRMT6 Expression and Subcellular Localization in Lung Cancer Tissue
3.3. PRMT6 Expression in Lung Cancer Tissue Using Digital Scoring
3.4. Concordance between Manual and Digital Scoring of Nuclear PRMT6 Expression in Lung Cancer Tissues
4. Discussion
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jaraj, S.J.; Camparo, P.; Boyle, H.; Germain, F.; Nilsson, B.; Petersson, F.; Egevad, L. Intra—And interobserver reproducibility of interpretation of immunohistochemical stains of prostate cancer. Virchows Arch. 2009, 455, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Weaver, D.L.; Krag, D.N.; Manna, E.A.; Ashikaga, T.; Harlow, S.P.; Bauer, K.D. Comparison of pathologist-detected and automated computer-assisted image analysis detected sentinel lymph node micrometastases in breast cancer. Mod. Pathol. 2003, 16, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Rimm, D.L.; Giltnane, J.M.; Moeder, C.; Harigopal, M.; Chung, G.G.; Camp, R.L.; Burtness, B. Bimodal population or pathologist artifact? J. Clin. Oncol. 2007, 25, 2487–2488. [Google Scholar] [CrossRef] [PubMed]
- Rimm, D.L. What brown cannot do for you. Nat. Biotechnol. 2006, 24, 914–916. [Google Scholar] [CrossRef] [PubMed]
- Feuchtinger, A.; Stiehler, T.; Jutting, U.; Marjanovic, G.; Luber, B.; Langer, R.; Walch, A. Image analysis of immunohistochemistry is superior to visual scoring as shown for patient outcome of esophageal adenocarcinoma. Histochem. Cell. Biol. 2015, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Harigopal, M.; Barlow, W.E.; Tedeschi, G.; Porter, P.L.; Yeh, I.T.; Haskell, C.; Livingston, R.; Hortobagyi, G.N.; Sledge, G.; Shapiro, C.; et al. Multiplexed assessment of the Southwest Oncology Group-directed Intergroup Breast Cancer Trial S9313 by AQUA shows that both high and low levels of HER2 are associated with poor outcome. Am. J. Pathol. 2010, 176, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Gavrielides, M.A.; Gallas, B.D.; Lenz, P.; Badano, A.; Hewitt, S.M. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch. Pathol. Lab. Med. 2011, 135, 233–242. [Google Scholar] [CrossRef]
- Bloom, K.; Harrington, D. Enhanced accuracy and reliability of HER-2/neu immunohistochemical scoring using digital microscopy. Am. J. Clin. Pathol. 2004, 121, 620–630. [Google Scholar] [CrossRef]
- Alvarenga, A.W.; Coutinho-Camillo, C.M.; Rodrigues, B.R.; Rocha, R.M.; Torres, L.F.; Martins, V.R.; da Cunha, I.W.; Hajj, G.N. A comparison between manual and automated evaluations of tissue microarray patterns of protein expression. J. Histochem. Cytochem. 2013, 61, 272–282. [Google Scholar] [CrossRef]
- FitzGerald, L.M.; Zhang, X.; Kolb, S.; Kwon, E.M.; Liew, Y.C.; Hurtado-Coll, A.; Knudsen, B.S.; Ostrander, E.A.; Stanford, J.L. Investigation of the relationship between prostate cancer and MSMB and NCOA4 genetic variants and protein expression. Hum. Mutat. 2013, 34, 149–156. [Google Scholar] [CrossRef]
- Laurinaviciene, A.; Dasevicius, D.; Ostapenko, V.; Jarmalaite, S.; Lazutka, J.; Laurinavicius, A. Membrane connectivity estimated by digital image analysis of HER2 immunohistochemistry is concordant with visual scoring and fluorescence in situ hybridization results: Algorithm evaluation on breast cancer tissue microarrays. Diagn. Pathol. 2011, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Faratian, D.; Kay, C.; Robson, T.; Campbell, F.M.; Grant, M.; Rea, D.; Bartlett, J.M. Automated image analysis for high-throughput quantitative detection of ER and PR expression levels in large-scale clinical studies: The TEAM Trial Experience. Histopathology 2009, 55, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Brugmann, A.; Eld, M.; Lelkaitis, G.; Nielsen, S.; Grunkin, M.; Hansen, J.D.; Foged, N.T.; Vyberg, M. Digital image analysis of membrane connectivity is a robust measure of HER2 immunostains. Breast Cancer Res. Treat. 2012, 132, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Bolton, K.L.; Garcia-Closas, M.; Pfeiffer, R.M.; Duggan, M.A.; Howat, W.J.; Hewitt, S.M.; Yang, X.R.; Cornelison, R.; Anzick, S.L.; Meltzer, P.; et al. Assessment of automated image analysis of breast cancer tissue microarrays for epidemiologic studies. Cancer Epidemiol. Biomark. Prev. 2010, 19, 992–999. [Google Scholar] [CrossRef]
- Braun, M.; Kirsten, R.; Rupp, N.J.; Moch, H.; Fend, F.; Wernert, N.; Kristiansen, G.; Perner, S. Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system. Histol. Histopathol. 2013, 28, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Messersmith, W.; Oppenheimer, D.; Peralba, J.; Sebastiani, V.; Amador, M.; Jimeno, A.; Embuscado, E.; Hidalgo, M.; Iacobuzio-Donahue, C. Assessment of Epidermal Growth Factor Receptor (EGFR) signaling in paired colorectal cancer and normal colon tissue samples using computer-aided immunohistochemical analysis. Cancer Biol. Ther. 2005, 4, 1381–1386. [Google Scholar] [CrossRef]
- Alexander, B.M.; Wang, X.Z.; Niemierko, A.; Weaver, D.T.; Mak, R.H.; Roof, K.S.; Fidias, P.; Wain, J.; Choi, N.C. DNA repair biomarkers predict response to neoadjuvant chemoradiotherapy in esophageal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, 164–171. [Google Scholar] [CrossRef]
- Sin, M.K. Lung cancer disparities and African-Americans. Public Health Nurs. 2017, 34, 359–362. [Google Scholar] [CrossRef]
- American Cancer Society. Key Statistics for Lung Cancer. Available online: https://www.cancer.org/cancer/non-small-cell-lung-cancer/about/key-statistics.html (accessed on 14 July 2023).
- Olaku, O.O.; Taylor, E.A. Cancer in the medically underserved population. Prim. Care Clin. Off. Pract. 2017, 44, 87–97. [Google Scholar] [CrossRef]
- Li, J.; Chung, S.; Wei, E.; Luft, H. New recommendation and coverage of low-dose computed tomography for lung cancer screening: Uptake has increased but is still low. BMC Health Serv. Res. 2018, 18, 528. [Google Scholar] [CrossRef]
- Orsi, J.; Margellos-Anast, H.; Whitman, S. Black–White Health Disparities in the United States and Chicago: A 15-Year Progress Analysis. Am. J. Public Health 2010, 100, 349–356. [Google Scholar] [CrossRef]
- Margellos, H.; Silva, A.; Whitman, S. Comparison of Health Status Indicators in Chicago: Are Black-White Disparities Worsening? Am. J. Public Health 2004, 94, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Hunt, B.; Whitman, S. Black:White Health Disparities in the United States and Chicago: 1990–2010. J. Racial Ethn. Health Disparities 2015, 2, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Raposo, A.; Piller, S. Protein arginine methylation: An emerging regulator of the cell cycle. Cell Div. 2018, 13, 1–16. [Google Scholar] [CrossRef]
- Lin, W.-J.; Gary, J.; Yang, M.; Clarke, S.; Herschman, H. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996, 271, 15034–15044. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, M.; Toyokawa, G.; Hayami, S.; Unoki, M.; Tsunoda, T.; Field, H.I.; Kelly, J.D.; Neal, D.E.; Maehara, Y.; Ponder, B.A.; et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 2011, 128, 562–573. [Google Scholar] [CrossRef]
- Mathioudaki, K.; Papadokostopoulou, A.; Scorilas, A.; Xynopoulos, D.; Agnanti, N.; Talieri, M. The PRMT1 gene expression pattern in colon cancer. Br. J. Cancer 2008, 99, 2094–2099. [Google Scholar] [CrossRef] [PubMed]
- Le Romancer, M.; Treilleux, I.; Leconte, N.; Robin-Lespinasse, Y.; Sentis, S.; Bouchekioua-Bouzaghou, K.; Goddard, S.; Gobert-Gosse, S.; Corbo, L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol. Cell 2008, 31, 212–221. [Google Scholar] [CrossRef]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef]
- Baldwin, R.M.; Morettin, A.; Cote, J. Role of PRMTs in cancer: Could minor isoforms be leaving a mark? World J. Biol. Chem. 2014, 5, 115–129. [Google Scholar]
- Avasarala, S.; Van Scoyk, M.; Karuppusamy Rathinam, M.K.; Zerayesus, S.; Zhao, X.; Zhang, W.; Pergande, M.R.; Borgia, J.A.; DeGregori, J.; Port, J.D.; et al. PRMT1 Is a Novel Regulator of Epithelial-Mesenchymal-Transition in Non-small Cell Lung Cancer. J. Biol. Chem. 2015, 290, 13479–13489. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Corbo, L.; Le Romancer, M. Protein arginine methylation/demethylation and cancer. Oncotarget 2016, 7, 67532–67550. [Google Scholar] [CrossRef] [PubMed]
- Avasarala, S.; Wu, P.Y.; Khan, S.Q.; Yanlin, S.; Van Scoyk, M.; Bao, J.; Di Lorenzo, A.; David, O.; Bedford, M.T.; Gupta, V.; et al. PRMT6 Promotes Lung Tumor Progression via the Alternate Activation of Tumor-Associated Macrophages. Mol. Cancer Res. 2020, 18, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Meng, Q.; Shi, R.; Xu, Y. PRMT6 serves an oncogenic role in lung adenocarcinoma via regulating p18. Mol. Med. Rep. 2020, 22, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gan, J.; Wei, Z.; Zhang, M.; Du, Y.; Xu, C.; Zhao, H. The Emerging Role of PRMT6 in Cancer. Front. Oncol. 2022, 12, 841381. [Google Scholar] [CrossRef]
- De Sousa, V.M.L.; Carvalho, L. Heterogeneity in Lung Cancer. Pathobiology 2018, 85, 96–107. [Google Scholar] [CrossRef]

| N | Mean Score | N (%) of High PRMT6 Cases a | |||||
|---|---|---|---|---|---|---|---|
| Pathologist 1 | Pathologist 2 | AI-Based | Pathologist 1 | Pathologist 2 | AI-Based | ||
| Total | 33 | 89 | 78 | 94 | 18 (54.6) | 16 (48.5) | 15 (45.5) |
| Self-reported race/ethnicity | |||||||
| Blacks | 20 | 101 | 90 | 101 | 15 (75.0) | 12 (60.0) | 12 (60.0) |
| Whites | 12 | 76 | 65 | 86 | 3 (25.0) | 4 (33.3) | 3 (25.0) |
| Other | 1 | 10 | 25 | 52 | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Age at diagnosis | |||||||
| ≤64 years | 19 | 88 | 75 | 96 | 9 (47.4) | 8 (42.1) | 8 (42.1) |
| >65 years | 14 | 90 | 82 | 92 | 9 (64.3) | 8 (57.1) | 7 (50.0) |
| Smoking status | |||||||
| Current smokers | 21 | 94 | 80 | 98 | 13 (61.9) | 12 (57.1) | 11 (52.4) |
| Former smokers | 8 | 92 | 79 | 95 | 5 (62.5) | 4 (50.0) | 4 (50.0) |
| Never smokers | 4 | 59 | 68 | 75 | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Histological subtypes | |||||||
| Squamous cell carcinoma | 8 | 73 | 70 | 3 (37.5) | 3 (37.5) | 2 (25.0) | |
| Basaloid squamous cell carcinoma | 1 | 165 | 145 | 201 | 1 (100.0) | 1 (100.0) | 1 (100.0) |
| Adenocarcinoma | 24 | 91 | 98 | 14 (58.3) | 12 (50.0) | 12 (50.0) | |
| Pathologist 1 | Pathologist 2 | AI-Based | |
|---|---|---|---|
| N | 33 | 31 | 33 |
| Mean | 89.1 | 78.2 | 94.3 |
| Standard Deviation | 53.5 | 54.9 | 41.7 |
| Range | 10.0–200.0 | 0–180.0 | 19.7–200.8 |
| Coefficient of Variation, % | 60.0 | 70.2 | 44.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoud, A.M.; Brister, E.; David, O.; Valyi-Nagy, K.; Sverdlov, M.; Gann, P.H.; Kim, S.J. Machine Learning for Digital Scoring of PRMT6 in Immunohistochemical Labeled Lung Cancer. Cancers 2023, 15, 4582. https://doi.org/10.3390/cancers15184582
Mahmoud AM, Brister E, David O, Valyi-Nagy K, Sverdlov M, Gann PH, Kim SJ. Machine Learning for Digital Scoring of PRMT6 in Immunohistochemical Labeled Lung Cancer. Cancers. 2023; 15(18):4582. https://doi.org/10.3390/cancers15184582
Chicago/Turabian StyleMahmoud, Abeer M., Eileen Brister, Odile David, Klara Valyi-Nagy, Maria Sverdlov, Peter H. Gann, and Sage J. Kim. 2023. "Machine Learning for Digital Scoring of PRMT6 in Immunohistochemical Labeled Lung Cancer" Cancers 15, no. 18: 4582. https://doi.org/10.3390/cancers15184582
APA StyleMahmoud, A. M., Brister, E., David, O., Valyi-Nagy, K., Sverdlov, M., Gann, P. H., & Kim, S. J. (2023). Machine Learning for Digital Scoring of PRMT6 in Immunohistochemical Labeled Lung Cancer. Cancers, 15(18), 4582. https://doi.org/10.3390/cancers15184582







