Deep Learning for the Pathologic Diagnosis of Hepatocellular Carcinoma, Cholangiocarcinoma, and Metastatic Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
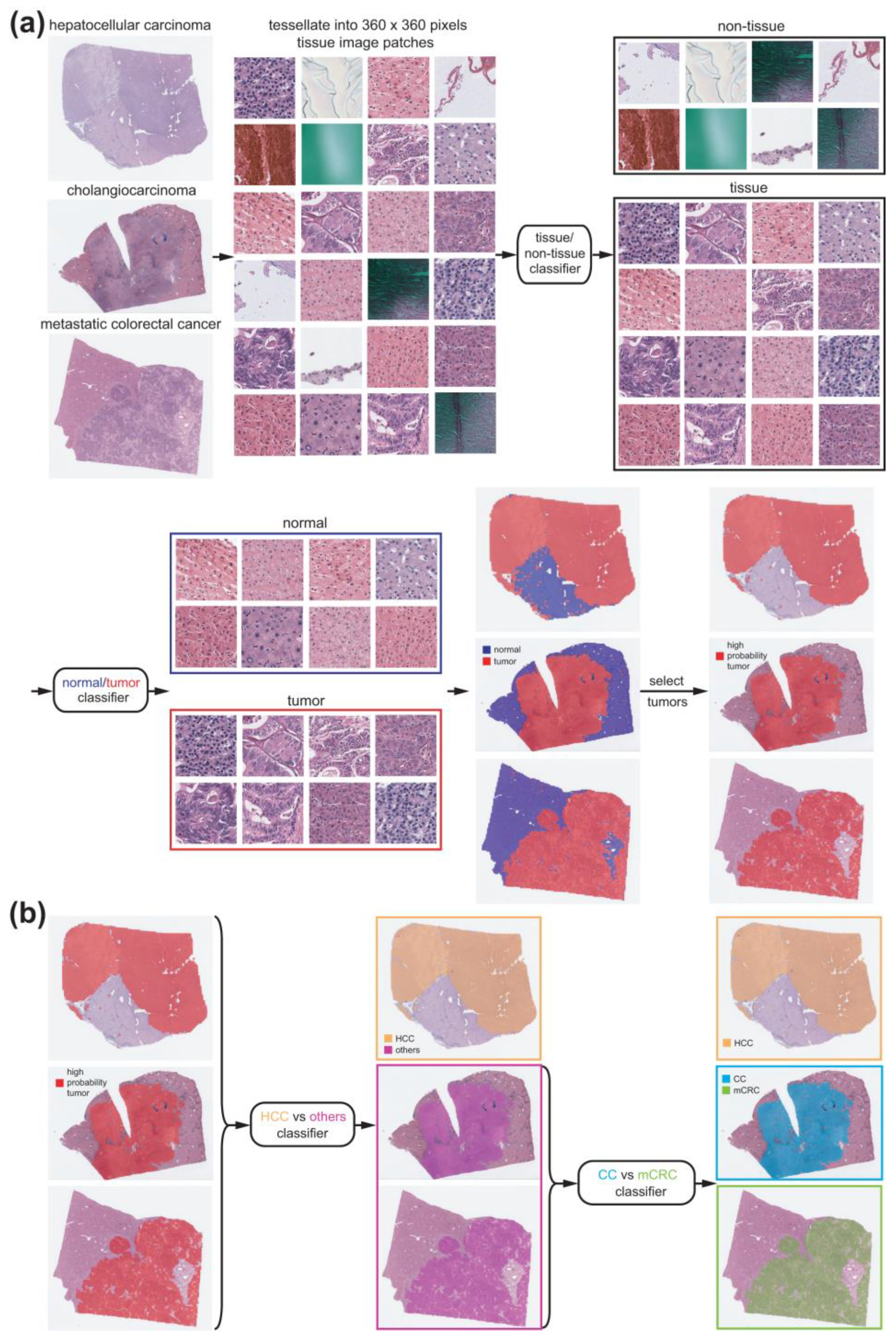
2.1. Datasets
2.2. Deep Learning Models
2.3. Visualization and Statistics
3. Results
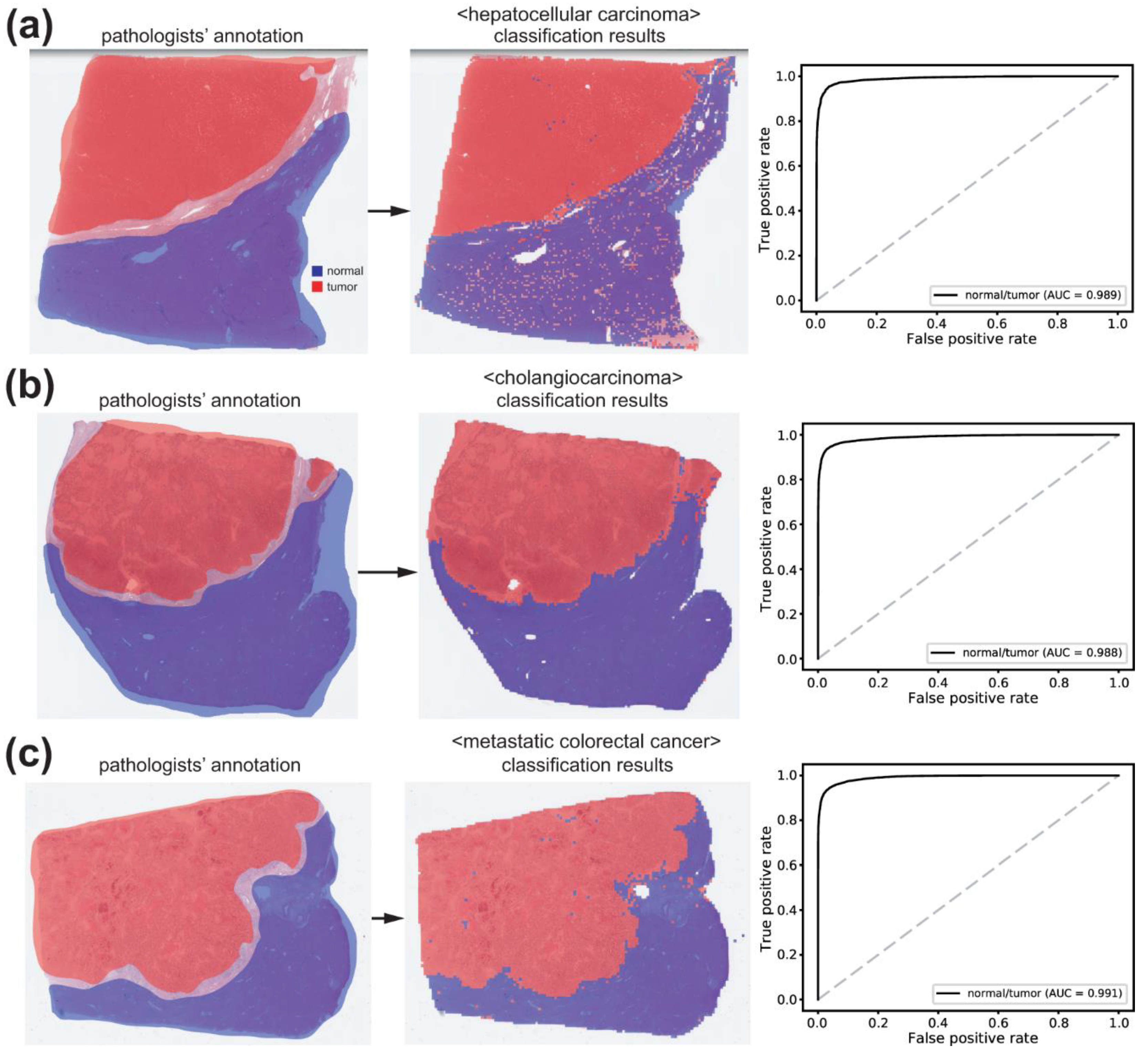
3.1. Normal/Tumor Classification
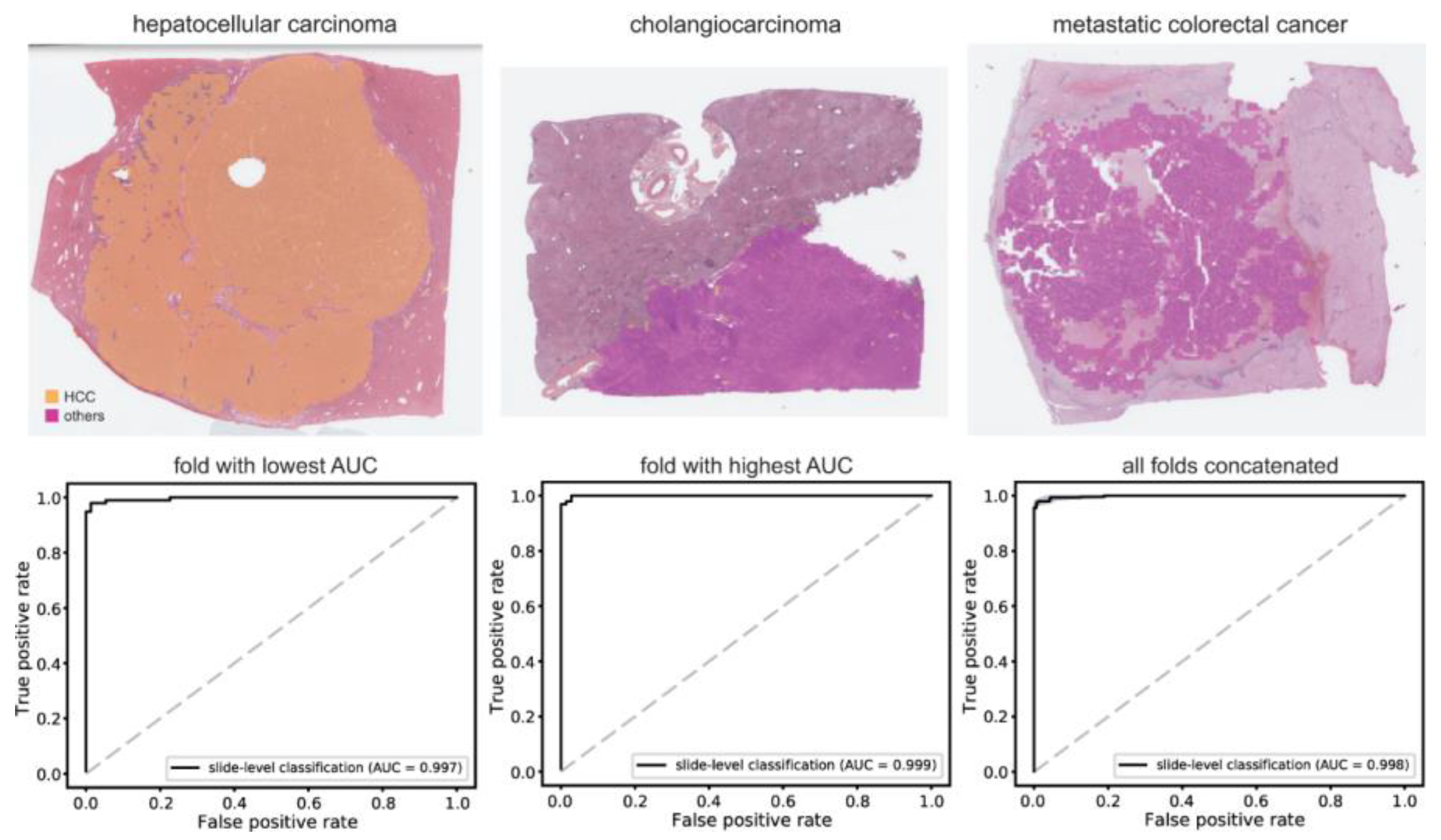
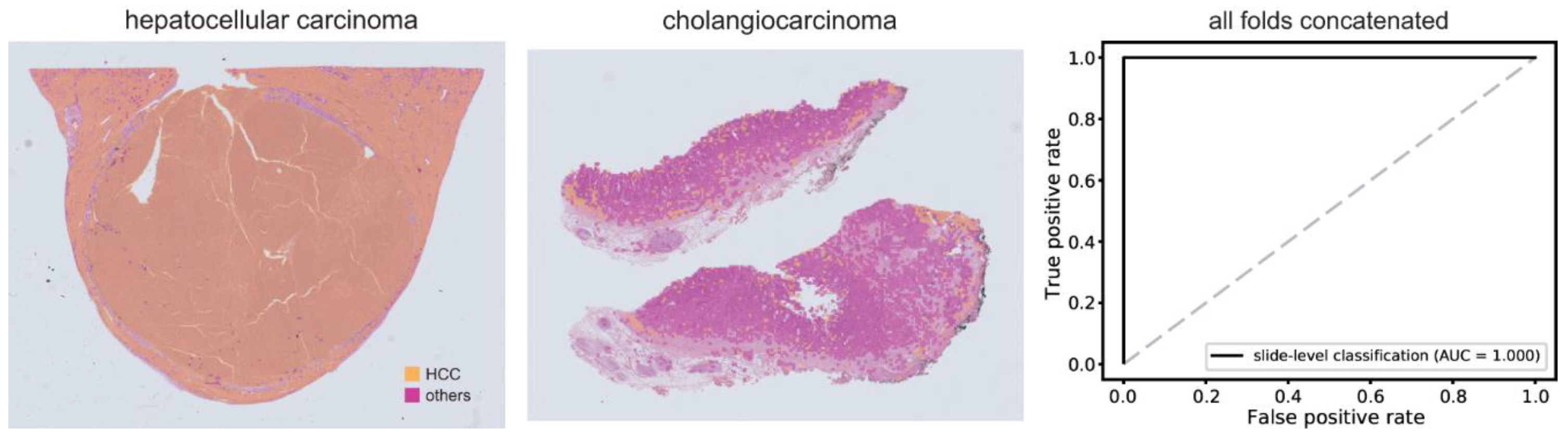
3.2. Classification of HCC/Other Cancer Types
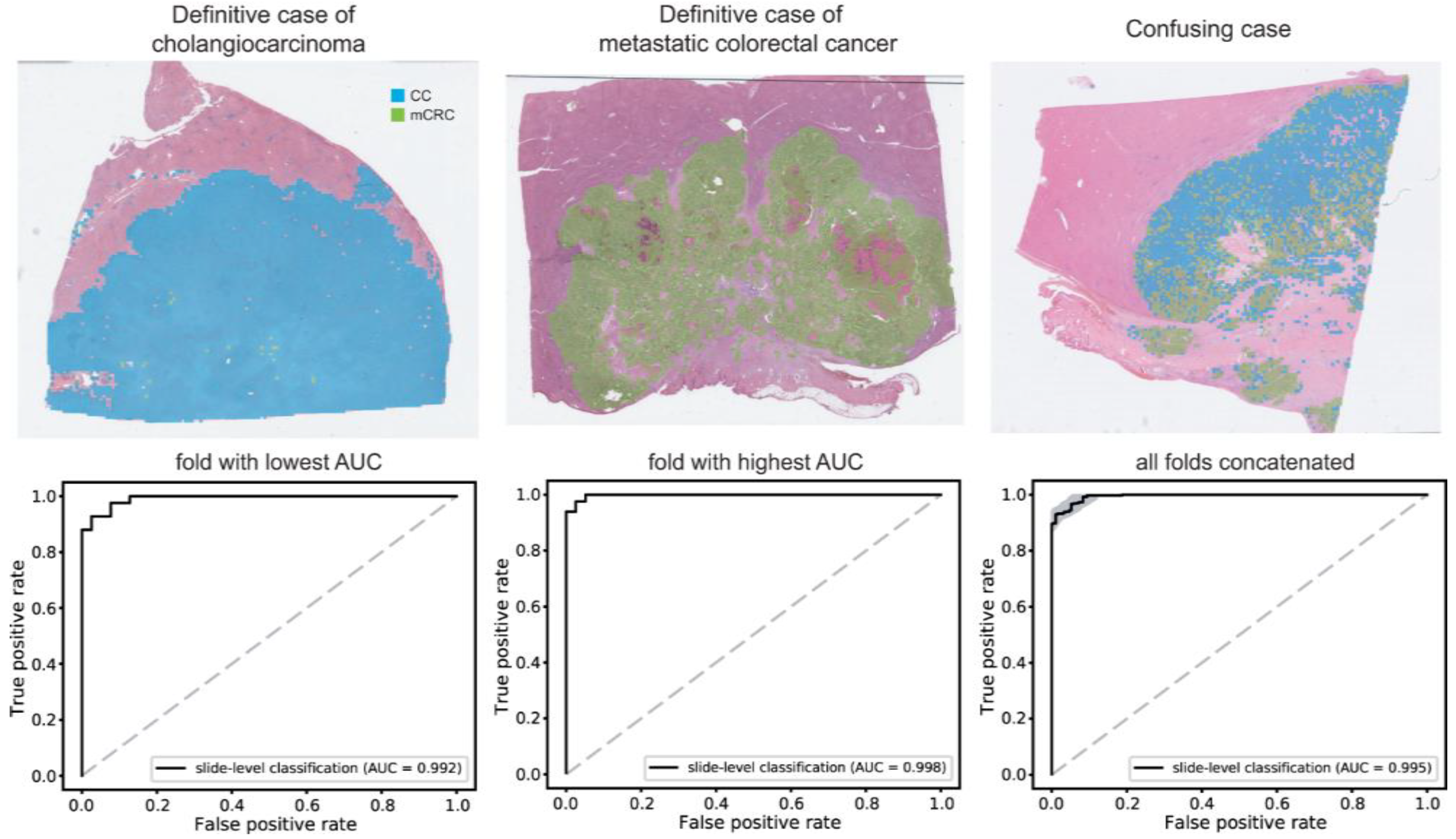
3.3. CC/mCRC Classification
3.4. Performance on an External Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Torbenson, M.; Ng, I.; Park, Y.; Roncalli, M.; Sakamato, M. WHO Classification of Tumours. Digestive System Tumours; International Agency for Research on Cancer: Lyon, France, 2019; pp. 229–239. [Google Scholar]
- Mak, L.Y.; Cruz-Ramon, V.; Chinchilla-Lopez, P.; Torres, H.A.; LoConte, N.K.; Rice, J.P.; Foxhall, L.E.; Sturgis, E.M.; Merrill, J.K.; Bailey, H.H.; et al. Global Epidemiology, Prevention, and Management of Hepatocellular Carcinoma. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 262–279. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.A.; Kong, J.; Gutman, D.A.; Dunn, W.D.; Nalisnik, M.; Brat, D.J. Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab. Investig. 2015, 95, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, C.; Ma, M.; Ma, X.; Lv, X.; Dong, X.; Yan, Z.; Zhu, M.; Chen, J. Classification of multi-differentiated liver cancer pathological images based on deep learning attention mechanism. BMC Med. Inform. Decis. Mak. 2022, 22, 176. [Google Scholar] [CrossRef]
- Friemel, J.; Rechsteiner, M.; Frick, L.; Bohm, F.; Struckmann, K.; Egger, M.; Moch, H.; Heikenwalder, M.; Weber, A. Intratumor heterogeneity in hepatocellular carcinoma. Clin. Cancer Res. 2015, 21, 1951–1961. [Google Scholar] [CrossRef]
- Massarweh, N.N.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control 2017, 24, 1073274817729245. [Google Scholar] [CrossRef]
- Chung, T.; Park, Y.N. Up-to-Date Pathologic Classification and Molecular Characteristics of Intrahepatic Cholangiocarcinoma. Front Med. 2022, 9, 857140. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C. AJCC Cancer Staging Manual; Springer: New York, NY, USA, 2017; Volume 1024. [Google Scholar]
- Altekruse, S.F.; Devesa, S.S.; Dickie, L.A.; McGlynn, K.A.; Kleiner, D.E. Histological classification of liver and intrahepatic bile duct cancers in SEER registries. J. Registry Manag. 2011, 38, 201–205. [Google Scholar]
- Lei, J.Y.; Bourne, P.A.; diSant’Agnese, P.A.; Huang, J. Cytoplasmic staining of TTF-1 in the differential diagnosis of hepatocellular carcinoma vs. cholangiocarcinoma and metastatic carcinoma of the liver. Am. J. Clin. Pathol. 2006, 125, 519–525. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, J.H. Pathologic differential diagnosis of metastatic carcinoma in the liver. Clin. Mol. Hepatol. 2019, 25, 12–20. [Google Scholar] [CrossRef]
- van der Geest, L.G.; Lam-Boer, J.; Koopman, M.; Verhoef, C.; Elferink, M.A.; de Wilt, J.H. Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin. Exp. Metastasis 2015, 32, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Riihimaki, M.; Hemminki, A.; Sundquist, J.; Hemminki, K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 2016, 6, 29765. [Google Scholar] [CrossRef] [PubMed]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef]
- Joshi, G.; Jain, A.; Adhikari, S.; Garg, H.; Bhandari, M. FDA approved Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices: An updated 2022 landscape. medRxiv 2022. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.; the, C.C.; Hermsen, M.; Manson, Q.F.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Song, Z.; Zou, S.; Zhou, W.; Huang, Y.; Shao, L.; Yuan, J.; Gou, X.; Jin, W.; Wang, Z.; Chen, X.; et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat. Commun. 2020, 11, 4294. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Feldman, M.D.; Abels, E.; Ashfaq, R.; Beltaifa, S.; Cacciabeve, N.G.; Cathro, H.P.; Cheng, L.; Cooper, K.; Dickey, G.E.; et al. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study). Am. J. Surg. Pathol. 2018, 42, 39–52. [Google Scholar] [CrossRef]
- Retamero, J.A.; Aneiros-Fernandez, J.; Del Moral, R.G. Complete Digital Pathology for Routine Histopathology Diagnosis in a Multicenter Hospital Network. Arch. Pathol. Lab. Med. 2020, 144, 221–228. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Vintch, B.; Zaharia, A.D.; Movshon, J.A.; Simoncelli, E.P. Efficient and direct estimation of a neural subunit model for sensory coding. Adv. Neural Inf. Process Syst. 2012, 25, 3113–3121. [Google Scholar] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e1129. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Yoshida, H.; Shimazu, T.; Kiyuna, T.; Marugame, A.; Yamashita, Y.; Cosatto, E.; Taniguchi, H.; Sekine, S.; Ochiai, A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer 2018, 21, 249–257. [Google Scholar] [CrossRef]
- Bandi, P.; Geessink, O.; Manson, Q.; Van Dijk, M.; Balkenhol, M.; Hermsen, M.; Ehteshami Bejnordi, B.; Lee, B.; Paeng, K.; Zhong, A.; et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans. Med. Imaging 2019, 38, 550–560. [Google Scholar] [CrossRef]
- Song, J.; Im, S.; Lee, S.H.; Jang, H.J. Deep Learning-Based Classification of Uterine Cervical and Endometrial Cancer Subtypes from Whole-Slide Histopathology Images. Diagnostics 2022, 12, 2623. [Google Scholar] [CrossRef]
- Venkatraman, E.S. A permutation test to compare receiver operating characteristic curves. Biometrics 2000, 56, 1134–1138. [Google Scholar] [CrossRef]
- Dimitriou, N.; Arandjelovic, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.J.; Reisenbuchler, D.; West, N.P.; Niehues, J.M.; Zhu, J.; Foersch, S.; Veldhuizen, G.P.; Quirke, P.; Grabsch, H.I.; van den Brandt, P.A.; et al. Transformer-based biomarker prediction from colorectal cancer histology: A large-scale multicentric study. Cancer Cell 2023, 41, 1650–1661.e1654. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Lee, A.; Kang, J.; Song, I.H.; Lee, S.H. Prediction of clinically actionable genetic alterations from colorectal cancer histopathology images using deep learning. World J. Gastroenterol. 2020, 26, 6207–6223. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Song, I.H.; Jang, H.J. Feasibility of deep learning-based fully automated classification of microsatellite instability in tissue slides of colorectal cancer. Int. J. Cancer 2021, 149, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Lee, A.; Kang, J.; Song, I.H.; Lee, S.H. Prediction of genetic alterations from gastric cancer histopathology images using a fully automated deep learning approach. World J. Gastroenterol. 2021, 27, 7687–7704. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, Y.; Jang, H.J. Deep learning captures selective features for discrimination of microsatellite instability from pathologic tissue slides of gastric cancer. Int. J. Cancer 2023, 152, 298–307. [Google Scholar] [CrossRef]
- Nam, S.; Chong, Y.; Jung, C.K.; Kwak, T.Y.; Lee, J.Y.; Park, J.; Rho, M.J.; Go, H. Introduction to digital pathology and computer-aided pathology. J. Pathol. Transl. Med. 2020, 54, 125–134. [Google Scholar] [CrossRef]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and Deep Learning in Diagnostic Pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef]
- Bousis, D.; Verras, G.-I.; Bouchagier, K.; Antzoulas, A.; Panagiotopoulos, I.; Katinioti, A.; Kehagias, D.; Kaplanis, C.; Kotis, K.; Anagnostopoulos, C.-N. The role of deep learning in diagnosing colorectal cancer. Gastroenterol. Rev./Przegląd Gastroenterol. 2023, 18, 266–273. [Google Scholar] [CrossRef]
- Yaqoob, A.; Musheer Aziz, R.; verma, N.K. Applications and Techniques of Machine Learning in Cancer Classification: A Systematic Review. Hum. Centric Intell. Syst. 2023, 1–28. [Google Scholar] [CrossRef]
- Afreen, S.; Bhurjee, A.K.; Aziz, R.M. Gene selection with Game Shapley Harris hawks optimizer for cancer classification. Chemom. Intell. Lab. Syst. 2023, 242, 104989. [Google Scholar] [CrossRef]
- Mulita, F.; Verras, G.I.; Anagnostopoulos, C.N.; Kotis, K. A Smarter Health through the Internet of Surgical Things. Sensors 2022, 22, 4577. [Google Scholar] [CrossRef]
- Evans, A.J.; Bauer, T.W.; Bui, M.M.; Cornish, T.C.; Duncan, H.; Glassy, E.F.; Hipp, J.; McGee, R.S.; Murphy, D.; Myers, C.; et al. US Food and Drug Administration Approval of Whole Slide Imaging for Primary Diagnosis: A Key Milestone Is Reached and New Questions Are Raised. Arch. Pathol. Lab. Med. 2018, 142, 1383–1387. [Google Scholar] [CrossRef] [PubMed]
- Kanavati, F.; Ichihara, S.; Rambeau, M.; Iizuka, O.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Gastric Signet Ring Cell Carcinoma Classification in Whole Slide Images. Technol. Cancer Res. Treat. 2021, 20, 15330338211027901. [Google Scholar] [CrossRef] [PubMed]
- Im, S.; Hyeon, J.; Rha, E.; Lee, J.; Choi, H.J.; Jung, Y.; Kim, T.J. Classification of Diffuse Glioma Subtype from Clinical-Grade Pathological Images Using Deep Transfer Learning. Sensors 2021, 21, 3500. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.H.; Wang, F.; Berry, G.J.; Re, C.; Altman, R.B.; Snyder, M.; Kohane, I.S. Classifying non-small cell lung cancer types and transcriptomic subtypes using convolutional neural networks. JAMA 2020, 27, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Song, I.H.; Lee, S.H. Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images. Cancers 2021, 13, 3811. [Google Scholar] [CrossRef]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, H.-J.; Go, J.-H.; Kim, Y.; Lee, S.H. Deep Learning for the Pathologic Diagnosis of Hepatocellular Carcinoma, Cholangiocarcinoma, and Metastatic Colorectal Cancer. Cancers 2023, 15, 5389. https://doi.org/10.3390/cancers15225389
Jang H-J, Go J-H, Kim Y, Lee SH. Deep Learning for the Pathologic Diagnosis of Hepatocellular Carcinoma, Cholangiocarcinoma, and Metastatic Colorectal Cancer. Cancers. 2023; 15(22):5389. https://doi.org/10.3390/cancers15225389
Chicago/Turabian StyleJang, Hyun-Jong, Jai-Hyang Go, Younghoon Kim, and Sung Hak Lee. 2023. "Deep Learning for the Pathologic Diagnosis of Hepatocellular Carcinoma, Cholangiocarcinoma, and Metastatic Colorectal Cancer" Cancers 15, no. 22: 5389. https://doi.org/10.3390/cancers15225389
APA StyleJang, H.-J., Go, J.-H., Kim, Y., & Lee, S. H. (2023). Deep Learning for the Pathologic Diagnosis of Hepatocellular Carcinoma, Cholangiocarcinoma, and Metastatic Colorectal Cancer. Cancers, 15(22), 5389. https://doi.org/10.3390/cancers15225389




