Study of Helicobacter pylori Isolated from a High-Gastric-Cancer-Risk Population: Unveiling the Comprehensive Analysis of Virulence-Associated Genes including Secretion Systems, and Genome-Wide Association Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sampling
2.2. H. pylori Isolation and Histological Evaluation
2.3. Genome Sequencing, De Novo Assembly, and Annotation
2.4. Identification of Virulence-Associated Genes and T4SS Genes
2.5. Characterization and Genotyping of Well-Known Virulence-Associated Genes
2.6. Population Structure and GWAS
2.7. Statistical Analysis
3. Results
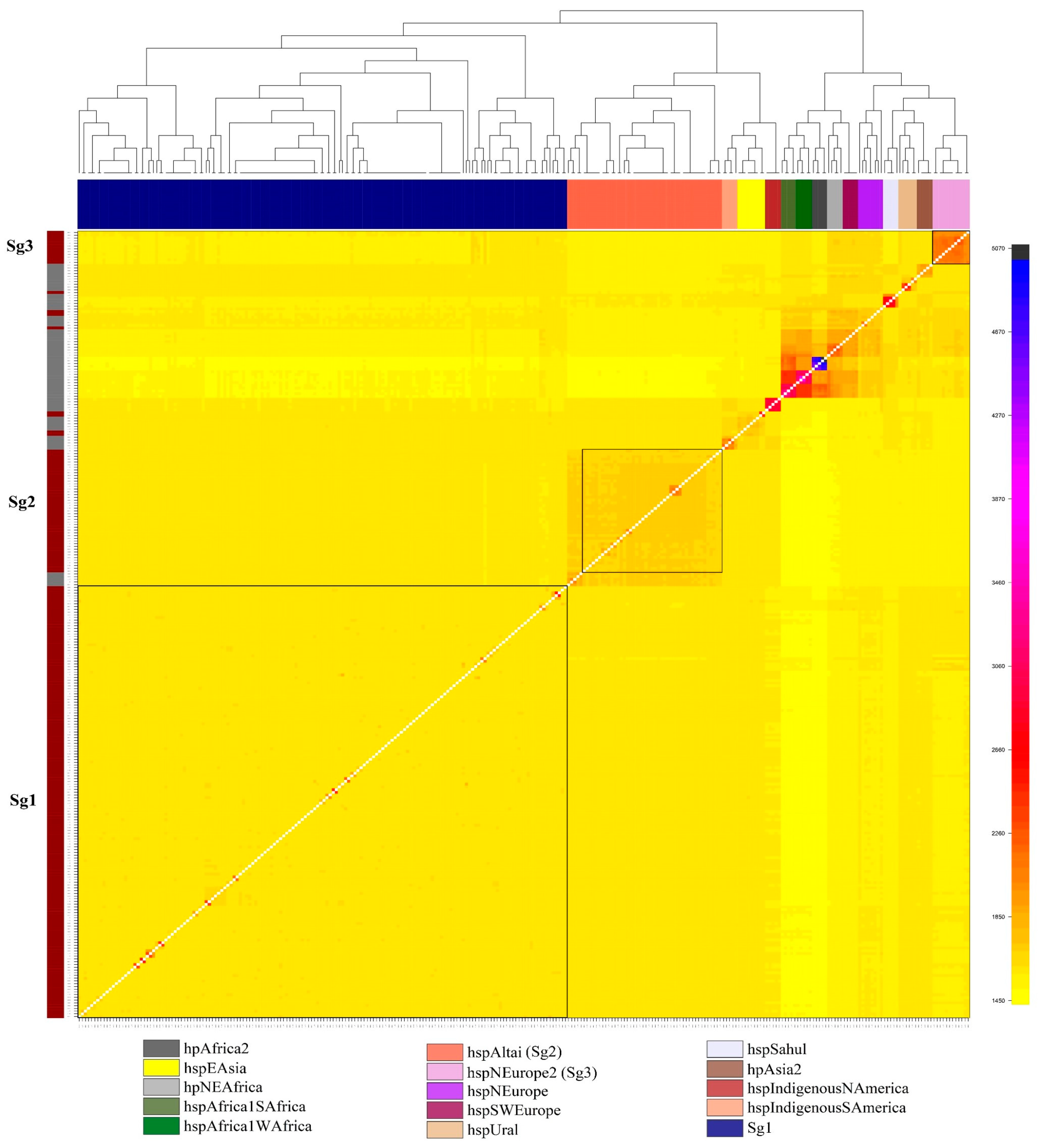
3.1. Characteristics of Clinical Isolates H. pylori, and Their Population Structure
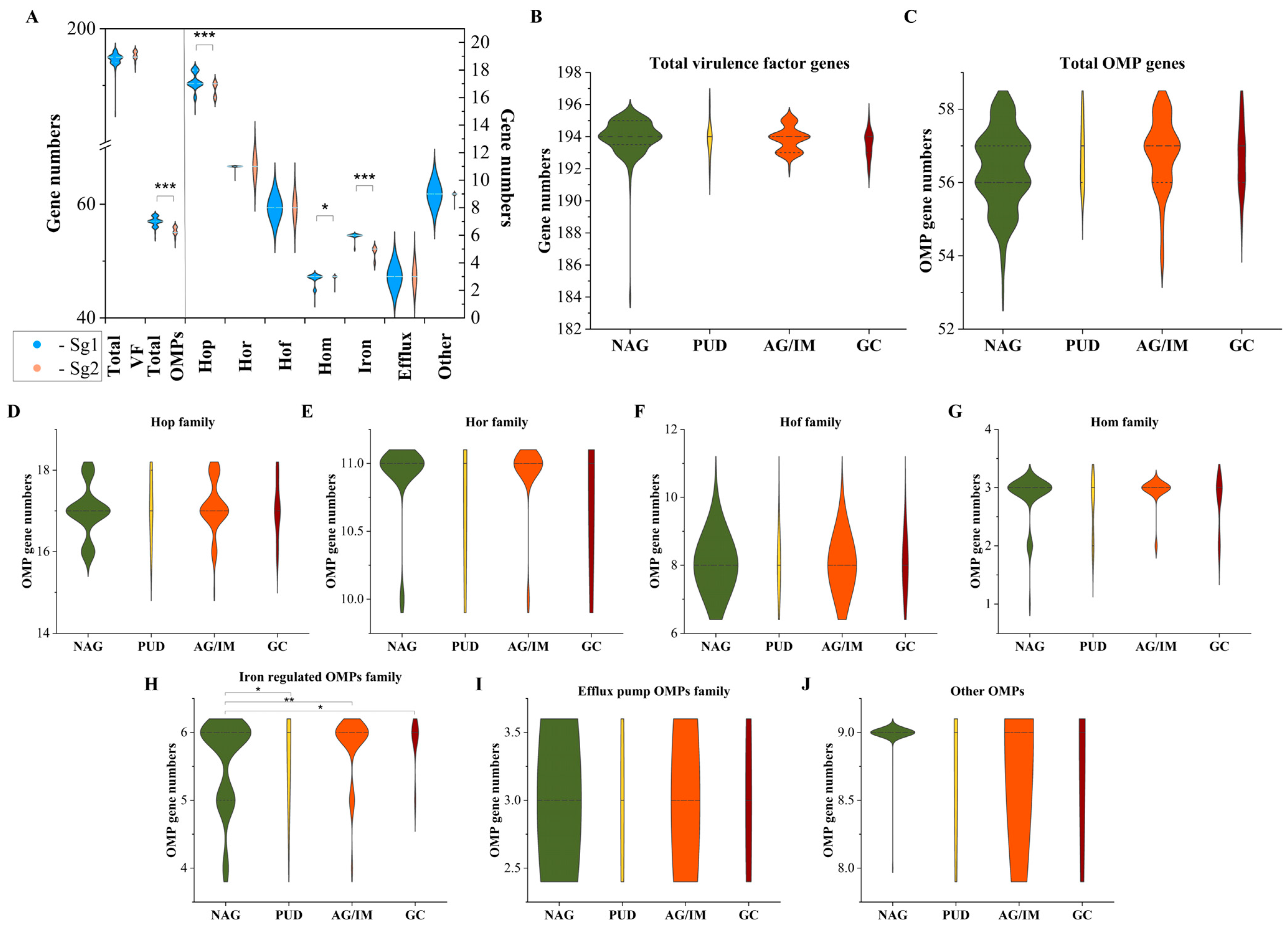
3.2. Diversity of OMP and Virulence Factor Genes Associated with Gastric Cancer and Other Diseases
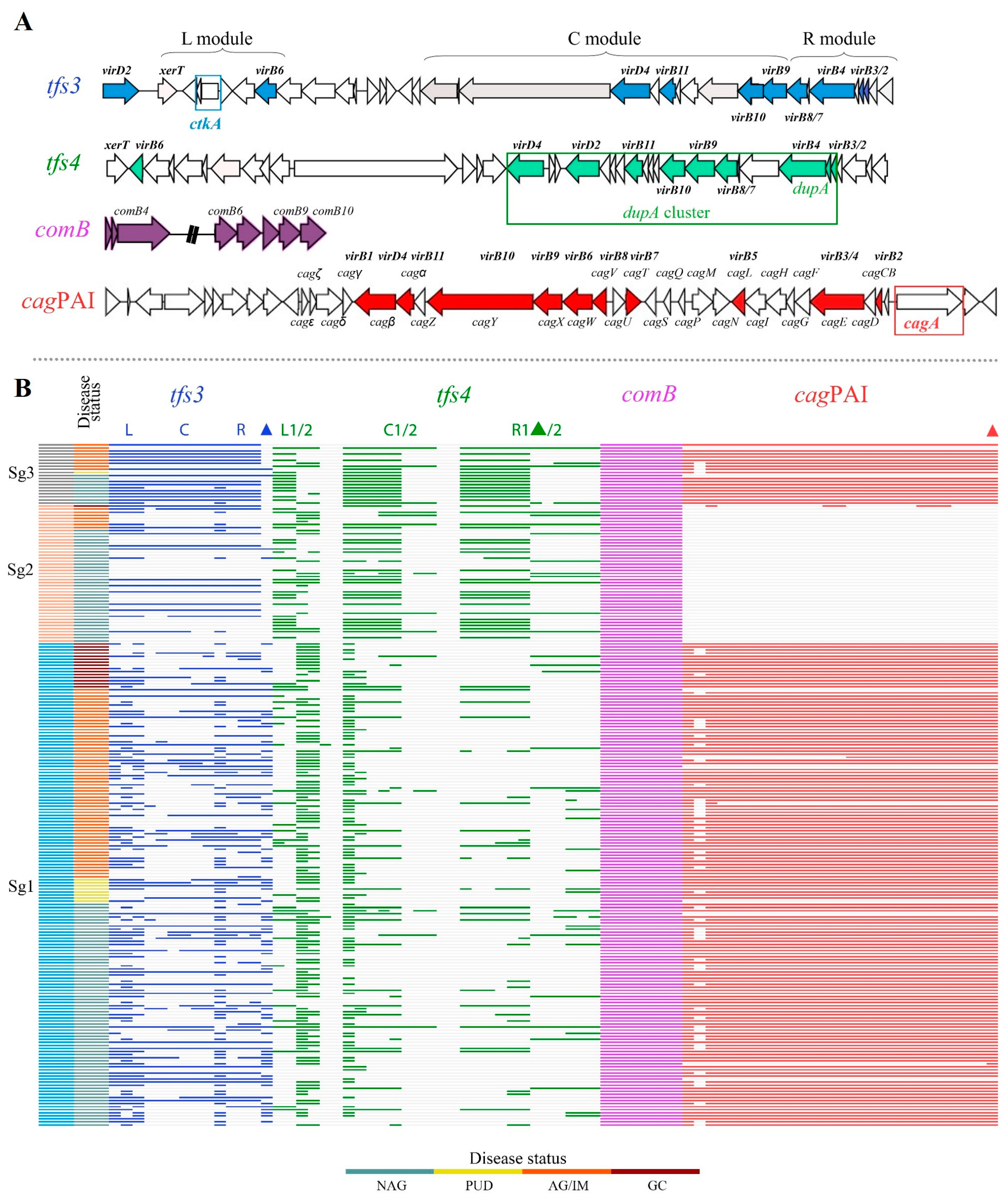
3.3. Diversity of ICEHptfs in Gastric Cancer and Other Diseases
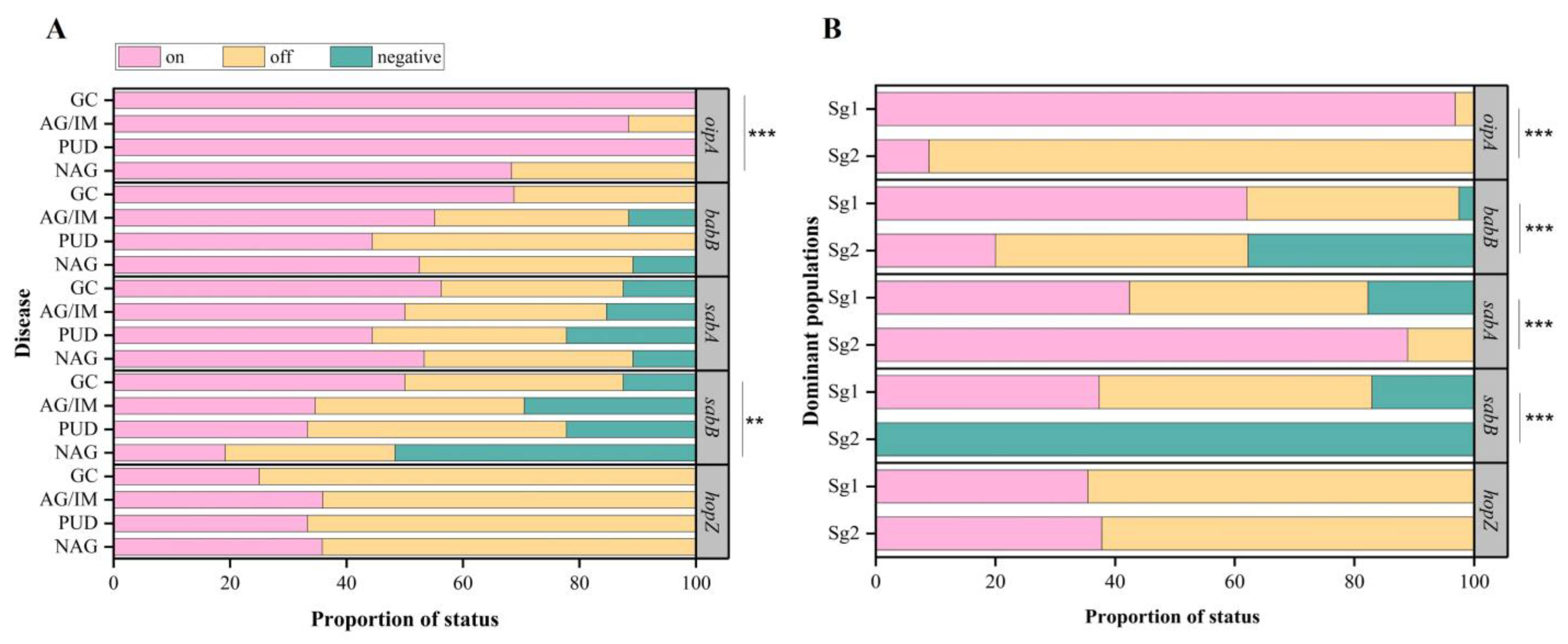
3.4. Risk Estimation of Significant Genes and Genotypes between Gastric Cancer and Other Diseases
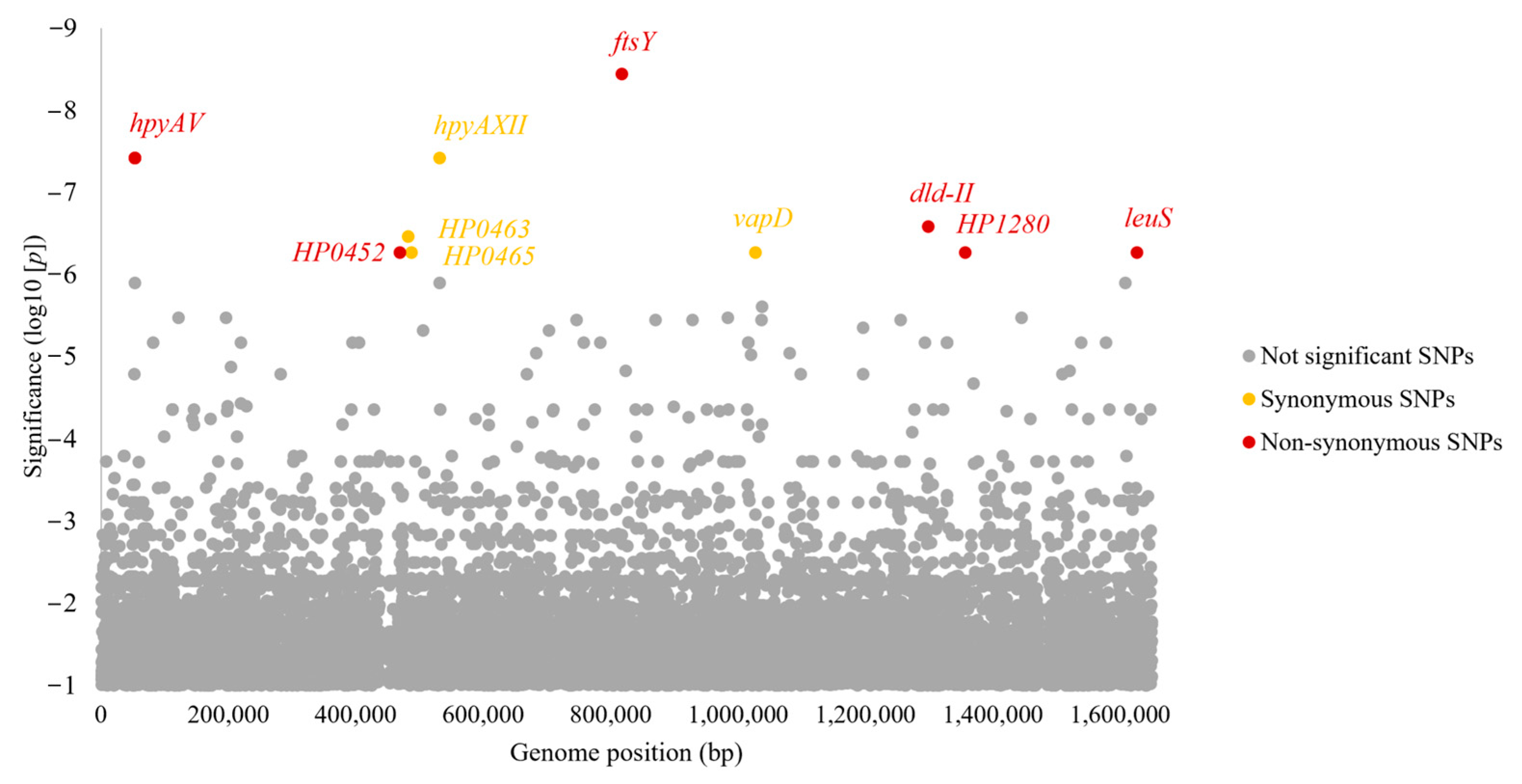
3.5. Identification of SNPs and Motifs Associated with Gastric Cancer and Other Diseases by GWAS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suerbaum, S.; Michetti, P. Helicobacter pylori infection. N. Engl. J. Med. 2002, 347, 1175–1186. [Google Scholar] [CrossRef]
- Marshall, B.J.; Goodwin, C.S.; Warren, J.R.; Murray, R.; Blincow, E.D.; Blackbourn, S.J.; Phillips, M.; Waters, T.E.; Sanderson, C.R. Prospective double-blind trial of duodenal ulcer relapse after eradication of Campylobacter pylori. Lancet 1988, 2, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, International Agency for Research on Cancer; World Health Organization. Schistosomes, Liver Flukes and Helicobacter pylori. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; World Health Organization: Geneva, Switzerland, 1994; Volume 61, pp. 1–241. [Google Scholar]
- Alfaray, R.I.; Saruuljavkhlan, B.; Ansari, S.; Fauzia, K.A.; Yamaoka, Y. Review: Epidemiology of Helicobacter pylori Infection. Microbiota Health Dis. 2022, 4, e733. [Google Scholar] [CrossRef]
- Kumar, S.; Metz, D.C.; Ellenberg, S.; Kaplan, D.E.; Goldberg, D.S. Risk Factors and Incidence of Gastric Cancer After Detection of Helicobacter pylori Infection: A Large Cohort Study. Gastroenterology 2020, 158, 527–536.e527. [Google Scholar] [CrossRef]
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [PubMed]
- Khasag, O.; Boldbaatar, G.; Tegshee, T.; Duger, D.; Dashdorj, A.; Uchida, T.; Matsuhisa, T.; Yamaoka, Y. The prevalence of Helicobacter pylori infection and other risk factors among Mongolian dyspeptic patients who have a high incidence and mortality rate of gastric cancer. Gut Pathog. 2018, 10, 14. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, Y.; Lu, N.H. Novel and Effective Therapeutic Regimens for Helicobacter pylori in an Era of Increasing Antibiotic Resistance. Front. Cell. Infect. Microbiol. 2017, 7, 168. [Google Scholar] [CrossRef]
- Hatakeyama, M.; Higashi, H. Helicobacter pylori CagA: A new paradigm for bacterial carcinogenesis. Cancer Sci. 2005, 96, 835–843. [Google Scholar] [CrossRef]
- Imai, S.; Ooki, T.; Murata-Kamiya, N.; Komura, D.; Tahmina, K.; Wu, W.; Takahashi-Kanemitsu, A.; Knight, C.T.; Kunita, A.; Suzuki, N.; et al. Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe 2021, 29, 941–958.e910. [Google Scholar] [CrossRef]
- Tserentogtokh, T.; Gantuya, B.; Subsomwong, P.; Oyuntsetseg, K.; Bolor, D.; Erdene-Ochir, Y.; Azzaya, D.; Davaadorj, D.; Uchida, T.; Matsuhisa, T.; et al. Western-Type Helicobacter pylori CagA are the Most Frequent Type in Mongolian Patients. Cancers 2019, 11, 725. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.Y.; Yan, J.J.; Yang, Y.C.; Wu, C.M.; Hu, Y.; Geng, J.L. Helicobacter pylori with East Asian-type cagPAI genes is more virulent than strains with Western-type in some cagPAI genes. Braz. J. Microbiol. 2017, 48, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Matsuhisa, T.; Yamaoka, Y.; Uchida, T.; Duger, D.; Adiyasuren, B.; Khasag, O.; Tegshee, T.; Tsogt-Ochir, B. Gastric mucosa in Mongolian and Japanese patients with gastric cancer and Helicobacter pylori infection. World J. Gastroenterol. 2015, 21, 8408–8417. [Google Scholar] [CrossRef]
- Alm, R.A.; Ling, L.S.; Moir, D.T.; King, B.L.; Brown, E.D.; Doig, P.C.; Smith, D.R.; Noonan, B.; Guild, B.C.; deJonge, B.L.; et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 1999, 397, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Rieder, G.; Fischer, W.; Haas, R. Interaction of Helicobacter pylori with host cells: Function of secreted and translocated molecules. Curr. Opin. Microbiol. 2005, 8, 67–73. [Google Scholar] [CrossRef]
- Falush, D.; Wirth, T.; Linz, B.; Pritchard, J.K.; Stephens, M.; Kidd, M.; Blaser, M.J.; Graham, D.Y.; Vacher, S.; Perez-Perez, G.I.; et al. Traces of human migrations in Helicobacter pylori populations. Science 2003, 299, 1582–1585. [Google Scholar] [CrossRef]
- Moodley, Y.; Brunelli, A.; Ghirotto, S.; Klyubin, A.; Maady, A.S.; Tyne, W.; Munoz-Ramirez, Z.Y.; Zhou, Z.; Manica, A.; Linz, B.; et al. Helicobacter pylori’s historical journey through Siberia and the Americas. Proc. Natl. Acad. Sci. USA 2021, 118, e2015523118. [Google Scholar] [CrossRef]
- Kodaman, N.; Pazos, A.; Schneider, B.G.; Piazuelo, M.B.; Mera, R.; Sobota, R.S.; Sicinschi, L.A.; Shaffer, C.L.; Romero-Gallo, J.; de Sablet, T.; et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc. Natl. Acad. Sci. USA 2014, 111, 1455–1460. [Google Scholar] [CrossRef]
- de Sablet, T.; Piazuelo, M.B.; Shaffer, C.L.; Schneider, B.G.; Asim, M.; Chaturvedi, R.; Bravo, L.E.; Sicinschi, L.A.; Delgado, A.G.; Mera, R.M.; et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut 2011, 60, 1189–1195. [Google Scholar] [CrossRef]
- You, Y.; Thorell, K.; He, L.; Yahara, K.; Yamaoka, Y.; Cha, J.H.; Murakami, K.; Katsura, Y.; Teamhp; Kobayashi, I.; et al. Genomic differentiation within East Asian Helicobacter pylori. Microb. Genom. 2022, 8, 000676. [Google Scholar] [CrossRef]
- Berthenet, E.; Yahara, K.; Thorell, K.; Pascoe, B.; Meric, G.; Mikhail, J.M.; Engstrand, L.; Enroth, H.; Burette, A.; Megraud, F.; et al. A GWAS on Helicobacter pylori strains points to genetic variants associated with gastric cancer risk. BMC Biol. 2018, 16, 84. [Google Scholar] [CrossRef] [PubMed]
- Tuan, V.P.; Yahara, K.; Dung, H.D.Q.; Binh, T.T.; Huu Tung, P.; Tri, T.D.; Thuan, N.P.M.; Khien, V.V.; Trang, T.T.H.; Phuc, B.H.; et al. Genome-wide association study of gastric cancer- and duodenal ulcer-derived Helicobacter pylori strains reveals discriminatory genetic variations and novel oncoprotein candidates. Microb. Genom. 2021, 7, 000680. [Google Scholar] [CrossRef]
- Sharaf, R.N.; Shergill, A.K.; Odze, R.D.; Krinsky, M.L.; Fukami, N.; Jain, R.; Appalaneni, V.; Anderson, M.A.; Ben-Menachem, T.; Chandrasekhara, V.; et al. Endoscopic mucosal tissue sampling. Gastrointest. Endosc. 2013, 78, 216–224. [Google Scholar] [CrossRef]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, G.; Cendron, L. Structural and functional aspects of the Helicobacter pylori secretome. World J. Gastroenterol. 2014, 20, 1402–1423. [Google Scholar] [CrossRef]
- Alm, R.A.; Bina, J.; Andrews, B.M.; Doig, P.; Hancock, R.E.; Trust, T.J. Comparative genomics of Helicobacter pylori: Analysis of the outer membrane protein families. Infect. Immun. 2000, 68, 4155–4168. [Google Scholar] [CrossRef]
- Delahay, R.M.; Croxall, N.J.; Stephens, A.D. Phylogeographic diversity and mosaicism of the Helicobacter pylori tfs integrative and conjugative elements. Mob. DNA 2018, 9, 5. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kim, A.; Servetas, S.L.; Kang, J.; Kim, J.; Jang, S.; Cha, H.J.; Lee, W.J.; Kim, J.; Romero-Gallo, J.; Peek, R.M., Jr.; et al. Helicobacter pylori bab Paralog Distribution and Association with cagA, vacA, and homA/B Genotypes in American and South Korean Clinical Isolates. PLoS ONE 2015, 10, e0137078. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.; Ortkamp, M.; Diehl, K.D.; Hundt, E.; Knapp, B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999, 27, 3325–3333. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Peek, R.M.; Pride, D.; Levine, S.M.; Takata, T.; Lee, Y.C.; Kusugami, K.; van der Ende, A.; Kuipers, E.J.; Kusters, J.G.; et al. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 2002, 40, 239–246. [Google Scholar] [CrossRef]
- de Jonge, R.; Pot, R.G.; Loffeld, R.J.; van Vliet, A.H.; Kuipers, E.J.; Kusters, J.G. The functional status of the Helicobacter pylori sabB adhesin gene as a putative marker for disease outcome. Helicobacter 2004, 9, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Holley, G.; Melsted, P. Bifrost: Highly parallel construction and indexing of colored and compacted de Bruijn graphs. Genome Biol. 2020, 21, 249. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629. [Google Scholar] [CrossRef]
- Sharafutdinov, I.; Tegtmeyer, N.; Linz, B.; Rohde, M.; Vieth, M.; Tay, A.C.; Lamichhane, B.; Tuan, V.P.; Fauzia, K.A.; Sticht, H.; et al. A single-nucleotide polymorphism in Helicobacter pylori promotes gastric cancer development. Cell Host Microbe 2023, 31, 1345–1358. [Google Scholar] [CrossRef]
- Cross, B.C.; Sinning, I.; Luirink, J.; High, S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 2009, 10, 255–264. [Google Scholar] [CrossRef]
- Steinberg, R.; Knüpffer, L.; Origi, A.; Asti, R.; Koch, H.G. Co-translational protein targeting in bacteria. FEMS Microbiol. Lett. 2018, 365, fny095. [Google Scholar] [CrossRef]
- Pinchuk, G.E.; Rodionov, D.A.; Yang, C.; Li, X.; Osterman, A.L.; Dervyn, E.; Geydebrekht, O.V.; Reed, S.B.; Romine, M.F.; Collart, F.R.; et al. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc. Natl. Acad. Sci. USA 2009, 106, 2874–2879. [Google Scholar] [CrossRef] [PubMed]
- Correa, P.; Houghton, J. Carcinogenesis of Helicobacter pylori. Gastroenterology 2007, 133, 659–672. [Google Scholar] [CrossRef]
- Toller, I.M.; Neelsen, K.J.; Steger, M.; Hartung, M.L.; Hottiger, M.O.; Stucki, M.; Kalali, B.; Gerhard, M.; Sartori, A.A.; Lopes, M.; et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA 2011, 108, 14944–14949. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629–641. [Google Scholar] [CrossRef]
- Hussein, N.R. Helicobacter pylori and gastric cancer in the Middle East: A new enigma? World J. Gastroenterol. 2010, 16, 3226–3234. [Google Scholar] [CrossRef]
- Rhead, J.L.; Letley, D.P.; Mohammadi, M.; Hussein, N.; Mohagheghi, M.A.; Eshagh Hosseini, M.; Atherton, J.C. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 2007, 133, 926–936. [Google Scholar] [CrossRef]
- Soyfoo, D.M.; Doomah, Y.H.; Xu, D.; Zhang, C.; Sang, H.-M.; Liu, Y.-Y.; Zhang, G.-X.; Jiang, J.-X.; Xu, S.-F. New genotypes of Helicobacter pylori VacA d-region identified from global strains. BMC Mol. Cell Biol. 2021, 22, 4. [Google Scholar] [CrossRef]
- O’Brien, V.P.; Jackson, L.K.; Frick, J.P.; Rodriguez Martinez, A.E.; Jones, D.S.; Johnston, C.D.; Salama, N.R. Helicobacter pylori Chronic Infection Selects for Effective Colonizers of Metaplastic Glands. mBio 2023, 14, e0311622. [Google Scholar] [CrossRef]
- Liu, J.; He, C.; Chen, M.; Wang, Z.; Xing, C.; Yuan, Y. Association of presence/absence and on/off patterns of Helicobacter pylori oipA gene with peptic ulcer disease and gastric cancer risks: A meta-analysis. BMC Infect. Dis. 2013, 13, 555. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Kudo, T.; Lu, H.; Casola, A.; Brasier, A.R.; Graham, D.Y. Role of interferon-stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology 2004, 126, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef] [PubMed]
- Javaheri, A.; Kruse, T.; Moonens, K.; Mejías-Luque, R.; Debraekeleer, A.; Asche, C.I.; Tegtmeyer, N.; Kalali, B.; Bach, N.C.; Sieber, S.A.; et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016, 2, 16189. [Google Scholar] [CrossRef] [PubMed]
- Leylabadlo, H.E.; Yekani, M.; Ghotaslou, R. Helicobacter pylori hopQ alleles (type I and II) in gastric cancer. Biomed. Rep. 2016, 4, 601–604. [Google Scholar] [CrossRef]
- Lu, H.; Hsu, P.-I.; Graham, D.Y.; Yamaoka, Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology 2005, 128, 833–848. [Google Scholar] [CrossRef]
- Jung, S.W.; Sugimoto, M.; Shiota, S.; Graham, D.Y.; Yamaoka, Y. The intact dupA cluster is a more reliable Helicobacter pylori virulence marker than dupA alone. Infect. Immun. 2012, 80, 381–387. [Google Scholar] [CrossRef]
- Phuc, B.H.; Tuan, V.P.; Dung, H.D.Q.; Binh, T.T.; Tung, P.H.; Tri, T.D.; Thuan, N.P.M.; Van Khien, V.; Trang, T.T.H.; Akada, J.; et al. Helicobacter pylori type 4 secretion systems as gastroduodenal disease markers. Sci. Rep. 2021, 11, 4584. [Google Scholar] [CrossRef]
- Waskito, L.A.; Miftahussurur, M.; Lusida, M.I.; Syam, A.F.; Suzuki, R.; Subsomwong, P.; Uchida, T.; Hamdan, M.; Nasronudin; Yamaoka, Y. Distribution and clinical associations of integrating conjugative elements and cag pathogenicity islands of Helicobacter pylori in Indonesia. Sci. Rep. 2018, 8, 6073. [Google Scholar] [CrossRef]
- Alandiyjany, M.N.; Croxall, N.J.; Grove, J.I.; Delahay, R.M. A role for the tfs3 ICE-encoded type IV secretion system in pro-inflammatory signalling by the Helicobacter pylori Ser/Thr kinase, CtkA. PLoS ONE 2017, 12, e0182144. [Google Scholar] [CrossRef]
- Iwatani, S.; Nagashima, H.; Reddy, R.; Shiota, S.; Graham, D.Y.; Yamaoka, Y. Identification of the Genes That Contribute to Lactate Utilization in Helicobacter pylori. PLoS ONE 2014, 9, e103506. [Google Scholar] [CrossRef][Green Version]
- Matsumoto, T.; Takahashi, T.; Yamada, H. A novel approach for screening of new anti-Helicobacter pylori substances. Biol. Pharm. Bull. 2008, 31, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Keilberg, D.; Steele, N.; Fan, S.; Yang, C.; Zavros, Y.; Ottemann, K.M. Gastric Metabolomics Detects Helicobacter pylori Correlated Loss of Numerous Metabolites in Both the Corpus and Antrum. Infect. Immun. 2021, 89, e00690-20. [Google Scholar] [CrossRef]
- Foegeding, N.J.; Caston, R.R.; McClain, M.S.; Ohi, M.D.; Cover, T.L. An Overview of Helicobacter pylori VacA Toxin Biology. Toxins 2016, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Morales-Espinosa, R.; Delgado, G.; Serrano, L.-R.; Castillo, E.; Santiago, C.A.; Hernández-Castro, R.; Gonzalez-Pedraza, A.; Mendez, J.L.; Mundo-Gallardo, L.F.; Manzo-Merino, J.; et al. High expression of Helicobacter pylori VapD in both the intracellular environment and biopsies from gastric patients with severity. PLoS ONE 2020, 15, e0230220. [Google Scholar] [CrossRef] [PubMed]
- Radin, J.N.; Gaddy, J.A.; González-Rivera, C.; Loh, J.T.; Algood, H.M.; Cover, T.L. Flagellar localization of a Helicobacter pylori autotransporter protein. mBio 2013, 4, e00613-00612. [Google Scholar] [CrossRef]
- Haley, K.P.; Gaddy, J.A. Metalloregulation of Helicobacter pylori physiology and pathogenesis. Front. Microbiol. 2015, 6, 911. [Google Scholar] [CrossRef]
- Gantuya, B.; El Serag, H.B.; Matsumoto, T.; Ajami, N.J.; Uchida, T.; Oyuntsetseg, K.; Bolor, D.; Yamaoka, Y. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment. Pharmacol. Ther. 2020, 51, 770–780. [Google Scholar] [CrossRef]
| H. pylori. Populations | Non-GC Group | GC group (n = 16) | OR a (95% CI) | p b | ||
|---|---|---|---|---|---|---|
| NAG (n = 120) | PUD (n = 9) | AG/IM (n = 78) | ||||
| Sg1 (n = 158) | 73 (60.8%) | 8 (88.9%) | 62 (79.5%) | 15 (93.8%) | 6.7 (0.9–51.9) | 0.044 |
| Other (n = 65) | 47 (39.2%) | 1 (11.1%) | 16 (20.5%) | 1 (6.2%) | ||
| Genes | Genotypes | NAG (n = 120) | PUD (n = 9) | AG/IM (n = 78) | GC (n = 16) | Total (n = 223) | p a |
|---|---|---|---|---|---|---|---|
| cagA | AB 1 | 4 (3.3%) | 0 | 0 | 0 | 4 (1.8%) | 0.006 |
| ABC 2 | 49 (40.8%) | 3 (33.3%) | 43 (55.1%) | 8 (50.0%) | 103 (46.2%) | ||
| ABCn 3 | 25 (20.8%) | 4 (44.4%) | 19 (24.4%) | 7 (43.8%) | 55 (24.7%) | ||
| ABD 4 | 2 (1.7%) | 1 (11.1%) | 5 (6.4%) | 0 | 8 (3.6%) | ||
| Negative | 40 (33.3%) | 1 (11.1%) b | 11 (14.1%) b | 1 (6.3%) b | 53 (23.8%) | ||
| vacA | s1i1m1 | 52 (43.3%) | 6 (66.7%) b | 53 (67.9%) b | 13 (81.3%) b | 124 (55.6%) | 0.014 |
| s1i1m2 | 12 (10.0%) | 1 (11.1%) | 4 (5.1%) | 1 (6.3%) | 18 (8.1%) | ||
| s1i3m2 | 19 (15.8%) | 1 (11.1%) | 10 (12.8%) | 1 (6.3%) | 31 (13.9%) | ||
| s2i3m2 | 31 (25.8%) | 0 b | 7 (9.0%) b | 1 (6.3%) b | 39 (17.5%) | ||
| other | 6 (5.0%) | 1 (11.1%) | 4 (5.1%) | 0 | 11 (4.9%) | ||
| htrA | S variant | 96 (80.0%) | 9 (100%) | 69 (88.5%) | 14 (87.5%) | 188 (84.3%) | 0.118 |
| L variant | 24 (20.0%) | 0 | 9 (11.5%) | 2 (12.5%) | 35 (15.7%) | ||
| iceA | Type 1 | 57 (47.5%) | 7 (77.8%) | 41 (52.6%) | 4 (25.0%) | 109 (48.9%) | 0.055 |
| Type 2A | 2 (1.7%) | 0 | 4 (5.1%) | 2 (12.5%) | 8 (3.6%) | ||
| Type 2B | 16 (13.3%) | 0 | 7 (9.0%) | 0 | 23 (10.3%) | ||
| Type 2C | 20 (16.7%) | 1 (11.1%) | 9 (11.5%) | 1 (6.3%) | 31 (13.9%) | ||
| Type 2D | 24 (20.0%) | 1 (11.1%) b | 17 (21.8%) | 9 (56.3%) b | 51 (22.9%) | ||
| Negative | 1 (0.8%) | 0 | 0 | 0 | 1 (0.4%) | ||
| babA | AD2 | 2 (1.7%) | 0 | 1 (1.3%) | 0 | 3 (1.3%) | 0.417 |
| AD3 | 36 (30.0%) | 5 (55.6%) | 25 (32.1%) | 4 (25.0%) | 70 (31.4%) | ||
| AD4 | 77 (64.2%) | 4 (44.4%) | 52 (66.7%) | 11 (68.8%) | 144 (64.6%) | ||
| Negative | 5 (4.2%) | 0 | 0 | 1 (6.3%) | 6 (2.7%) | ||
| babB | BD1 | 22 (18.3%) | 2 (22.2%) | 19 (24.4%) | 5 (31.3%) | 48 (21.5%) | 0.516 |
| BD2 | 46 (38.3%) | 4 (44.4%) | 30 (38.5%) | 9 (56.3%) | 89 (39.9%) | ||
| BD3 | 19 (15.8%) | 2 (22.2%) | 10 (12.8%) | 1 (6.3%) | 32 (14.3%) | ||
| unclassified | 21 (17.5%) | 1 (11.1%) | 10 (12.8%) | 1 (6.3%) | 33 (14.8%) | ||
| Negative | 12 (10.0%) | 0 | 9 (11.5%) | 0 | 21 (9.4%) | ||
| hopQ | Type I | 74 (61.7%) | 8 (88.9%) b | 65 (83.3%) b | 15 (93.8%) b | 162 (72.6%) | <0.001 |
| Type II | 42 (35.0%) | 0 b | 9 (11.5%) b | 1 (6.3%) b | 52 (23.3%) | ||
| Type I/II | 4 (3.3%) | 1 (11.1%) | 4 (5.1%) | 0 | 9 (4.0%) | ||
| hopZ | Type I | 97 (80.8%) | 8 (88.9%) | 73 (93.6%) | 14 (87.5%) | 192 (86.1%) | 0.121 |
| Type II | 23 (19.2%) | 1 (11.1%) | 5 (6.4%) | 2 (12.5%) | 31 (13.9%) | ||
| sabA | Present | 107 (89.2%) | 7 (77.8%) | 66 (84.6%) | 14 (87.5%) | 194 (87.0%) | 0.688 |
| Negative | 13 (10.8%) | 2 (22.2%) | 12 (15.4%) | 2 (12.5%) | 29 (13.0%) | ||
| sabB | Present | 58 (48.3%) | 7 (77.8%) b | 55 (70.5%) b | 14 (87.5%) b | 134 (60.1%) | 0.001 |
| Negative | 62 (51.7%) | 2 (22.2%) b | 23 (29.5%) b | 2 (12.5%) b | 89 (39.9%) | ||
| homA/B | homA | 67 (55.8%) | 3 (33.3%) b | 32 (41.0%) b | 3 (18.8%) b | 105 (47.1%) | 0.115 |
| homB | 45 (37.5%) | 6 (66.7%) | 41 (52.6%) | 10 (62.5%) | 102 (45.7%) | ||
| homA/B | 5 (4.2%) | 0 | 3 (3.8%) | 2 (12.5%) | 10 (4.5%) | ||
| homI | 3 (2.5%) | 0 | 2 (2.6%) | 1 (6.3%) | 6 (2.7%) |
| T4SS | Genotypes | NAG (n = 120) | PUD (n = 9) | AG/IM (n = 78) | GC (n = 16) | Total (n = 223) | p a | ||
|---|---|---|---|---|---|---|---|---|---|
| cagPAI | Presence § | Y | 80 (66.7%) | 8 (88.9%) b | 68 (87.2%) b | 16 (100.0%) b | 172 (77.1%) | <0.001 | |
| N | 40 (33.3%) | 1 (11.1%) b | 10 (12.8%) b | 0 (0.0%) b | 51 (22.9%) | ||||
| Completeness §§ | Y | 79 (65.8%) | 8 (88.9%) b | 67 (85.9%) b | 15 (93.8%) b | 169 (75.8%) | <0.001 | ||
| N | 1 (0.8%) | 0 b | 1 (1.3%) b | 1 (6.3%) b | 3 (1.3%) | ||||
| cagA presence | Y | 80 (66.7%) | 8 (88.9%) b | 67 (85.9%) b | 15 (93.7%) b | 170 (98.8%) | 0.274 | ||
| N | 40 (33.3%) | 1 (11.1%) b | 11 (14.1%) b | 1 (6.3%) b | 2 (1.2%) | ||||
| comB | Presence § and Completeness §§ | Y | 120 (100%) | 9 (100%) | 78 (100%) | 16 (100%) | 223 (100%) | - | |
| N | 0 | 0 | 0 | 0 | 0 | ||||
| tfs3 | Presence § | Y | 84 (70.0%) | 7 (77.8%) | 57 (73.1%) | 11 (68.8%) | 159 (71.3%) | 0.927 | |
| N | 36 (30.0%) | 2 (22.2%) | 21 (26.9%) | 5 (31.3%) | 64 (28.7%) | ||||
| Completeness §§ | Y | 28 (23.3%) | 1 (11.1%) | 12 (15.4%) | 1 (6.3%) | 42 (18.8%) | 0.132 | ||
| N | 56 (46.7%) | 6 (66.7%) | 45 (57.7%) | 10 (62.5%) | 117 (52.5%) | ||||
| ctkA | Y | 43 (35.8%) | 5 (55.6%) | 31 (39.7%) | 4 (25.0%) | 83 (37.2%) | 0.451 | ||
| N | 77 (64.2%) | 4 (44.4%) | 47 (60.3%) | 12 (75.0%) | 140 (62.8%) | ||||
| tfs4 | Presence § | Y | 101 (84.2%) | 8 (88.9%) | 74 (94.9%) | 16 (100.0%) b | 199 (89.2%) | 0.022 | |
| N | 19 (15.8%) | 1 (11.1%) | 4 (5.1%) | 0 b | 24 (10.8%) | ||||
| Completeness §§ | Y | 42 (35.0%) | 1 (11.1%) | 16 (20.5%) b | 2 (12.5%) b | 61 (27.4%) | 0.005 | ||
| N | 59 (49.2%) | 7 (77.8%) | 58 (74.4%) b | 14 (87.5%) b | 138 (61.9%) | ||||
| dupA | Y | 37 (30.8%) | 2 (22.2%) | 15 (19.2%) | 2 (12.5%) | 56 (25.1%) | 0.163 | ||
| N | 83 (69.2%) | 7 (77.8%) | 63 (80.8%) | 14 (87.5%) | 167 (74.9%) | ||||
| Type | tfs4a | 7 (5.8%) | 0 | 1 (1.3%) | 1 (6.3%) | 9 (4.0%) | 0.012 | ||
| tfs4b | 31 (25.8%) | 1 (11.1%) | 12 (15.4%) | 1 (6.3%) | 45 (20.2%) | ||||
| tfs4c | 1 (0.8%) | 0 | 2 (2.6%) | 1 (6.3%) | 4 (1.8%) | ||||
| tfs4-other §§§ | 62 (51.7%) | 7 (77.8%) b | 59 (75.6%) b | 13 (81.3%) b | 141 (63.2%) | ||||
| Combinations | cagPAI+tfs3 | 69 (57.5%) | 6 (66.7%) | 51 (62.4%) | 11 (68.8%) | 137 (61.4%) | 0.224 | ||
| cagPAI+tfs4 | 72 (60.0%) | 7 (77.8%) | 64 (82.1%) | 16 (100%) | 159 (71.3%) | 0.229 | |||
| tfs3+tfs4 | 75 (62.5%) | 6 (66.7%) | 54 (69.2%) | 11 (68.8%) | 146 (65.5%) | 0.313 | |||
| tfs3+tfs4a | 2 (1.7%) | 0 | 1 (1.3%) | 0 | 3 (1.3%) | 0.386 | |||
| tfs3+tfs4b | 19 (15.8%) | 0 | 10 (12.8%) | 1 (6.3%) | 30 (7.4%) | ||||
| tfs3+tfs4c | 0 | 0 | 0 | 0 | 0 | ||||
| All T4SS | 62 (51.7%) | 5 (55.6%) | 48 (61.5%) | 11 (68.8%) | 126 (56.5%) | 0.398 | |||
| Univariate Logistic Regression | |||
|---|---|---|---|
| OR a | OR 95% CI | p | |
| cagA presence/oipA “on” vs. cagA absence/oipA “off” status | 5.0 | 0.6–39.0 | 0.122 |
| vacA s1i1m1 vs. vacA other alleles | 3.7 | 1.0–13.5 | 0.044 |
| hopQ-I vs. II or I/II alleles | 6.1 | 0.8–47.4 | 0.083 |
| sabB presence vs. absence | 5.1 | 1.1–22.9 | 0.035 |
| sabB “on” vs. “off” status | 2.9 | 1.0–8.1 | 0.042 |
| tfs4 complete vs. incomplete | 0.4 | 0.1–1.6 | 0.183 |
| tfs4b vs. tfs4 other types | 0.3 | 0.0–1.9 | 0.182 |
| ceuE presence vs. absence | 0.14 | 0.0–1.1 | 0.062 |
| hopD presence vs. absence | 0.27 | 0.1–0.8 | 0.014 |
| frpB2 presence vs. absence | 4.3 | 0.6–33.3 | 0.164 |
| Iron regulated OMP genes (numbers) many vs. less | 2.3 | 0.6–8.9 | 0.240 |
| Types | p | Gene Locus in 26,695 | Gene Name | Position in the 26,695 Genome | Position in the Gene | Safe Genotype | Risk Genotype | Frequency of Non-GC/GC, % | Effect on Amino Acid Sequence | Predicted Function |
|---|---|---|---|---|---|---|---|---|---|---|
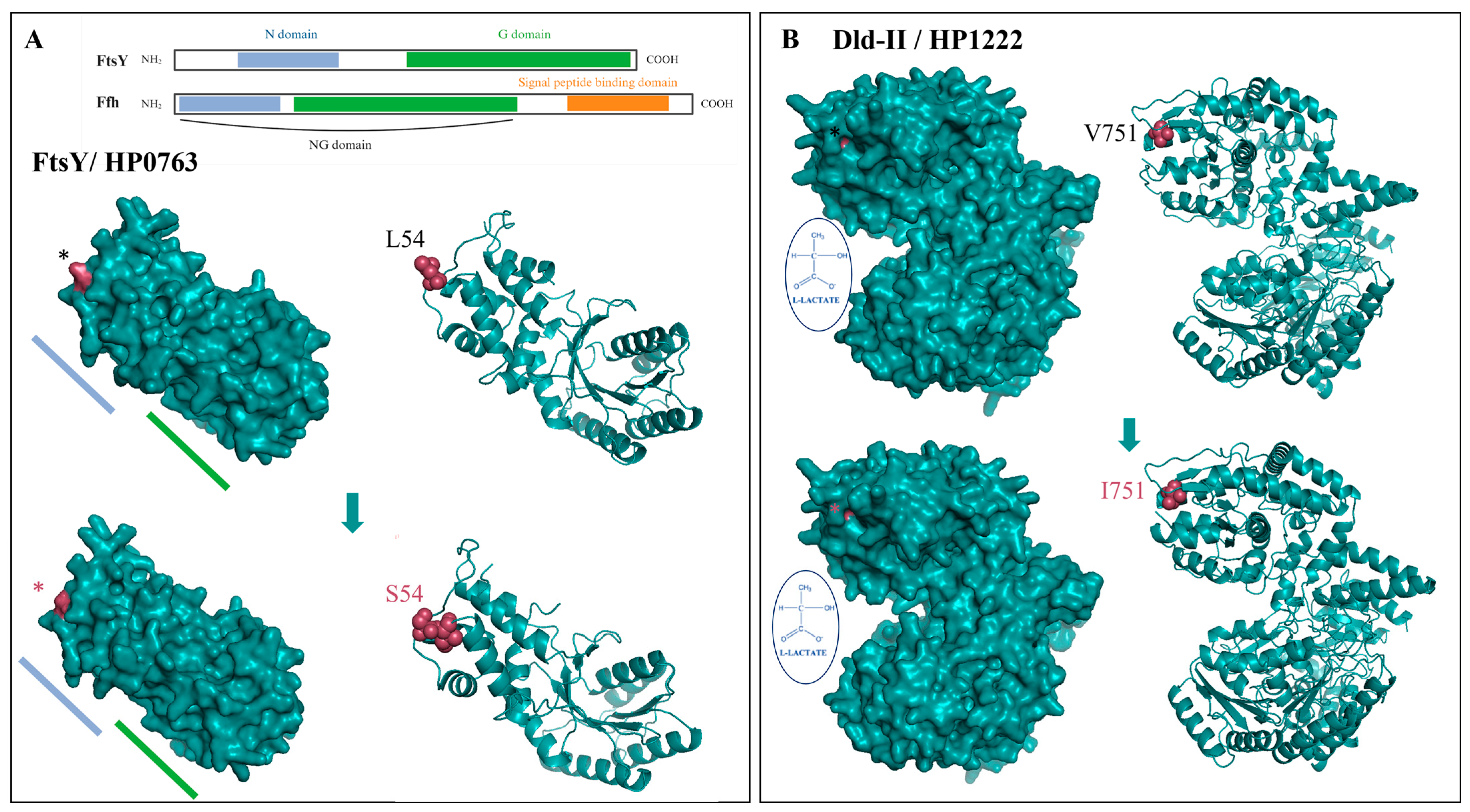
| SNP | 3.62 × 10−9 | HP0763 | ftsY | 817,043 | 161 | T | C | 2.8/40.0 | Leu54Ser | Signal recognition particle-docking protein |
| SNP | 3.80 × 10−8 | HP0053 | hpyAV | 52,671 | 1048 | G | A | 3.5/40.0 | Leu350Phe | R-M system type II |
| SNP | 3.80 × 10−8 | 52,679 | 1039 | G | T | 3.5/40.0 | Thr347Asn | |||
| SNP | 3.80 × 10−8 | 52,681 | 1041 | C | T | 3.5/40.0 | ||||
| SNP | 2.57 × 10−7 | HP1222 | dld-II | 1,297,719 | 2251 | C | T | 4.2/40.0 | Val751Ile | FAD-binding and (Fe-S)-binding domain-containing protein |
| SNP | 5.39 × 10−7 | HP0452 | hyp a | 469,160 | 405 | G | T | 2.8/33.3 | Glu135Asp | (McrB family protein) |
| SNP | 5.39 × 10−7 | HP1280 | hyp a | 1,355,874 | 808 | C | T | 2.8/33.3 | Thr270Val | (Anthranilate phosphoribosyltransferase) |
| SNP | 5.39 × 10−7 | HP1547 | leuS a | 1,625,904 | 2014 | C | T | 2.8/33.3 | Ala672Thr | Leuci-tRNA ligase |
| k-mer | 3.62 × 10−9 | HP1220 | yhcG | 1,295,987–1,296,020 | 283–316 | other b | TTTTACAAGGATTTTTTTAGCGATTTTGATCCAT | 2.8/40.0 | multiple | ABC transporter ATP-binding protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saruuljavkhlan, B.; Alfaray, R.I.; Oyuntsetseg, K.; Gantuya, B.; Khangai, A.; Renchinsengee, N.; Matsumoto, T.; Akada, J.; Azzaya, D.; Davaadorj, D.; et al. Study of Helicobacter pylori Isolated from a High-Gastric-Cancer-Risk Population: Unveiling the Comprehensive Analysis of Virulence-Associated Genes including Secretion Systems, and Genome-Wide Association Study. Cancers 2023, 15, 4528. https://doi.org/10.3390/cancers15184528
Saruuljavkhlan B, Alfaray RI, Oyuntsetseg K, Gantuya B, Khangai A, Renchinsengee N, Matsumoto T, Akada J, Azzaya D, Davaadorj D, et al. Study of Helicobacter pylori Isolated from a High-Gastric-Cancer-Risk Population: Unveiling the Comprehensive Analysis of Virulence-Associated Genes including Secretion Systems, and Genome-Wide Association Study. Cancers. 2023; 15(18):4528. https://doi.org/10.3390/cancers15184528
Chicago/Turabian StyleSaruuljavkhlan, Batsaikhan, Ricky Indra Alfaray, Khasag Oyuntsetseg, Boldbaatar Gantuya, Ayush Khangai, Namsrai Renchinsengee, Takashi Matsumoto, Junko Akada, Dashdorj Azzaya, Duger Davaadorj, and et al. 2023. "Study of Helicobacter pylori Isolated from a High-Gastric-Cancer-Risk Population: Unveiling the Comprehensive Analysis of Virulence-Associated Genes including Secretion Systems, and Genome-Wide Association Study" Cancers 15, no. 18: 4528. https://doi.org/10.3390/cancers15184528
APA StyleSaruuljavkhlan, B., Alfaray, R. I., Oyuntsetseg, K., Gantuya, B., Khangai, A., Renchinsengee, N., Matsumoto, T., Akada, J., Azzaya, D., Davaadorj, D., & Yamaoka, Y. (2023). Study of Helicobacter pylori Isolated from a High-Gastric-Cancer-Risk Population: Unveiling the Comprehensive Analysis of Virulence-Associated Genes including Secretion Systems, and Genome-Wide Association Study. Cancers, 15(18), 4528. https://doi.org/10.3390/cancers15184528











