Non-Invasive Assessment of Isocitrate Dehydrogenase-Mutant Gliomas Using Optimized Proton Magnetic Resonance Spectroscopy on a Routine Clinical 3-Tesla MRI
Simple Summary
Abstract
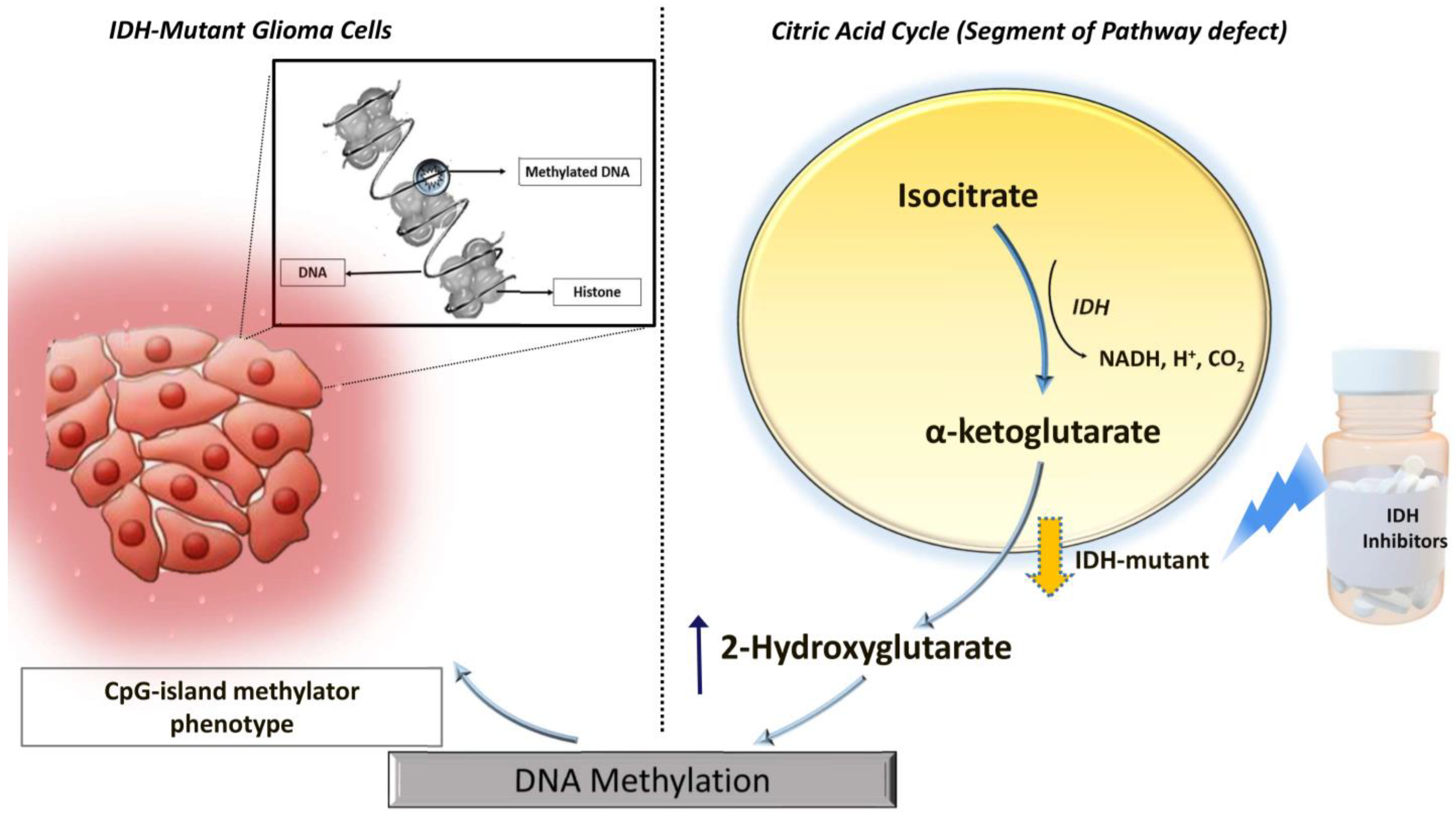
1. Introduction
2. Methods
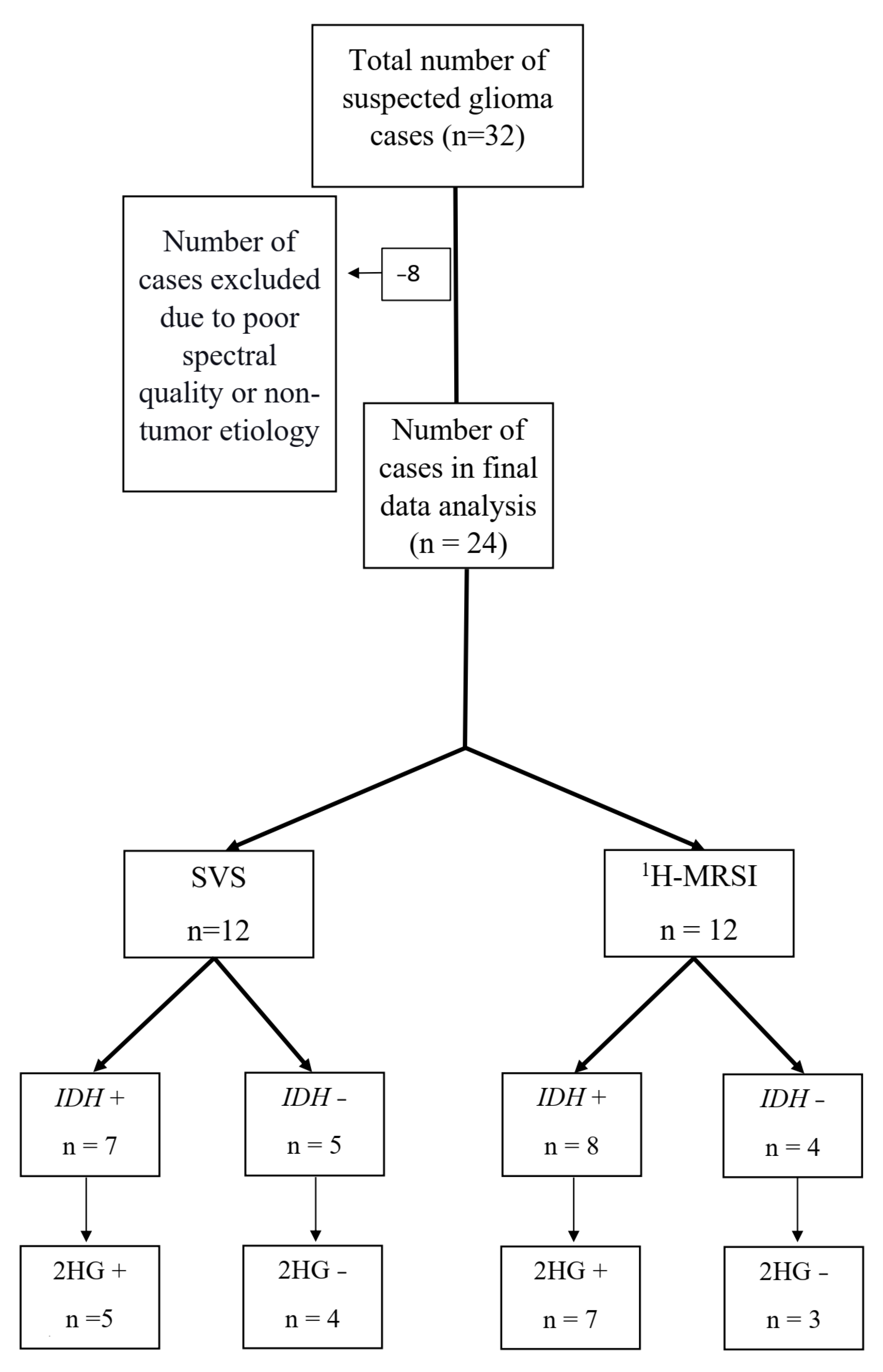
2.1. Subjects
2.2. Data Acquisition
2.3. Data Processing
2.4. Determination of IDH Status Using Immunohistochemistry
2.5. Statistical Analysis
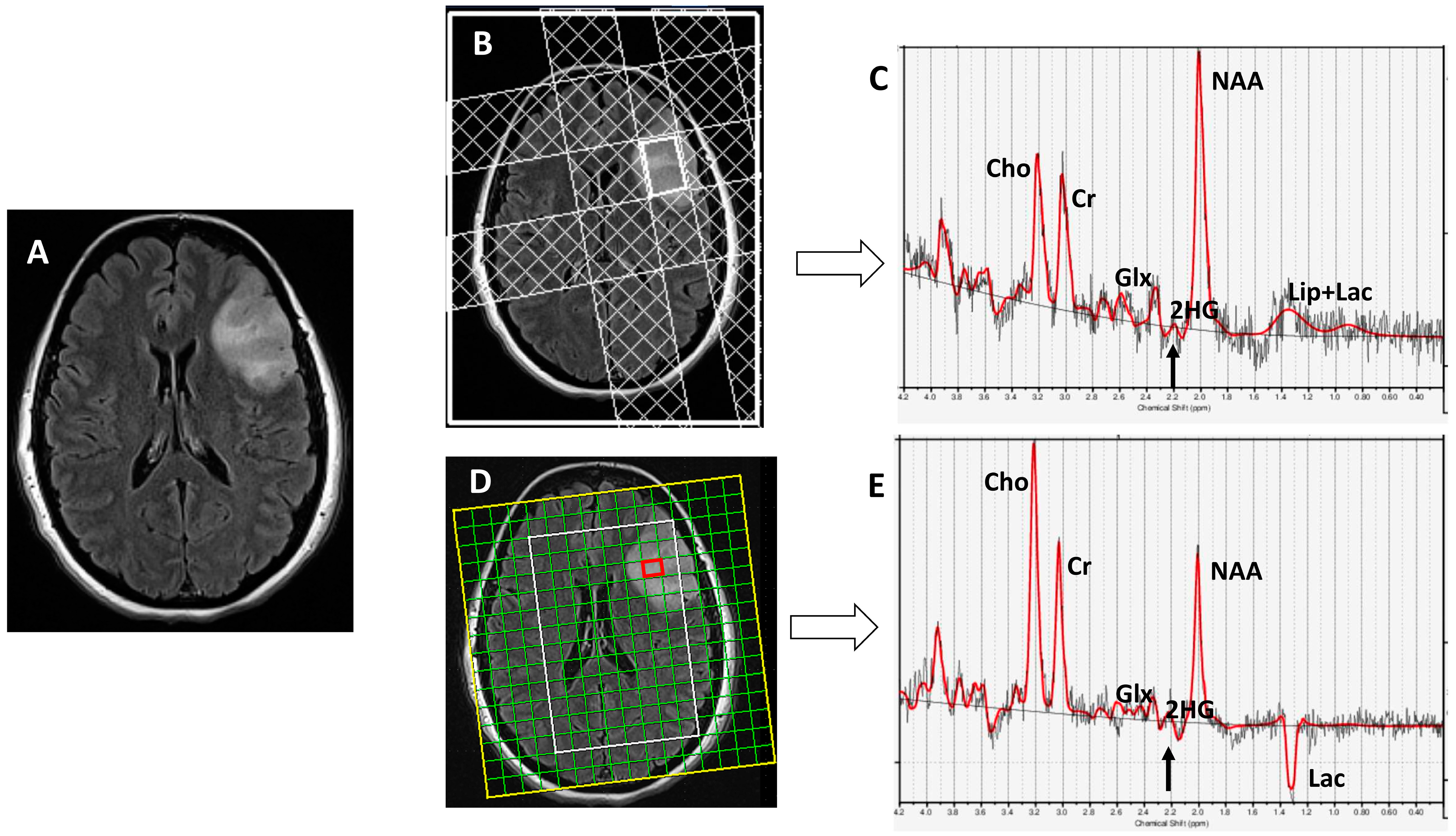
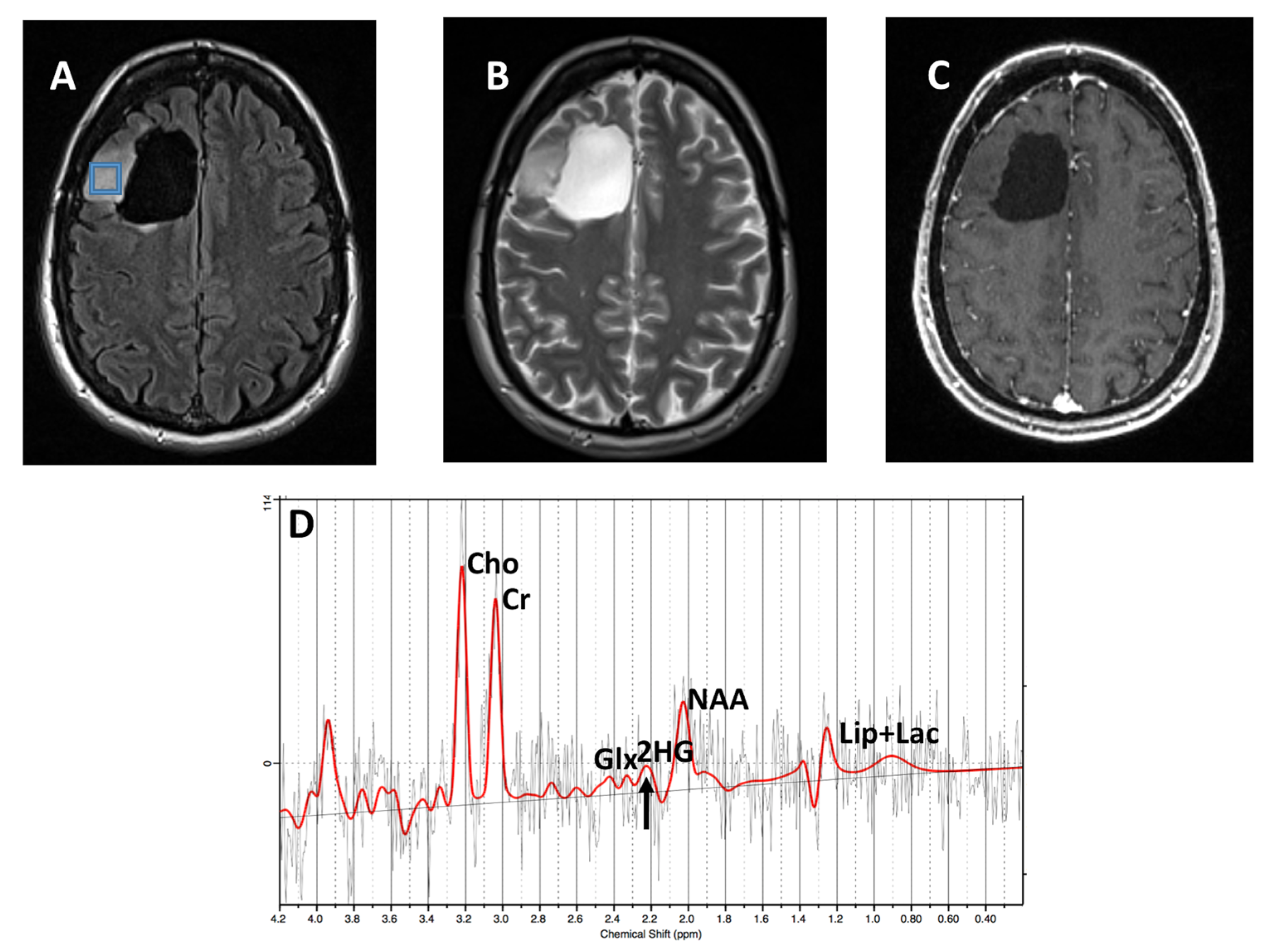
3. Results
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zhang, W.; You, G.; Bao, Z.; Wang, Y.; Liu, Y.; Kang, C.; You, Y.; Wang, L.; Jiang, T. Correlation of IDH1 Mutation with Clinicopathologic Factors and Prognosis in Primary Glioblastoma: A Report of 118 Patients from China. PLoS ONE 2012, 7, e30339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-B.; Bao, Z.-S.; Wang, H.-J.; Yan, W.; Liu, Y.-W.; Li, M.-Y.; Zhang, W.; Chen, L.; Jiang, T. Correlation of IDH1/2 Mutation with Clinicopathologic Factors and Prognosis in Anaplastic Gliomas: A Report of 203 Patients from China. J. Cancer Res. Clin. Oncol. 2014, 140, 45–51. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Bertholdo, D.; Watcharakorn, A.; Castillo, M. Brain Proton Magnetic Resonance Spectroscopy: Introduction and Overview. Neuroimaging Clin. N. Am. 2013, 23, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Posse, S.; Otazo, R.; Dager, S.R.; Alger, J. MR Spectroscopic Imaging: Principles and Recent Advances. J. Magn. Reson. Imaging JMRI 2013, 37, 1301–1325. [Google Scholar] [CrossRef]
- Salzillo, T.C.; Hu, J.; Nguyen, L.; Whiting, N.; Lee, J.; Weygand, J.; Dutta, P.; Pudakalakatti, S.; Millward, N.Z.; Gammon, S.T.; et al. Interrogating Metabolism in Brain Cancer. Magn. Reson. Imaging Clin. N. Am. 2016, 24, 687–703. [Google Scholar] [CrossRef]
- Martín Noguerol, T.; Sánchez-González, J.; Martínez Barbero, J.P.; García-Figueiras, R.; Baleato-González, S.; Luna, A. Clinical Imaging of Tumor Metabolism with 1H Magnetic Resonance Spectroscopy. Magn. Reson. Imaging Clin. N. Am. 2016, 24, 57–86. [Google Scholar] [CrossRef] [PubMed]
- Chawla, S.; Krejza, J.; Vossough, A.; Zhang, Y.; Kapoor, G.S.; Wang, S.; O’Rourke, D.M.; Melhem, E.R.; Poptani, H. Differentiation between Oligodendroglioma Genotypes Using Dynamic Susceptibility Contrast Perfusion-Weighted Imaging and Proton MR Spectroscopy. AJNR Am. J. Neuroradiol. 2013, 34, 1542–1549. [Google Scholar] [CrossRef] [PubMed]
- Chawla, S.; Oleaga, L.; Wang, S.; Krejza, J.; Wolf, R.L.; Woo, J.H.; O’Rourke, D.M.; Judy, K.D.; Grady, M.S.; Melhem, E.R.; et al. Role of Proton Magnetic Resonance Spectroscopy in Differentiating Oligodendrogliomas from Astrocytomas. J. Neuroimaging Off. J. Am. Soc. Neuroimaging 2010, 20, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nanga, R.P.R.; Verma, G.; Wilson, N.; Brisset, J.C.; Nath, K.; Chawla, S. Emerging MR Imaging and Spectroscopic Methods to Study Brain Tumor Metabolism. Front. Neurol. 2022, 13, 789355. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Ganji, S.; Hulsey, K.; Madan, A.; Kovacs, Z.; Dimitrov, I.; Zhang, S.; Pichumani, K.; Mendelsohn, D.; Mickey, B.; et al. A Comparative Study of Short- and Long-TE 1H MRS at 3 T for in Vivo Detection of 2-Hydroxyglutarate in Brain Tumors. NMR Biomed. 2013, 26, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Preusser, M.; Wöhrer, A.; Stary, S.; Höftberger, R.; Streubel, B.; Hainfellner, J.A. Value and Limitations of Immunohistochemistry and Gene Sequencing for Detection of the IDH1-R132H Mutation in Diffuse Glioma Biopsy Specimens. J. Neuropathol. Exp. Neurol. 2011, 70, 715–723. [Google Scholar] [CrossRef]
- Choi, C.; Raisanen, J.M.; Ganji, S.K.; Zhang, S.; McNeil, S.S.; An, Z.; Madan, A.; Hatanpaa, K.J.; Vemireddy, V.; Sheppard, C.A.; et al. Prospective Longitudinal Analysis of 2-Hydroxyglutarate Magnetic Resonance Spectroscopy Identifies Broad Clinical Utility for the Management of Patients With IDH-Mutant Glioma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 4030–4039. [Google Scholar] [CrossRef]
- Suh, C.H.; Kim, H.S.; Jung, S.C.; Choi, C.G.; Kim, S.J. 2-Hydroxyglutarate MR Spectroscopy for Prediction of Isocitrate Dehydrogenase Mutant Glioma: A Systemic Review and Meta-Analysis Using Individual Patient Data. Neuro-Oncology 2018, 20, 1573–1583. [Google Scholar] [CrossRef] [PubMed]
- Provencher, S.W. Estimation of Metabolite Concentrations from Localized in Vivo Proton NMR Spectra. Magn. Reson. Med. 1993, 30, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Batsios, G.; Viswanath, P.; Subramani, E.; Najac, C.; Gillespie, A.M.; Santos, R.D.; Molloy, A.R.; Pieper, R.O.; Ronen, S.M. PI3K/MTOR Inhibition of IDH1 Mutant Glioma Leads to Reduced 2HG Production That Is Associated with Increased Survival. Sci. Rep. 2019, 9, 10521. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.; Lee, H.H.; Heo, H. In-Vivo Proton Magnetic Resonance Spectroscopy of 2-Hydroxyglutarate in Isocitrate Dehydrogenase-Mutated Gliomas: A Technical Review for Neuroradiologists. Korean J. Radiol. 2016, 17, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Tietze, A.; Choi, C.; Mickey, B.; Maher, E.A.; Parm Ulhøi, B.; Sangill, R.; Lassen-Ramshad, Y.; Lukacova, S.; Østergaard, L.; von Oettingen, G. Noninvasive Assessment of Isocitrate Dehydrogenase Mutation Status in Cerebral Gliomas by Magnetic Resonance Spectroscopy in a Clinical Setting. J. Neurosurg. 2018, 128, 391–398. [Google Scholar] [CrossRef]
- Verma, G.; Mohan, S.; Nasrallah, M.P.; Brem, S.; Lee, J.Y.K.; Chawla, S.; Wang, S.; Nagarajan, R.; Thomas, M.A.; Poptani, H. Non-Invasive Detection of 2-Hydroxyglutarate in IDH-Mutated Gliomas Using Two-Dimensional Localized Correlation Spectroscopy (2D L-COSY) at 7 Tesla. J. Transl. Med. 2016, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.G.; Alves, C.A.P.F.; Vossough, A. Updates in Pediatric Malignant Gliomas. Top. Magn. Reson. Imaging TMRI 2020, 29, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, E.; De Biase, D.; Di Nunno, V.; Pession, A.; Tosoni, A.; Gatto, L.; Tallini, G.; Visani, M.; Lodi, R.; Bartolini, S.; et al. IDH1 Non-Canonical Mutations and Survival in Patients with Glioma. Diagnostics 2021, 11, 342. [Google Scholar] [CrossRef]
- Izquierdo-Garcia, J.L.; Viswanath, P.; Eriksson, P.; Chaumeil, M.M.; Pieper, R.O.; Phillips, J.J.; Ronen, S.M. Metabolic Reprogramming in Mutant IDH1 Glioma Cells. PLoS ONE 2015, 10, e0118781. [Google Scholar] [CrossRef]
- Bisdas, S.; Chadzynski, G.L.; Braun, C.; Schittenhelm, J.; Skardelly, M.; Hagberg, G.E.; Ethofer, T.; Pohmann, R.; Shajan, G.; Engelmann, J.; et al. MR Spectroscopy for in Vivo Assessment of the Oncometabolite 2-Hydroxyglutarate and Its Effects on Cellular Metabolism in Human Brain Gliomas at 9.4T. J. Magn. Reson. Imaging JMRI 2016, 44, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Natsumeda, M.; Motohashi, K.; Igarashi, H.; Nozawa, T.; Abe, H.; Tsukamoto, Y.; Ogura, R.; Okada, M.; Kobayashi, T.; Aoki, H.; et al. Reliable Diagnosis of IDH-Mutant Glioblastoma by 2-Hydroxyglutarate Detection: A Study by 3-T Magnetic Resonance Spectroscopy. Neurosurg. Rev. 2018, 41, 641–647. [Google Scholar] [CrossRef]
- Nagashima, H.; Tanaka, K.; Sasayama, T.; Irino, Y.; Sato, N.; Takeuchi, Y.; Kyotani, K.; Mukasa, A.; Mizukawa, K.; Sakata, J.; et al. Diagnostic Value of Glutamate with 2-Hydroxyglutarate in Magnetic Resonance Spectroscopy for IDH1 Mutant Glioma. Neuro-Oncology 2016, 18, 1559–1568. [Google Scholar] [CrossRef] [PubMed]
- Ohka, F.; Ito, M.; Ranjit, M.; Senga, T.; Motomura, A.; Motomura, K.; Saito, K.; Kato, K.; Kato, Y.; Wakabayashi, T.; et al. Quantitative Metabolome Analysis Profiles Activation of Glutaminolysis in Glioma with IDH1 Mutation. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 5911–5920. [Google Scholar] [CrossRef]
- Dekker, L.J.M.; Verheul, C.; Wensveen, N.; Leenders, W.; Lamfers, M.L.M.; Leenstra, S.; Luider, T.M. Effects of the IDH1 R132H Mutation on the Energy Metabolism: A Comparison between Tissue and Corresponding Primary Glioma Cell Cultures. ACS Omega 2022, 7, 3568–3578. [Google Scholar] [CrossRef]
- Reitman, Z.J.; Jin, G.; Karoly, E.D.; Spasojevic, I.; Yang, J.; Kinzler, K.W.; He, Y.; Bigner, D.D.; Vogelstein, B.; Yan, H. Profiling the Effects of Isocitrate Dehydrogenase 1 and 2 Mutations on the Cellular Metabolome. Proc. Natl. Acad. Sci. USA 2011, 108, 3270–3275. [Google Scholar] [CrossRef] [PubMed]
- Branzoli, F.; Marjańska, M. Magnetic Resonance Spectroscopy of Isocitrate Dehydrogenase Mutated Gliomas: Current Knowledge on the Neurochemical Profile. Curr. Opin. Neurol. 2020, 33, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, H.; Zhang, J.; Wu, C.; Zhu, W.; Li, F.; Chen, X.; Xu, B. The Diagnostic Performance of Magnetic Resonance Spectroscopy in Differentiating High-from Low-Grade Gliomas: A Systematic Review and Meta-Analysis. Eur. Radiol. 2016, 26, 2670–2684. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Ganji, S.K.; DeBerardinis, R.J.; Hatanpaa, K.J.; Rakheja, D.; Kovacs, Z.; Yang, X.-L.; Mashimo, T.; Raisanen, J.M.; Marin-Valencia, I.; et al. 2-Hydroxyglutarate Detection by Magnetic Resonance Spectroscopy in IDH-Mutated Patients with Gliomas. Nat. Med. 2012, 18, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, J.A.; Lai, A.; Nghiemphu, P.L.; Kim, H.J.; Phillips, H.S.; Kharbanda, S.; Moftakhar, P.; Lalaezari, S.; Yong, W.; Ellingson, B.M.; et al. Relationship between Tumor Enhancement, Edema, IDH1 Mutational Status, MGMT Promoter Methylation, and Survival in Glioblastoma. AJNR Am. J. Neuroradiol. 2012, 33, 1349–1355. [Google Scholar] [CrossRef]
- Lasocki, A.; Tsui, A.; Gaillard, F.; Tacey, M.; Drummond, K.; Stuckey, S. Reliability of Noncontrast-Enhancing Tumor as a Biomarker of IDH1 Mutation Status in Glioblastoma. J. Clin. Neurosci. Off. J. Neurosurg. Soc. Australas. 2017, 39, 170–175. [Google Scholar] [CrossRef]
- Zhou, H.; Vallières, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI Features Predict Survival and Molecular Markers in Diffuse Lower-Grade Gliomas. Neuro-Oncology 2017, 19, 862–870. [Google Scholar] [CrossRef]
- Leu, K.; Ott, G.A.; Lai, A.; Nghiemphu, P.L.; Pope, W.B.; Yong, W.H.; Liau, L.M.; Cloughesy, T.F.; Ellingson, B.M. Perfusion and Diffusion MRI Signatures in Histologic and Genetic Subtypes of WHO Grade II-III Diffuse Gliomas. J. Neurooncol. 2017, 134, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Hiwatashi, A.; Togao, O.; Kikuchi, K.; Hatae, R.; Yoshimoto, K.; Mizoguchi, M.; Suzuki, S.O.; Yoshiura, T.; Honda, H. MR Imaging-Based Analysis of Glioblastoma Multiforme: Estimation of IDH1 Mutation Status. AJNR Am. J. Neuroradiol. 2016, 37, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Yang, X.; She, D.; Lin, Y.; Zhang, Y.; Cao, D. Noninvasive Assessment of IDH Mutational Status in World Health Organization Grade II and III Astrocytomas Using DWI and DSC-PWI Combined with Conventional MR Imaging. AJNR Am. J. Neuroradiol. 2017, 38, 1138–1144. [Google Scholar] [CrossRef]
- Zhang, Y.; Taub, E.; Salibi, N.; Uswatte, G.; Maudsley, A.A.; Sheriff, S.; Womble, B.; Mark, V.W.; Knight, D.C. Comparison of Reproducibility of Single Voxel Spectroscopy and Whole-Brain Magnetic Resonance Spectroscopy Imaging at 3T. NMR Biomed. 2018, 31, e3898. [Google Scholar] [CrossRef] [PubMed]
- Andronesi, O.C.; Kim, G.S.; Gerstner, E.; Batchelor, T.; Tzika, A.A.; Fantin, V.R.; Vander Heiden, M.G.; Sorensen, A.G. Detection of 2-Hydroxyglutarate in IDH-Mutated Glioma Patients by in Vivo Spectral-Editing and 2D Correlation Magnetic Resonance Spectroscopy. Sci. Transl. Med. 2012, 4, 116ra4. [Google Scholar] [CrossRef] [PubMed]
- Emir, U.E.; Larkin, S.J.; de Pennington, N.; Voets, N.; Plaha, P.; Stacey, R.; Al-Qahtani, K.; Mccullagh, J.; Schofield, C.J.; Clare, S.; et al. Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations. Cancer Res. 2016, 76, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Beiko, J.; Suki, D.; Hess, K.R.; Fox, B.D.; Cheung, V.; Cabral, M.; Shonka, N.; Gilbert, M.R.; Sawaya, R.; Prabhu, S.S.; et al. IDH1 Mutant Malignant Astrocytomas Are More Amenable to Surgical Resection and Have a Survival Benefit Associated with Maximal Surgical Resection. Neuro-Oncology 2014, 16, 81–91. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, M.I.; Young, R.J.; Rubel, J.; Rosenblum, M.; Tisnado, J.; Briggs, S.; Arevalo-Perez, J.; Cross, J.R.; Campos, C.; Straley, K.; et al. Integration of 2-Hydroxyglutarate-Proton Magnetic Resonance Spectroscopy into Clinical Practice for Disease Monitoring in Isocitrate Dehydrogenase-Mutant Glioma. Neuro-Oncology 2016, 18, 283–290. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Arrillaga-Romany, I.C.; Ly, K.I.; Bogner, W.; Ratai, E.M.; Reitz, K.; Iafrate, A.J.; Dietrich, J.; Gerstner, E.R.; Chi, A.S.; et al. Pharmacodynamics of Mutant-IDH1 Inhibitors in Glioma Patients Probed by in Vivo 3D MRS Imaging of 2-Hydroxyglutarate. Nat. Commun. 2018, 9, 1474. [Google Scholar] [CrossRef]
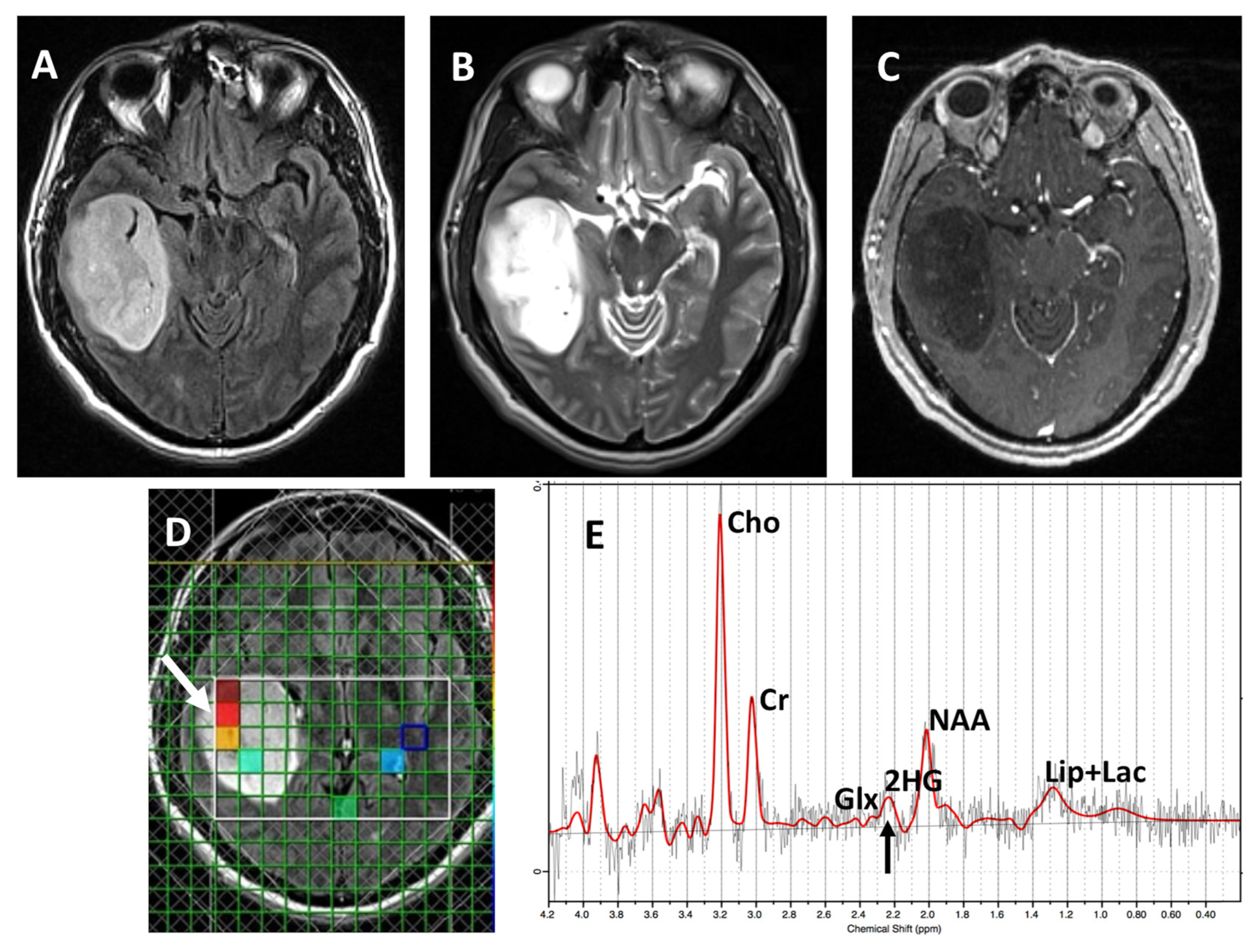
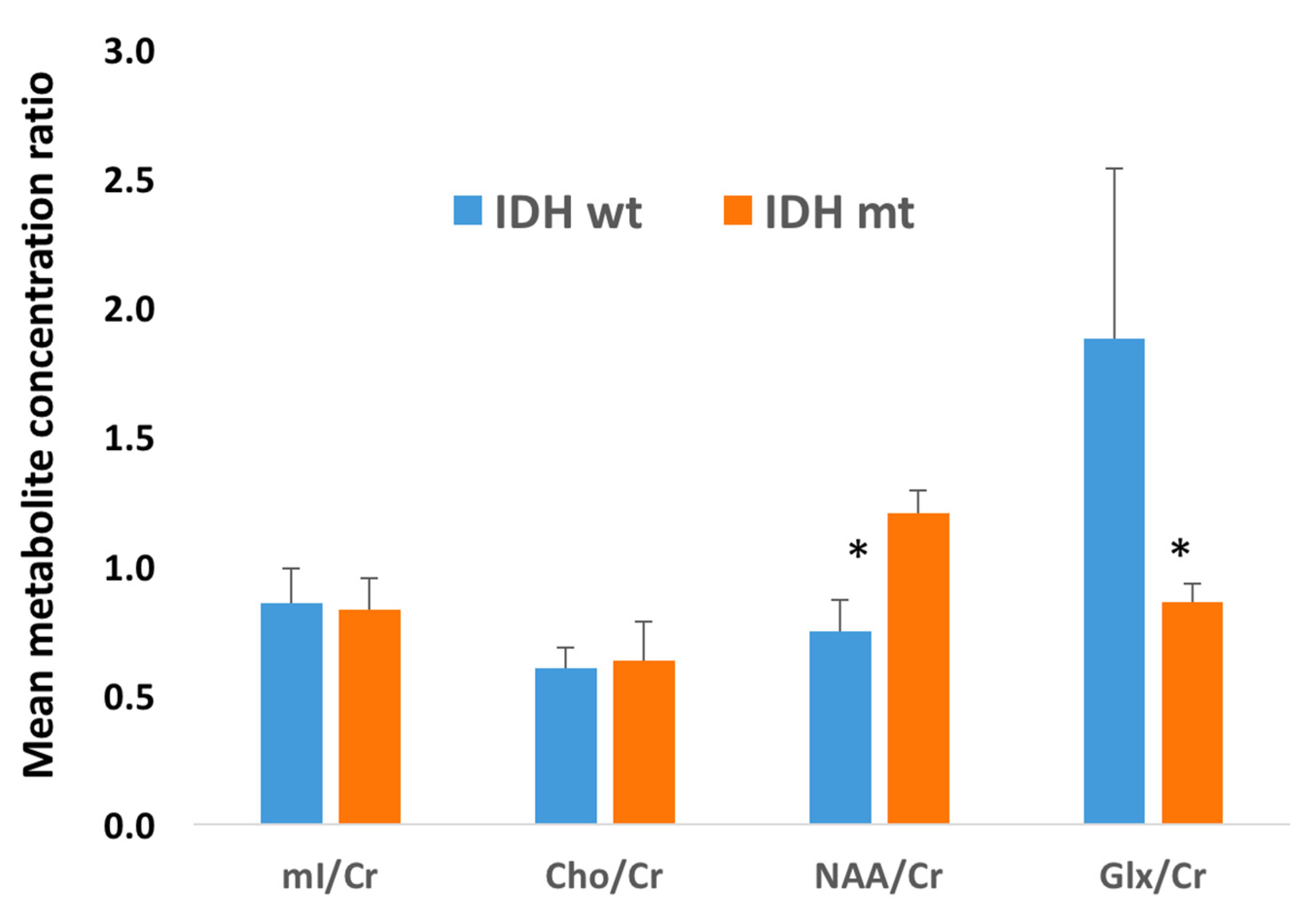
| Patient | Gender | Age (Years) | 1H-MRS Modality | 1H-MRS 2HG Results | IDH1 Status | Histopathology Results | Status at Time of 1H-MRS |
|---|---|---|---|---|---|---|---|
| 1 | F | 54 | SVS | Negative | IDH1-R132H negative | Glioblastoma, WHO grade 4 | New |
| 2 | M | 63 | SVS | Positive | IDH1-R132H mutant | Oligodendroglioma, WHO grade 2 | New |
| 3 | F | 32 | SVS | Positive | IDH1-mutant | Mixed Oligoastrocytoma, WHO grade 3 | Recurrent |
| 4 | F | 34 | SVS | Positive | IDH1-R132H mutant | Astrocytoma, WHO grade 3 | New |
| 5 | M | 38 | SVS | Negative | IDH1-R132H negative | Glioblastoma (RTK1 subclass), WHO grade 4 | New |
| 6 | F | 53 | SVS | Negative | IDH1-R132H negative | Glioblastoma, WHO grade 4 | New |
| 7 | F | 72 | SVS | Negative | IDH1-R132H negative | Molecular Glioblastoma, WHO grade 4 | New |
| 8 | M | 24 | SVS | Positive | IDH1-R132H mutant | Anaplastic Astrocytoma, WHO grade 3 | New |
| 9 | F | 36 | SVS | Positive | IDH1-R132H mutant | Astrocytoma, WHO grade 3 | New |
| 10 | F | 48 | SVS | Positive | IDH1-R132H mutant | Astrocytoma, WHO grade 3 | New |
| 11 | F | 46 | SVS | Positive | IDH1-R132H mutant | Oligodendroglioma, WHO grade 2 | New |
| 12 | M | 64 | SVS | Negative | IDH1-R132H negative | Glioblastoma, WHO grade 4 | New |
| 13 | F | 28 | 1H-MRSI | Positive | IDH1-R132H mutant | Astrocytoma, WHO grade 3 | New |
| 14 | M | 36 | 1H-MRSI | Positive | IDH1-R132H mutant | Anaplastic Astrocytoma, WHO grade 3 | New |
| 15 | M | 69 | 1H-MRSI | Negative | IDH1-R132H negative | Anaplastic Astrocytoma, WHO grade 3 | New |
| 16 | F | 40 | 1H-MRSI | Positive | IDH1-R132H mutant | Anaplastic Astrocytoma, WHO grade 3 | New |
| 17 | F | 36 | 1H-MRSI | Positive | IDH1-R132H mutant | Anaplastic Oligoastrocytoma, WHO Grade 3 | New |
| 18 | M | 39 | 1H-MRSI | Negative | IDH1-R132H negative | Glioblastoma, WHO grade 4 | New |
| 19 | M | 53 | 1H-MRSI | Negative | IDH1-R132H negative | Glioblastoma with sarcomatous features, WHO Grade 4 | New |
| 20 | M | 35 | 1H-MRSI | Positive | IDH1-mutant | Oligodendroglioma, WHO grade 2 | New |
| 21 | M | 30 | 1H-MRSI | Positive | IDH1-mutant | Anaplastic astrocytoma, WHO Grade 3 | New |
| 22 | M | 38 | 1H-MRSI | Positive | IDH1-R132H mutant | Diffuse Astrocytoma, WHO Grade 2 | New |
| 23 | F | 51 | 1H-MRSI | Positive | IDH1-mutant | Recurrent Astrocytoma, progression to WHO grade 4 | Recurrent |
| 24 | F | 80 | 1H-MRSI | Negative | IDH1-R132H negative | Infiltrating astrocytoma, WHO grade 2 | New |
| 2HG Status | |||
|---|---|---|---|
| Negative | Positive | ||
| IDH Status | Wild-Type | 7 | 2 |
| Mutant | 3 | 12 | |
| Accuracy (%) | 79 | ||
| Sensitivity (%) | 80 | ||
| Specificity (%) | 77 | ||
| Positive Predictive Value (%) | 86 | ||
| Negative Predictive Value (%) | 70 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Godoy, L.L.; Lim, K.C.; Rajan, A.; Verma, G.; Hanaoka, M.; O’Rourke, D.M.; Lee, J.Y.K.; Desai, A.; Chawla, S.; Mohan, S. Non-Invasive Assessment of Isocitrate Dehydrogenase-Mutant Gliomas Using Optimized Proton Magnetic Resonance Spectroscopy on a Routine Clinical 3-Tesla MRI. Cancers 2023, 15, 4453. https://doi.org/10.3390/cancers15184453
de Godoy LL, Lim KC, Rajan A, Verma G, Hanaoka M, O’Rourke DM, Lee JYK, Desai A, Chawla S, Mohan S. Non-Invasive Assessment of Isocitrate Dehydrogenase-Mutant Gliomas Using Optimized Proton Magnetic Resonance Spectroscopy on a Routine Clinical 3-Tesla MRI. Cancers. 2023; 15(18):4453. https://doi.org/10.3390/cancers15184453
Chicago/Turabian Stylede Godoy, Laiz Laura, Kheng Choon Lim, Archith Rajan, Gaurav Verma, Mauro Hanaoka, Donald M. O’Rourke, John Y. K. Lee, Arati Desai, Sanjeev Chawla, and Suyash Mohan. 2023. "Non-Invasive Assessment of Isocitrate Dehydrogenase-Mutant Gliomas Using Optimized Proton Magnetic Resonance Spectroscopy on a Routine Clinical 3-Tesla MRI" Cancers 15, no. 18: 4453. https://doi.org/10.3390/cancers15184453
APA Stylede Godoy, L. L., Lim, K. C., Rajan, A., Verma, G., Hanaoka, M., O’Rourke, D. M., Lee, J. Y. K., Desai, A., Chawla, S., & Mohan, S. (2023). Non-Invasive Assessment of Isocitrate Dehydrogenase-Mutant Gliomas Using Optimized Proton Magnetic Resonance Spectroscopy on a Routine Clinical 3-Tesla MRI. Cancers, 15(18), 4453. https://doi.org/10.3390/cancers15184453






