Simple Summary
A tiny human sample is enough to uncover the complete genome sequence of that individual with the advances in biomedical technologies and data analysis. Jumping genes constituting about half of the human genome, have been implicated in cancer and predisposition to inflammatory reactions. Inflammation may restrict the activity of these genes and reduce the tumor burden. This article summarizes related literature on factors regulating jumping genes and discusses their immune-related evidence made available by genome-wide studies.
Abstract
Advances in sequencing technologies and the bioinformatic analysis of big data facilitate the study of jumping genes’ activity in the human genome in cancer from a broad perspective. Retrotransposons, which move from one genomic site to another by a copy-and-paste mechanism, are regulated by various molecular pathways that may be disrupted during tumorigenesis. Active retrotransposons can stimulate type I IFN responses. Although accumulated evidence suggests that retrotransposons can induce inflammation, the research investigating the exact mechanism of triggering these responses is ongoing. Understanding these mechanisms could improve the therapeutic management of cancer through the use of retrotransposon-induced inflammation as a tool to instigate immune responses to tumors.
Keywords:
transposable elements; mobile genome; insertions; tumorigenesis; immunity; type I IFN; jumping genes 1. Background
“You just know sooner or later, it will come out in the wash, but you may have to wait sometime.” Dr. Barbara McClintock conveyed this statement upon receiving the Nobel Prize recognizing her discovery of transposable elements (TEs) [1]. TEs are mobile DNA sequences that can move from one genomic location to another in a process called “transposition” [2]. Transposition in the genome is facilitated by one or more proteins encoded by a TE [3]. In this review, we shed light on the regulatory mechanisms affecting the active classes of TEs and their immunological impact on human cancer using evidence from recent genome-wide studies. As illustrated in Figure 1, TEs are categorized into two broad classes: DNA transposons and retrotransposons, based on their transposition intermediate and mobility mechanisms [3]. DNA transposons are sequences that use element-encoded transposases to move from one genomic location to another by a cut-and-paste mechanism [3]. A retrotransposon element inserts into a new genomic location by a copy-and-paste mechanism using an RNA intermediate [3,4]. This article’s focus is on retrotransposons since there is no evidence of DNA transposon insertion into the human genome in the last 37 million years [5].
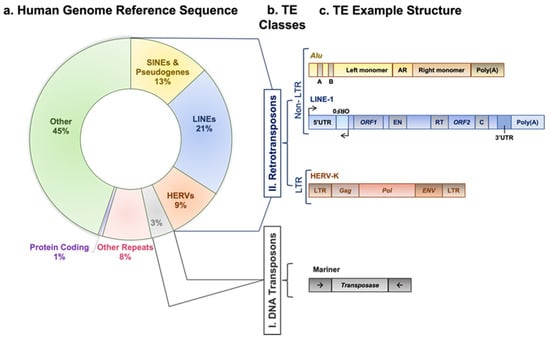
Figure 1.
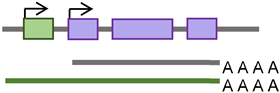
Transposable element classes, structure, and activity. (a) A doughnut chart represents fractions of human genome reference sequence constituents, as described in the recent telomere-to-telomere (T2T) assembly [6]. (b) TE main categories: first class, DNA transposons; second class, retrotransposons. The latter is subcategorized into elements having or lacking LTRs. LTR-containing elements include HERVs family, and non-LTR elements include SINEs, LINEs, and pseudogenes families. (c) Outline diagrams to represent structure of example elements per classes of transposable elements; Alu element from SINEs family is composed of two monomers separated by adenosine-rich (AR) linker. The left monomer contains an internal RNA polymerase III promoter (bars labeled A and B), and the right monomer is followed by a poly (A) tail. L1 element is a protein-coding element of the LINEs family; it has an internal promoter in its 5′ untranslated region (5′UTR) followed by a primate-specific antisense region (ORF0) and regions encoding L1 proteins (ORF1 and ORF2). ORF1p is a nuclear binding protein, and ORF2p has EN, RT, and cysteine-rich (C) domains. L1 element is ended by a poly (A) tail in its 3′ untranslated region (3′UTR). HERV-K element of the HERVs family contains two LTR regions separated by gag, pol, and env regions. Mariner of the DNA transposons class encodes transposase, an enzyme that binds and cuts near inverted repeats flanking the element (denoted by little arrows).
According to the presence or absence of long terminal repeats (LTR) in their sequences, retrotransposons are subdivided into LTR- and non-LTR-containing elements [3]. Human endogenous retroviruses (HERVs) are autonomous protein-encoding LTR-containing elements [6]. Most HERV elements are non-functional due to accumulated mutations or internal recombination, resulting in solitary LTRs [7]. However, the evidence suggests the recent insertion of HERV elements within the human population in polymorphic loci [8]. Retrotransposons lacking LTR include long interspersed elements (LINEs) and short interspersed elements (SINEs) [6]. Of these, the most active elements retrotransposing in the human genome include autonomous LINE-1 (L1) from the LINEs and non-autonomous Alu from the SINEs (reviewed in [3]). L1 has two promoters (sense and antisense) to transcribe three different open-reading frame (ORF) regions. The sense promotor transcribes ORF1 and ORF2 [9,10]. At the same time, the antisense promoter transcribes a primate-specific ORF (ORF0) in the opposite orientation to that of L1 [11]. ORF1 encodes a 40 kDa protein (ORF1p) with a nucleic acid chaperone and RNA binding activities [12]. ORF2 encodes a 150 kDa protein (ORF2p) that has endonuclease (EN) and reverse transcriptase (RT) activities [13,14]. The Alu elements are primate-specific retrotransposons, with the most recent amplification in lineages attributed to a series of Y subfamilies (Ya5 and Yb8 dominate in humans) [15]. Each Alu element comprises two dimers ancestrally derived from the 7SL RNA and separated by a short polyA sequence. A longer polyA tail occupies its 3′ end [15]. Alu elements do not encode proteins; instead, they hijack L1 proteins to mediate their retrotransposition [15], which occurs through the life cycle of L1, starting with the transcription of L1 mRNA from its genomic copy [3]. L1 RNA is exported to the cytoplasm, where the ORF1p and ORF2p proteins are translated [16]. These proteins bind the retrotransposon RNA (L1/Alu) to form a ribonucleoprotein particle (RNP) [17,18]. The RNP is imported into the nucleus to facilitate L1/Alu retrotransposition via two distinct pathways [19]. The canonical pathway is called target-primed reverse transcription (TPRT) [20,21], in which the L1 EN activity produces a nick at a target site in the genomic DNA [13,22]. It preferentially cuts DNA at the consensus sequence 5′-TTTT/A-3′ or its variants [22]. Then, using the retrotransposon RNA as a template, the L1 RT moiety extends the unbound 3′-OH group from DNA to begin reverse transcription, starting within the polyA tail of the retrotransposon RNA [3,22]. Retrotransposition can also occur via an endonuclease-independent pathway or non-classical L1 insertion. Endonuclease cleavage is not required in this pathway, and the reverse transcription is initiated at pre-existing DNA break regions [23,24].
De novo retrotransposon insertions can occur in exons, introns, or the regulatory regions of the genome, disrupting their function, providing new promoter and enhancer regions, and contributing to disease [25,26]. These insertions can exert deleterious, “disruptive,” or beneficial “exaptation” effects on the host [27]. Retrotransposition in introns can affect the splicing process by different mechanisms [28,29]. It can provide alternative (donor or acceptor) splice sites, cause exonization (a process by which genes acquire new exons from intronic DNA sequences), or promote exon skipping [28,29]. Alternative splicing and exon-acquisition events of the CHRM3 gene, a muscarinic acetylcholine receptor family member, are examples of TE integrations into the host genome that are naturally selected and conserved over generations [30]. About 62% of exonizations in the human genome are Alu-derived [31]. The insertion of Alu into one of the Factor VIII gene introns resulted in exon skipping and the consequent onset of hemophilia A [32]. Table 1 outlines the mechanisms by which retrotransposons can impact genomic structure and function.

Table 1.
Mechanisms by which retrotransposons can affect the genome structure.
2. Regulation of Retrotransposons and Their Association with Tumorigenesis
Reports have demonstrated that retrotransposon expression and activity occur primarily in cells associated with the germline, with little expression in most somatic tissues under physiological conditions [51]. L1 retrotranspositions can occur during early human embryonic development [52]. They were identified in neuronal precursor cells [53] and have been observed in various cell lines when a tagged L1 construct was employed. However, limited data are available on whether retrotranspositions occur in normal somatic adult tissues other than the brain [54,55,56]. A few findings indicated that somatic insertions in hepatocytes and the esophagus, stomach, and colon may have occurred during embryogenesis [57,58,59]. This lack of evidence could be related to the somatic insertions occurring in a few cells within the tissue that are challenging to identify in whole-tissue sequencing.
De novo somatic insertions were identified in different tumor tissues of epithelial origin at varying frequencies [60,61]. These insertions are characterized by the fact that they have more 5′ truncations and exist with less dependence on L1-encoded EN cleavage than germline insertions [62]. Retrotransposon activity was associated with tumorigenesis in the early observations of Miki et al., who detected that a novel L1 insertion impacted the tumor suppressor APC in colon cancer but not in normal colon tissues from affected individuals [63]. More than two decades later, another L1 insertion was found to disrupt the other allele of the APC gene, which contributed to colon tumorigenesis [64]. ST18 (suppression of tumorigenicity 18) and PTEN genes were other tumor suppressors interrupted by new L1 insertions in hepatocellular carcinoma and endometrial cancer, respectively [65,66]. There is much evidence to show that retrotransposons are crucial contributors to tumorigenesis, especially with the global epigenetic dysregulation that characterizes tumorigenesis [67].
By introducing the high-throughput L1-sequencing assay, Iskow et al. could identify a hypomethylation signature that characterized lung tumors, which made them more L1-permissive and had a higher frequency of L1 somatic insertion than the brain tumors included in that study [68]. Tumors of epithelial origin, such as colorectal, prostate, and ovarian cancers, showed more pronounced L1 activity than the brain and blood cancer types with the performance of single-nucleotide resolution analysis of TE insertions in whole-genome sequencing datasets [61]. In agreement with these findings, colorectal and lung cancers were the most frequently affected by L1 somatic insertions exhibiting hypomethylated promotors by tracking down the L1 insertion sources via the identification of 3′ transductions [69]. The preference for retrotransposon activity in specific tumor types could be related to a range of transcription factors activated in specific cell types over others. The activation of transcription factors in epithelial tumors might modulate retrotransposon expression and activity. For example, epithelial tumors such as breast, colorectal, prostate, and cervical cancers are characterized by Oct1 (Octamer transcription factor 1, POU2F1) protein upregulation [70,71,72,73]. Oct1 controls stem cell phenotypes in normal and tumor cells [73]. In epithelial cells, high Oct1 protein expression was spatially correlated with stem cell niches and the increased expression of stem cell markers such as ALDH1 [73]. Transcription factors like Oct1 may play a role in epithelial cell de-differentiation into a more stem-like phenotype [74]. These cells may be more disposed to L1 retrotransposition than other populations of cancer cells [74].
Several transcription factors were demonstrated to regulate the transcription of retrotransposons by binding their promoters. These factors include YY1, RUNX3, p53, Oct4, Sox2, Nanog, KLF4, MYC, and CTCF [75,76,77,78,79,80,81,82,83]. Although the L1 5′UTR promoter region is prone to higher mutation rates than the L1 ORF regions, the evolutionary analysis showed conservation in the transcription factor binding sites among human-specific L1 elements [84]. The transcription factors regulating retrotransposon expression are not isolated from other regulators that modulate retrotransposon activity in the cell. Each of these regulators is a part of different pathways that make up an interconnected network of factors controlling retrotransposon expression and activity.
Retrotransposons have long been considered genomic threats to somatic cellular functions and are under control mechanisms that restrict their activity [85]. These regulation mechanisms sometimes fail in cases of age or disease [85]. The factors restricting retrotransposons fall into one of two categories: cytoplasmic or nuclear—most factors acting in the cytoplasm limit the retrotransposon’s expression by post-transcriptional mechanisms. The suppressing nuclear factors either restrict the transcription of retrotransposons or interfere with their genomic integration (see Table 2). These factors (being cytoplasmic or nuclear) are illustrated in Figure 2 based on the retrotransposon’s life cycle.

Table 2.
Regulators of retrotransposon activity and their mechanism of regulation.
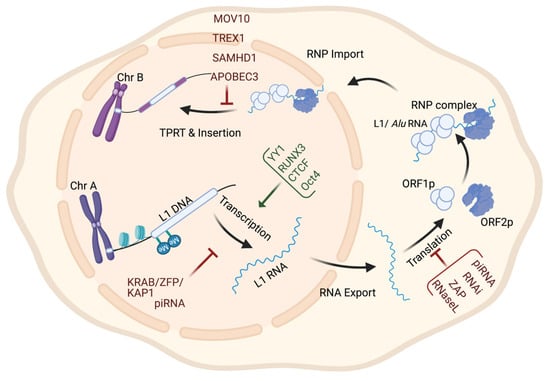
Figure 2.
Retrotransposon levels of regulation throughout its life cycle. The regulation of retrotransposon activity can occur at the transcriptional level by histone modification or DNA methylation; at the post-transcriptional level by targeting RNA for degradation; and at the genomic insertion level by interfering with RNP complexes integrity or inhibiting TPRT.
3. Retrotransposons in Cancer from a Genome-Wide Perspective
Recent advances in bioinformatics tools have paved the way for studying retrotransposons. It is a significant challenge to precisely determine their insertion sites using standard DNA sequencing technologies. This difficulty can be related to the retrotransposon sequence characteristics or the available data quality. The L1 sequence, for example, differs among genomic copies in terms of the polyadenylation signal and 3′ UTR, with most copies being 5′ truncated [104,105,106]. Most available whole-genome sequencing (WGS) data consist of single- or paired-end short reads of about 100–250 nt in length [107]. Using these reads to detect 6000 kbp L1 insertions requires methods to identify the sequences overlapping TE elements and new genomic locations. Filters and measures are needed to reduce the number of false-positive insertions detected while maintaining reasonable sensitivity in detecting new TE insertion events [107].
Large-scale sequencing projects include data from thousands of individuals deposited in public databases such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC). In addition, bioinformatics tools and pipelines have facilitated the comprehensive detection and analysis of retrotransposons in cancer [108]. The available tools that accelerate research in the TE field can range from data repositories to insertion detecting tools and strategies to investigate the TEs’ biological impacts. Databases such as RepBase Update and the European database of L1HS retrotransposon insertions (EUL1Db) were developed as repositories focused on assembling TE consensus sequences with the reference genome and identifying common polymorphic TE insertions [109,110].
Two factors are required to identify TE polymorphisms in an individual sequenced genome: an available reference genome and the annotated TE sequences in that genome; both are made accessible in public databases. TE polymorphisms are detected using reads that span the borders of retrotransposons and new genomic locations in the search for retrotransposons not yet included in the reference sequence [107]. Some identified polymorphic insertions were linked to diseases such as hemophilia [37] and Rett syndrome [111]. Many TE detection software tools have been developed to identify germline and somatic TE insertions using short-read sequencing, as in the TCGA [108]. Short reads do not frequently span the entire interval affected by retrotransposon-mediated genomic rearrangement [107]. Therefore, computational tools were developed to utilize up to three strategies in detecting TE insertions: inference from discordant read pair (DRP) mapping; clustering of split reads (SR); and sequence re-alignment through the identification of TE-specific motifs [112]. DRP methods detect a pair of reads from the same TE insert whose alignment to the reference sequence has an orientation or distance that differs from the expected range [113]. No identification of exact junctions between TEs and the reference genome is possible using DRP methods alone [107].
On the other hand, the SR methods detect reads that map partially with the surrounding genome and partially in a TE sequence [113] (Figure 3A). Non-reference SRs are clipped to align with the reference sequence and can be used to identify the junctions between the TE and reference genome sequence [107]. Therefore, SR strategies provide a higher positional accuracy by identifying the junction between the TE and host sequence. DRP strategies, on the other hand, offer higher sensitivity, providing more reads to support TE insertions [108]. However, another strategy is required to refine the DRP mapping by requiring an SR- or TE-specific motif detection to exclude TE-unrelated rearrangements [107]. In the TE-specific motif detection strategy, tools were developed to identify insertions by looking for common TE signatures, such as target site duplications (TSDs) flanking most TE insertions, long stretches of poly (A) tails, and 3′ transduction in L1-mediated insertions [108].
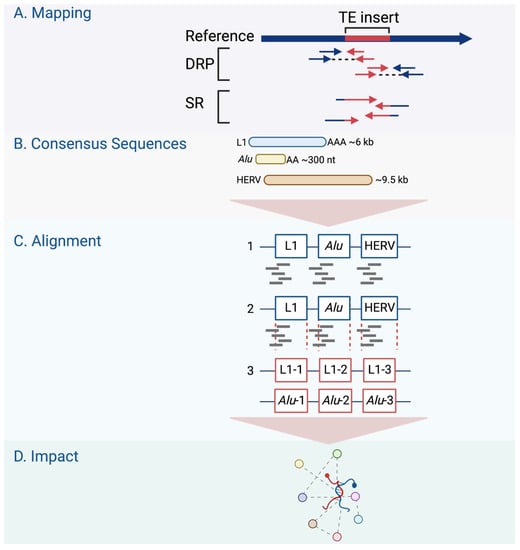
Figure 3.
Genome-wide retrotransposon studies workflow. (A) TE novel insertions are detected in the human genome using sequencing reads using different mapping strategies, including DRP and SR. (B–D) The consensus sequences of active TE classes are obtained from repository databases and aligned to identify their differential expression in the genome.
The TE field advances have been extended to offer tools that predict the impacts of TEs on gene regulation, such as measuring the overlap with other genomic regions, looking for associations with transcription regulation datasets, or considering signs for negative or positive selection [108]. In searching for active TEs and studying the effect of these elements on the expression of nearby genes, alignment tools were developed, such as RepEnrich [114] and SQuIRE [115]. These tools are designed to identify the differential expression analysis of TEs in chromatin immunoprecipitation (ChIP) sequencing and/or RNA sequencing data [114]. The RepEnrich tool creates a series of contiguous segments representing all TE instances of each TE subfamily annotated in the TE repository (e.g., Repbase, Figure 3B) [114]. These series are then used to identify reads that map only to one subfamily of TEs, such as L1HS (Figure 3C). The reads identified using this tool can be described as unique to a particular subfamily in the genome. The SQuIRE tool quantifies the TE subfamily expression and performs differential analyses on TEs and genes at the locus level [115] (Figure 3D). As summarized in Table 3, genome-wide research follows one of two strategies used to study retrotransposon activity in cancer: targeted resequencing assays and bioinformatics analysis of WGS or whole-exome sequencing (WES) data.

Table 3.
Retrotransposon activity in cancer genome-wide studies.
4. Immune Signature of Retrotransposons in Cancer
Most of the (above-mentioned) genome-wide studies were focused on identifying new insertions and characterizing their effect on tumor-modulating genes. There is also a growing interest in identifying the factors controlling retrotransposon RNA expression or the factors triggered by its activation, such as the emerging data demonstrating that retrotransposon activation can be immunogenic and may instigate IFN and apoptosis signaling [118,121,122,123,124].
Tumors with high immune activity, such as those associated with the Epstein–Barr virus (EBV) infection, demonstrated a low number of L1 insertions [118]. Reports also indicate high retrotransposon activity in head and neck squamous cell carcinoma (HNSCC) patients. The overexpression of retrotransposons in HNSCC was shown to be associated with robust DNA CpG demethylation of tumor tissue [125]. A high expression of the long terminal repeat (LTR) retrotransposon HERVs in HNSCC cases was accompanied by high cytolytic effectors, which correlated positively with cytolytic immune activity [126]. This activity could be related to the oncogenic human papillomavirus (HPV), whose infection is among the etiological factors contributing to a subset of HNSCC tumors. HPV-positive cases often present with better outcomes [127] and are less likely to have TP53 mutation [66]. These tumors have also demonstrated less retrotransposon somatic insertions (i.e., activity) [66]. The examples above suggest the involvement of a defense mechanism against retrotransposons resembling antiviral actions.
Many retrotransposon regulation mechanisms are similarly used to protect cells from exogenous viral infections. When nucleic acids of foreign origin are detected by endosomal or pattern recognition receptors (PRR), an IFN-driven immune response is initiated to eliminate the affected cell populations [128]. The cell is equipped with a heterogeneous group of PRRs that includes but is not limited to Toll-like receptors (TLR3, TLR7, TLR8, and TLR9); the RNA sensors RIG-I (retinoic acid-inducible gene I), MDA5 (melanoma differentiation-associated protein 5), and LGP2 (RIG-I-like receptor LGP2); and the DNA sensors cGAS (cyclic GMP-AMP synthase) and AIM2 (absent in melanoma 2) [129].
Specific criteria, including location, nucleic acid sequence pattern, and threshold quantity, determine which nucleic acid each PRR senses [129]. TLR nucleic acid binding domains face the lumen of endosomal compartments, and the other PRRs are present in the cytoplasm [129]. TLR3 binds dsRNA of >40 bp size; TLR7/8 bind fragmented RNA with unmodified nucleosides; and TLR9 binds ssDNA of >11 nt size with a high affinity to the unmethylated cytosine CpG motif [129]. RIG-I binds >20 bp dsRNA with blunt end conformation; MDA5 binds >1–2 Kb dsRNA; cGAS binds dsDNA of >20–40 bp size; and AIM2 binds dsDNA of >50–80 bp size [129]. The quantity of detected nucleic acid can be affected by the increased supply that causes the accumulation of nucleic acids and the defective mechanisms of their clearance.
The failure of one or more of the (above-described) retrotransposon regulatory mechanisms (due to aging, tumorigenesis, or autoimmune disease) can result in retrotransposon activation. This activity promotes dsRNA or dsDNA (sequences of different sizes and motifs) release into the cytoplasm and their detection by cGAS or MDA5, respectively [123,124,130]. Most ADAR-mediated A-to-I RNA editing sites are found in close proximity to retrotransposons. Upon the depletion of ADAR1 in conditions such as Aicardi–Goutières syndrome and some cancers, unedited endogenous RNAs trigger a chronic type I IFN response via MDA5 facilitated by the LGP2 RNA sensor [131]. The activation of L1 during cellular senescence triggered the release of L1 dsDNA in the cytoplasm and promoted type I IFN responses and sterile inflammation [122].
In addition to the evidence summarized in Table 4 below, many examples suggest the retrotransposon activation of innate immune response in cancer. By analyzing TCGA RNA sequencing data, specific HERV elements were highly enriched in tumor samples compared to their normal counterparts, and this enrichment was associated with an increased immune response [126]. Another piece of evidence showed that cytosolic ssDNA and dsDNA in several tumor cell lines were mainly retrotransposon-derived and associated with the cGAS-activated STING and type I IFN response [130]. Activating HERV expression using DNMT inhibitors (DNMTi) in cancer cells triggered cytosolic dsRNA release, and MDA5 stimulated immune response [124]. In addition, expressing ERV sequences in TLR3, TLR7, and TLR9 triple-deficient mice failed to induce a sufficient immune response, resulting in their development of T-cell acute lymphoblastic leukemia and their early death [132]. Blood samples from individuals with the autoimmune disease SLE, systemic lupus erythematosus, were enriched in Alu RNA associated with high levels of type I IFN response [133]. Although the triggers of retrotransposon activation in the disorders mentioned above may differ, their induction of TEs is likely to be the cause of the IFN responses as a means of protection. A feedback loop may be generated to inhibit L1 activity, as suggested by specific interferon-stimulated proteins directly interacting with its encoded ORF1p [134].

Table 4.
Retrotransposon activity and associated immune response in cancer.
Tumor-specific characteristics may alter the tumor microenvironment and play a role in retrotransposon expression and its associated immune response. TP53, for example, has immunomodulatory roles, and its dysfunction associates with immunosuppression [126,146], which is consistent with the evidence of gastrointestinal tumors with TP53 mutations showing low immune activity and higher loads of L1 insertions than tumors with wild-type TP53 [118]. Also, evidence from colon cancer shows that in response to viral infection in cells, TP53 induces an IFN-dependent antiviral response by activating IFN-stimulated genes [147]. Another piece of evidence showed that TP53 cooperates with DNA methylation to maintain the silencing of SINEs and other non-coding RNAs [148]. The TP53-deficient cells in this study exhibited high SINE element expression accompanied by a high type I IFN response [148]. However, not all tumors exhibit the same type of TP53 mutation, and not all mutations result in TP53 protein deficiency [149]. TP53 mutation can contribute to tumorigenesis by losing TP53 function and gaining mutant functions [149]. Whereas frequent TP53 loss of function mutations in basal-like breast cancer could increase retrotransposon expression and the associated IFN response, the TP53 gain of function mutations in high-grade serous ovarian tumors could reduce retrotransposon expression and its associated IFN response [86].
Apart from TP53, gastrointestinal tumors had strong associations between retrotransposons and TLRs or IFN-induced mRNAs, which was not the case in breast and ovarian cancers [86,118]. Also, IFNε, which is hormonally regulated and expressed in the cells of reproductive organs [150], presented high associations with retrotransposon expression in breast and ovarian cancers [86]. It could be that because retrotransposons contain several binding motifs for estrogen response elements (ERE) [151], they may play a role in IFNε expression in the tumors of reproductive organs. Therefore, the effect of IFN on retrotransposons could be related to the hormone-regulated microenvironment and might be tumor type-specific. The context-dependent IFN signaling associated with the ER+ and ER-negative breast cancer subtypes, which impacts their response to therapy and overall outcomes, reinforces the above notion [152]. It could be interesting to extend these experiments to identify the levels of retrotransposon expression among ER+ and ER- breast tumors. The examples above support the assertion that the tumor type and specific characteristics could affect the retrotransposon’s expression and linked immune response. These variabilities should be considered when studying the retrotransposon’s activity in different types of cancer.
5. Therapeutic Opportunities for Retrotransposon Activity in Cancer
Throughout their evolutionary timeline, significant retrotransposon-related activities at the genomic and cellular levels have been attributed to their RT [14]. However, retrotransposon genomic insertions in cancer have drawn considerable attention beyond the attention given to retrotransposon RT activity [153]. RT activity was shown to increase during tumorigenesis. Anti-retroviral non-nucleoside reverse transcriptase inhibitors (NNRTIs), such as efavirenz and nevirapine, reduced RT activity significantly by inducing conformational changes in the enzyme [154,155]. The NNRTIs reduced tumor growth by decreasing cellular proliferation and promoting differentiation [156,157]. The effect of inhibiting RT using NNRTIs was similar to that of the L1 siRNA suppressing effect; therefore, they were assumed to target L1 activity [158]. Other lines of evidence suggest that another class of RT inhibitors, nucleoside reverse transcriptase inhibitors (NRTIs), are capable of inhibiting L1 activity and having anticancer effects in cells [159,160]. This evidence suggests that L1-encoded RT is a potential marker for diagnostic purposes and a potential target for therapeutic intervention. However, further work is still required to understand the exact mechanism of the observed effect of RT inhibitors on cancer [161]. Although both NRTIs and NNRTIs could inhibit cancer cell growth, only NRTIs inhibited telomerase RT in vitro [162], which may suggest a mechanism related to L1 RT particularly to affect cancer growth.
Among the mechanisms that activate retrotransposons, demethylating agents such as DNMTi act by releasing the epigenetic restriction placed on retrotransposons [123,125,163]. Activating various TE classes in glioblastoma cells triggered type I and II IFN responses [125]. TE-derived peptides were processed and presented on MHC class I molecules that activated adaptive immunity [125]. Activation of HERVs resulted in a viral mimicry response of dsRNAs, inducing the MDA5/MAVS RNA recognition pathway and the downstream activation of interferon response factor 7 (IRF7) [123]. Recent evidence (based on TCGA data analysis and in vitro DNMTi treatment of ovarian cancer cells) suggested that high HERV expression in patients was associated with better survival and correlated with the infiltration of cytotoxic T cells [164]. The use of DNA-hypomethylating agent 5-azacitidine (AZA) in colon and ovarian cancer cell models was associated with the increased expression of HERV and L1 RNA [124,165]. HERV expression was linked to regulatory T cell tumor infiltrates and predicted cytolytic activity in AZA-treated cells [165].
In contrast, L1 expression correlated with TP53 status and predicted AZA drug sensitivity [165]. A dinitroazetidine derivative (RRx-001), another hypomethylating drug less toxic than AZA, is currently in phase II clinical trials [166]. RRx-001 induced antitumorigenic effects by activating the expression of HERV and IFN-responsive genes [166]. Similarly, treating colon cancer cells and tumor organoids with another derivative of a hypomethylating agent (5-aza-2′-deoxycytidine) was sufficient to induce a growth-inhibiting immune response by triggering retrotransposon expression [123,163]. Interestingly, the combination of DNMTi and HDACi selectively induced LTR retrotransposons more efficiently than using each drug individually [167]. The treatment-activated TSS of LTR elements induced them de novo from non-annotated TSS [167]. This activation resulted in chimeric products with predicted immunogenic functions [167].
In addition, some targeted cancer therapeutics and chemotherapeutic agents were shown to activate retrotransposon expression in cancer cells [121,168]. Cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitors repressed DNMT1 and caused activation of repeat elements, including retrotransposons in breast cancer [168]. This activation promoted cytotoxic T-cell-mediated clearance of tumor cells and increased tumor immunogenicity [168]. However, some cells within a heterogeneous cancer population may develop adaptation mechanisms to survive the challenging tumor microenvironment conditions [121]. These cells could modulate retrotransposon expression with lethal drug exposures by maintaining their epigenetic repression [121]. This evidence suggests combining HDACi with other targeted therapeutics may enhance their efficacy in treating cancer [128].
The examples mentioned above support the notion that retrotransposon activation in tumors may contribute to their turning into ‘hot tumors’, which are inflamed and T-cell- infiltrated tumors [169]. In such a microenvironment, the antitumor immune response will reduce the tumor burden and sensitize it to other targeted therapies and immunotherapy [169]. Retrotransposon activity in cancer probably occurs more in specific tumor types than in others [60,61]. It is unclear whether this is related to a more vigorous immune defense or a higher level of cellular adaptation by implementing changes in their epigenome or transcriptome [10].
Tumor-derived extracellular vesicles (EVs) are enriched in retrotransposon RNA and involved in the horizontal transfer of retrotransposons to normal cells. They may broadly influence the tumor microenvironment and immune response [170,171]. This evidence suggests that EVs facilitate the release and transfer of retrotransposons to other cells, contributing to tumor evolution or metastasis (if derived from tumor cells). Also, retrotransposon RNA transfer can influence recipient cells’ transcriptional and post-transcriptional regulation. For example, the increased L1-derived RNA transcripts in recipient cells after the EVs transfer activate members of the APOBEC3 [171]. EVs are currently subject to multiple clinical trials at different phases and are to be used as non-invasive tools for diagnosis and therapeutics. They can serve as cargo for drug delivery in cancer and other conditions (as referred to https://clinicaltrials.gov/, accessed on 18 June 2023). The increased expression of retrotransposons in EVs derived from tumor cells compared to those derived from normal cells [170] could potentially serve as a valuable biomarker for diagnostic purposes. Studies to characterize the origin, biogenesis, and destination of EVs containing retrotransposon RNA and protein in cancer patients are currently needed to understand their potential fully.
6. Closing Remarks
Overall, the advances in sequencing technologies and bioinformatic analysis made studying the activity of retrotransposons in cancer more accessible than before. However, these advances are accompanied by the complexities of dealing with big data. Therefore, tools are being developed to study retrotransposons to cope with these concerns and bring rigorous methods and strategies to keep the field moving forward.
Different cellular and molecular mechanisms regulate the activity of retrotransposons in the human genome. The deregulation of these mechanisms can activate retrotransposons and contribute to the process of tumorigenesis. Accumulating evidence indicates strong associations between retrotransposons and type I IFN immune responses. Retrotransposons could be carried in the extracellular space by tumor-derived EVs, which facilitate their release in the cytosol of surrounding cells, where different PRRs detect them. This detection can activate IRF-mediated type I IFN responses. An inflammatory response could be generated from IFN signaling, leading to a negative feedback loop to inhibit further retrotransposon activity (Figure 4). Extensive research to validate these assumptions is required in different types of tumors; this research is currently more accessible due to the advances in sequencing technologies and the strategies of bioinformatic data analysis.
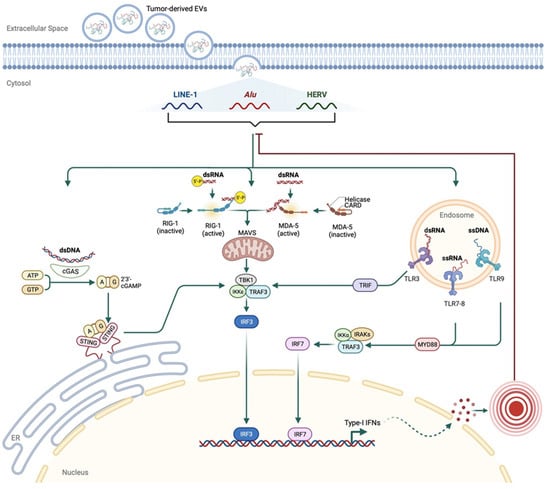
Figure 4.
A proposed mechanism for retrotransposon-induced immune response in cancer. Tumor-derived EVs containing retrotransposons could be imported through the plasma membranes of other cells. Released cytosolic retrotransposons bind one of the PRRs, such as cGAS, RIG-I, MDA-5, or endosomal TLRs. The activated PRRs induce an IRF-mediated type I IFN response, stimulating inflammatory responses. These responses could create negative feedback on the retrotransposons to inhibit their activity.
Prospectively, these retrotransposon-induced inflammatory responses could be used as tools to improve options for cancer treatment by considering the variations between different types of cancer and tailoring the therapeutic choices to the associated response.
Author Contributions
All authors contributed to this work equally. M.I.A. prepared the artwork. All authors have read and agreed to the published version of the manuscript.
Funding
Not applicable.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Figures were created with BioRender.com. Apologies to all authors whose work was not cited due to the limited scope and space constraints. Open Access funding is provided by the Qatar National Library.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ravindran, S. Barbara McClintock and the discovery of jumping genes. Proc. Natl. Acad. Sci. USA 2012, 109, 20198–20199. [Google Scholar] [CrossRef] [PubMed]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements 06 Biological Sciences 0604 Genetics. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Boeke, J.D.; Garfinkel, D.J.; Styles, C.A.; Fink, G.R. Ty elements transpose through an RNA intermediate. Cell 1985, 40, 491–500. [Google Scholar] [CrossRef]
- Pace, J.K.; Feschotte, C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007, 17, 422–432. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 378, 44–53. [Google Scholar] [CrossRef]
- Jern, P.; Coffin, J.M. Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef]
- Wildschutte, J.H.; Williams, Z.H.; Montesion, M.; Subramanian, R.P.; Kidd, J.M.; Coffin, J.M. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. USA 2016, 113, E2326–E2334. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H., Jr. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203. [Google Scholar] [CrossRef]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.L.; Branciforte, D.; Keller, D.; Bain, D.L. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. 2), 13815–13820. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Moran, J.V.; Kazazian, H.H.; Boeke, J.D. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H.; Boeke, J.D.; Gabriel, A. Reverse Transcriptase Encoded by a Human Transposable Element. Science 1991, 254, 1808–1810. [Google Scholar] [CrossRef]
- Deininger, P.L. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef]
- McMillan, J.P.; Singer, M.F. Translation of the human LINE-1 element, L1Hs. Proc. Natl. Acad. Sci. USA 1993, 90, 11533–11537. [Google Scholar] [CrossRef]
- Kulpa, A.D.; Moran, J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006, 13, 655–660. [Google Scholar] [CrossRef]
- Doucet, A.J.; Hulme, A.E.; Sahinovic, E.; Kulpa, D.A.; Moldovan, J.B.; Kopera, H.C.; Athanikar, J.N.; Hasnaoui, M.; Bucheton, A.; Moran, J.V.; et al. Characterization of LINE-1 Ribonucleoprotein Particles. PLoS Genet. 2010, 6, e1001150. [Google Scholar] [CrossRef]
- Viollet, S.; Monot, C.; Cristofari, G. L1 retrotransposition. Mob. Genet. Elem. 2014, 4, e28907. [Google Scholar] [CrossRef][Green Version]
- Cost, G.J.; Feng, Q.; Jacquier, A.; Boeke, J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002, 21, 5899–5910. [Google Scholar] [CrossRef]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Cost, G.J.; Boeke, J.D. Targeting of Human Retrotransposon Integration Is Directed by the Specificity of the L1 Endonuclease for Regions of Unusual DNA Structure. Biochemistry 1998, 37, 18081–18093. [Google Scholar] [CrossRef] [PubMed]
- Morrish, T.A.; Gilbert, N.; Myers, J.S.; Vincent, B.J.; Stamato, T.D.; Taccioli, G.E.; Batzer, M.A.; Moran, J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002, 31, 159–165. [Google Scholar] [CrossRef]
- Sen, S.K.; Huang, C.T.; Han, K.; Batzer, M.A. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007, 35, 3741–3751. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Szak, S.T.; Boeke, J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 2004, 429, 268–274. [Google Scholar] [CrossRef]
- Mita, P.; Boeke, J.D. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 2016, 37, 90–100. [Google Scholar] [CrossRef]
- Lev-Maor, G.; Sorek, R.; Shomron, N.; Ast, G. The Birth of an Alternatively Spliced Exon: 3’ Splice-Site Selection in Alu Exons. Science 2003, 300, 1288–1291. [Google Scholar] [CrossRef]
- Sela, N.; Mersch, B.; Gal-Mark, N.; Lev-Maor, G.; Hotz-Wagenblatt, A.; Ast, G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007, 8, R127. [Google Scholar] [CrossRef]
- Huh, J.W.; Kim, Y.H.; Lee, S.R.; Kim, H.; Kim, D.S.; Kim, H.S.; Kang, H.-S.; Chang, K.-T. Gain of new exons and promoters by lineage-specific transposable elements-integration and conservation event on CHRM3 gene. Mol. Cells 2009, 28, 111–117. [Google Scholar] [CrossRef]
- Zhang, X.H.F.; Chasin, L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl. Acad. Sci. USA 2006, 103, 13427–13432. [Google Scholar]
- Ganguly, A.; Dunbar, T.; Chen, P.; Godmilow, L.; Ganguly, T. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum. Genet. 2003, 113, 348–352. [Google Scholar] [PubMed]
- Conley, A.B.; Piriyapongsa, J.; Jordan, I.K. Retroviral promoters in the human genome. Bioinformatics 2008, 24, 1563–1567. [Google Scholar] [CrossRef]
- Emera, D.; Casola, C.; Lynch, V.J.; Wildman, D.E.; Agnew, D.; Wagner, G.P. Convergent Evolution of Endometrial Prolactin Expression in Primates, Mice, and Elephants Through the Independent Recruitment of Transposable Elements. Mol. Biol. Evol. 2012, 29, 239–247. [Google Scholar] [PubMed]
- Bejerano, G.; Lowe, C.B.; Ahituv, N.; King, B.; Siepel, A.; Salama, S.R.; Rubin, E.M.; Kent, W.J.; Haussler, D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 2006, 441, 87–90. [Google Scholar] [PubMed]
- Franchini, L.F.; López-Leal, R.; Nasif, S.; Beati, P.; Gelman, D.M.; Low, M.J.; de Souza, F.J.S.; Rubinstein, M. Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons. Proc. Natl. Acad. Sci. USA 2011, 108, 15270–15275. [Google Scholar]
- Kazazian, H.H.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988, 332, 164–166. [Google Scholar]
- Maksakova, I.A.; Romanish, M.T.; Gagnier, L.; Dunn, C.A.; Van De Lagemaat, L.N.; Mager, D.L. Retroviral Elements and Their Hosts: Insertional Mutagenesis in the Mouse Germ Line. PLoS Genet. 2006, 2, e2. [Google Scholar]
- Shen, S.; Lin, L.; Cai, J.J.; Jiang, P.; Kenkel, E.J.; Stroik, M.R.; Sato, S.; Davidson, B.L.; Xing, Y. Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl. Acad. Sci. USA 2011, 108, 2837–2842. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ji, Z.; Bin Tian, B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008, 36, 5581–5590. [Google Scholar]
- Tang, W.; Gunn, T.M.; McLaughlin, D.F.; Barsh, G.S.; Schlossman, S.F.; Duke-Cohan, J.S. Secreted and membrane attractin result from alternative splicing of the human ATRN gene. Proc. Natl. Acad. Sci. USA 2000, 97, 6025–6230. [Google Scholar]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [PubMed]
- Piriyapongsa, J.; Mariño-Ramírez, L.; Jordan, I.K. Origin and Evolution of Human microRNAs From Transposable Elements. Genetics 2007, 176, 1323–1337. [Google Scholar] [PubMed]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar]
- Dupressoir, A.; Lavialle, C.; Heidmann, T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 2012, 33, 663–671. [Google Scholar] [CrossRef]
- Medstrand, P.; van de Lagemaat, L.N.; Mager, D.L. Retroelement Distributions in the Human Genome: Variations Associated With Age and Proximity to Genes. Genome Res. 2002, 12, 1483–1495. [Google Scholar] [CrossRef]
- Boissinot, S.; Davis, J.; Entezam, A.; Petrov, D.; Furano, A.V. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl. Acad. Sci. USA 2006, 103, 9590–9594. [Google Scholar]
- Song, M.; Boissinot, S. Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene 2007, 390, 206–213. [Google Scholar]
- Wang, J.; Lunyak, V.V.; Jordan, I.K. Genome-wide prediction and analysis of human chromatin boundary elements. Nucleic Acids Res. 2012, 40, 511–529. [Google Scholar]
- Wang, J.; Vicente-García, C.; Seruggia, D.; Moltó, E.; Fernandez-Miñán, A.; Neto, A.; Lee, E.; Gómez-Skarmeta, J.L.; Montoliu, L.; Lunyak, V.V.; et al. MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl. Acad. Sci. USA 2015, 112, E4428–E4437. [Google Scholar]
- Belancio, V.P.; Roy-Engel, A.M.; Pochampally, R.R.; Deininger, P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010, 38, 3909–3922. [Google Scholar]
- van den Hurk, J.A.; Meij, I.C.; del Carmen Seleme, M.; Kano, H.; Nikopoulos, K.; Hoefsloot, L.H.; Sistermans, E.A.; de Wijs, I.J.; Mukhopadhyay, A.; Plomp, A.S.; et al. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 2007, 16, 1587–1592. [Google Scholar] [PubMed]
- Muotri, A.R.; Chu, V.T.; Marchetto, M.C.N.; Deng, W.; Moran, J.V.; Gage, F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005, 435, 903–910. [Google Scholar]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 retrotransposition in human neural progenitor cells. Nature 2009, 460, 1127–1131. [Google Scholar] [PubMed]
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A.; et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 2015, 151, 4834–4896. [Google Scholar]
- Upton, K.R.; Gerhardt, D.J.; Jesuadian, J.S.; Richardson, S.R.; Sánchez-Luque, F.J.; Bodea, G.O.; Ewing, A.D.; Salvador-Palomeque, C.; Van Der Knaap, M.S.; Brennan, P.M.; et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 2015, 161, 228–239. [Google Scholar]
- Doucet-O’Hare, T.T.; Rodić, N.; Sharma, R.; Darbari, I.; Abril, G.; Choi, J.A.; Ahn, J.Y.; Cheng, Y.; Anders, R.A.; Burns, K.H.; et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. USA 2015, 112, E4894–E4900. [Google Scholar]
- Ewing, A.D.; Gacita, A.; Wood, L.D.; Ma, F.; Xing, D.; Kim, M.-S.; Manda, S.S.; Abril, G.; Pereira, G.; Makohon-Moore, A.; et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015, 25, 1536–1545. [Google Scholar]
- Doucet-O’Hare, T.T.; Sharma, R.; Rodić, N.; Anders, R.A.; Burns, K.H.; Kazazian, H.H. Somatically Acquired LINE-1 Insertions in Normal Esophagus Undergo Clonal Expansion in Esophageal Squamous Cell Carcinoma. Hum. Mutat. 2016, 37, 942–954. [Google Scholar]
- Scott, E.C.; Devine, S.E. The Role of Somatic L1 Retrotransposition in Human Cancers. Viruses 2017, 9, 131. [Google Scholar]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J., 3rd; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of Somatic Retrotransposition in Human Cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Solyom, S.; Ewing, A.D.; Rahrmann, E.P.; Doucet, T.; Nelson, H.H.; Burns, M.B.; Harris, R.S.; Sigmon, D.F.; Casella, A.; Erlanger, B.; et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012, 22, 2328–2338. [Google Scholar] [CrossRef]
- Miki, Y.; Nishisho, I.; Horii, A.; Miyoshi, Y.; Utsunomiya, J.; Kinzler, K.W.; Vogelstein, B.; Nakamura, Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992, 52, 643–645. [Google Scholar]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef]
- Shukla, R.; Upton, K.R.; Muñoz-Lopez, M.; Gerhardt, D.J.; Fisher, M.E.; Nguyen, T.; Brennan, P.M.; Baillie, J.K.; Collino, A.; Ghisletti, S.; et al. Endogenous Retrotransposition Activates Oncogenic Pathways in Hepatocellular Carcinoma. Cell 2013, 153, 101–111. [Google Scholar] [CrossRef]
- Helman, E.; Lawrence, M.S.; Stewart, C.; Sougnez, C.; Getz, G.; Meyerson, M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014, 24, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Iskow, R.C.; McCabe, M.T.; Mills, R.E.; Torene, S.; Pittard, W.S.; Neuwald, A.F.; Van Meir, E.G.; Vertino, P.M.; Devine, S.E. Natural Mutagenesis of Human Genomes by Endogenous Retrotransposons. Cell 2010, 141, 1253–1261. [Google Scholar] [CrossRef]
- Tubio, J.M.; Li, Y.; Ju, Y.S.; Martincorena, I.; Cooke, S.L.; Tojo, M.; Gundem, G.; Pipinikas, C.P.; Zamora, J.; Raine, K.; et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014, 345, 1251343. [Google Scholar] [CrossRef]
- Wang, Y.P.; Song, G.H.; Chen, J.; Xiao, C.; Li, C.; Zhong, L.; Sun, X.; Wang, Z.W.; Deng, G.L.; Yu, F.D.; et al. Elevated OCT1 participates in colon tumorigenesis and independently predicts poor prognoses of colorectal cancer patients. Tumor Biol. 2016, 37, 3247–3255. [Google Scholar] [CrossRef]
- Obinata, D.; Takayama, K.-I.; Urano, T.; Murata, T.; Kumagai, J.; Fujimura, T.; Ikeda, K.; Horie-Inoue, K.; Homma, Y.; Ouchi, Y.; et al. Oct1 regulates cell growth of LNCaP cells and is a prognostic factor for prostate cancer. Int. J. Cancer 2012, 130, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liao, S.; Zhou, Y.; Jiang, B.; Li, Y.; Xue, M. High expression of octamer transcription factor 1 in cervical cancer. Oncol. Lett. 2014, 7, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Maddox, J.; Shakya, A.; South, S.; Shelton, D.; Andersen, J.N.; Chidester, S.; Kang, J.; Gligorich, K.M.; Jones, D.A.; Spangrude, G.J.; et al. Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant. PLoS Genet. 2012, 8, e1003048. [Google Scholar] [CrossRef]
- Carreira, P.E.; Richardson, S.R.; Faulkner, G.J. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 2014, 281, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.G.; Swergold, G.; Ozato, K.; Thayer, R.E. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet. 1993, 2, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zhang, L.; Zhang, Y.; Kazazian, H.H., Jr. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003, 31, 4929–4940. [Google Scholar] [CrossRef] [PubMed]
- Athanikar, J.N.; Badge, R.M.; Moran, J.V. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004, 32, 3846–3855. [Google Scholar] [CrossRef]
- Harris, C.R.; DeWan, A.; Zupnick, A.; Normart, R.; Gabriel, A.; Prives, C.; Levine, A.J.; Hoh, J. p53 responsive elements in human retrotransposons. Oncogene 2009, 28, 3857–3865. [Google Scholar] [CrossRef]
- Kunarso, G.; Chia, N.-Y.; Jeyakani, J.; Hwang, C.; Lu, X.; Chan, Y.-S.; Ng, H.-H.; Bourque, G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010, 42, 631–634. [Google Scholar] [CrossRef]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Raskó, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516, 405–409. [Google Scholar] [CrossRef]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Mark, W.; Wesche, D.J.; Lance, M.; Ware, C.B.; Blish, C.A.; Chang, H.Y. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015, 522, 221–225. [Google Scholar] [CrossRef]
- Wylie, A.; Jones, A.E.; D’Brot, A.; Lu, W.-J.; Kurtz, P.; Moran, J.V.; Rakheja, D.; Chen, K.S.; Hammer, R.E.; Comerford, S.A.; et al. p53 genes function to restrain mobile elements. Genes Dev. 2016, 30, 64–77. [Google Scholar] [CrossRef]
- Sun, X.; Wang, X.; Tang, Z.; Grivainis, M.; Kahler, D.; Yun, C.; Mita, P.; Fenyö, D.; Boeke, J.D. Transcription factor profiling reveals molecular choreography and key regulators of human retrotransposon expression. Proc. Natl. Acad. Sci. USA 2018, 115, E5526–E5535. [Google Scholar] [CrossRef]
- Thornburg, B.G.; Gotea, V.; Makałowski, W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006, 365, 104–110. [Google Scholar] [CrossRef]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014, 5, 5011. [Google Scholar] [CrossRef]
- Alkailani, M.; Palidwor, G.; Poulin, A.; Mohan, R.; Pepin, D.; Vanderhyden, B.; Gibbings, D. A genome-wide strategy to identify causes and consequences of retrotransposon expression finds activation by BRCA1 in ovarian cancer. NAR Cancer 2021, 3, zcaa040. [Google Scholar] [CrossRef]
- Grandi, F.C.; Rosser, J.M.; Newkirk, S.J.; Yin, J.; Jiang, X.; Xing, Z.; Whitmore, L.; Bashir, S.; Ivics, Z.; Izsvák, Z.; et al. Retrotransposition creates sloping shores: A graded influence of hypomethylated CpG islands on flanking CpG sites. Genome Res. 2015, 25, 1135–1146. [Google Scholar] [CrossRef]
- Fazzari, M.J.; Greally, J.M. Epigenomics: Beyond CpG islands. Nat. Rev. Genet. 2004, 5, 446–455. [Google Scholar] [CrossRef]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Wolf, G.; Yang, P.; Füchtbauer, A.C.; Füchtbauer, E.M.; Silva, A.M.; Park, C.; Wu, W.; Nielsen, A.L.; Pedersen, F.S.; Macfarlan, T.S. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015, 29, 538–554. [Google Scholar] [CrossRef]
- Imbeault, M.; Helleboid, P.Y.; Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017, 543, 550–554. [Google Scholar] [CrossRef]
- Milewska, A.; Kindler, E.; Vkovski, P.; Zeglen, S.; Ochman, M.; Thiel, V.; Rajfur, Z.; Pyrc, K. APOBEC3-mediated restriction of RNA virus replication. Sci. Rep. 2018, 8, 5960. [Google Scholar] [CrossRef]
- Esnault, C. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006, 34, 1522–1531. [Google Scholar] [CrossRef]
- Richardson, S.R.; Narvaiza, I.; Planegger, R.A.; Weitzman, M.D.; Moran, J. APOBEC3A Deaminates Transiently Exposed Single-Strand DNA during LINE-1 Retrotransposition. Elife 2014, 3, e02008. [Google Scholar] [CrossRef]
- Horn, A.V.; Klawitter, S.; Held, U.; Berger, A.; Jaguva Vasudevan, A.A.; Bock, A.; Hofmann, H.; Hanschmann, K.M.; Trösemeier, J.H.; Flory, E.; et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2014, 42, 396–416. [Google Scholar] [CrossRef]
- Liang, W.; Xu, J.; Yuan, W.; Song, X.; Zhang, J.; Wei, W.; Yu, X.-F.; Yang, Y. APOBEC3DE Inhibits LINE-1 Retrotransposition by Interacting with ORF1p and Influencing LINE Reverse Transcriptase Activity. PLoS ONE 2016, 11, e0157220. [Google Scholar] [CrossRef]
- Herrmann, A.; Wittmann, S.; Thomas, D.; Shepard, C.N.; Kim, B.; Ferreirós, N.; Gramberg, T. The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mob. DNA 2018, 28, 11. [Google Scholar] [CrossRef]
- Li, P.; Du, J.; Goodier, J.L.; Hou, J.; Kang, J.; Kazazian, H.H.; Zhao, K.; Yu, X.-F. Aicardi-Goutières syndrome protein TREX1 suppresses L1 and maintains genome integrity through exonuclease-independent ORF1p depletion. Nucleic Acids Res. 2017, 45, 4619–4631. [Google Scholar] [CrossRef]
- Lee, Y.C. The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster. PLoS Genet. 2015, 11, e1005269. [Google Scholar] [CrossRef]
- Dong, J.; Wang, X.; Cao, C.; Wen, Y.; Sakashita, A.; Chen, S.; Zhang, J.; Zhang, Y.; Zhou, L.; Luo, M.; et al. UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat Commun. 2019, 10, 4705. [Google Scholar] [CrossRef]
- Choi, J.; Hwang, S.-Y.; Ahn, K. Interplay between RNASEH2 and MOV10 controls LINE-1 retrotransposition. Nucleic Acids Res. 2018, 46, 1912–1926. [Google Scholar] [CrossRef]
- Zhang, A.; Dong, B.; Doucet, A.J.; Moldovan, J.B.; Moran, J.V.; Silverman, R.H. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 2014, 42, 3803–3820. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, J.B.; Moran, J.V. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015, 11, e1005121. [Google Scholar] [CrossRef] [PubMed]
- Boissinot, S.; Furano, A.V. Adaptive Evolution in LINE-1 Retrotransposons. Mol. Biol. Evol. 2001, 18, 2186–2194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scott, A.F.; Schmeckpeper, B.J.; Abdelrazik, M.; Comey, C.T.; O’Hara, B.; Rossiter, J.P.; Cooley, T.; Heath, P.; Smith, K.D.; Margolet, L. Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1987, 1, 113–125. [Google Scholar] [CrossRef]
- Szak, S.T.; Pickeral, O.K.; Makalowski, W.; Boguski, M.S.; Landsman, D.; Boeke, J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002, 3, research0052.1. [Google Scholar] [CrossRef] [PubMed]
- Ewing, A.D. Transposable element detection from whole genome sequence data. Mob. DNA 2015, 6, 24. [Google Scholar] [CrossRef]
- Goerner-Potvin, P.; Bourque, G. Computational tools to unmask transposable elements. Nat. Rev. Genet. 2018, 19, 688–704. [Google Scholar] [CrossRef]
- Mir, A.A.; Philippe, C.; Cristofari, G. euL1db: The European database of L1HS retrotransposon insertions in humans. Nucleic Acids Res. 2015, 43, D43–D47. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Yu, F.; Zingler, N.; Schumann, G.; Strätling, W.H. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001, 29, 4493–4501. [Google Scholar] [CrossRef] [PubMed]
- Rishishwar, L.; Mariño-Ramírez, L.; Jordan, I.K. Benchmarking computational tools for polymorphic transposable element detection. Brief. Bioinform. 2017, 18, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Elyanow, R.; Wu, H.T.; Raphael, B.J. Identifying structural variants using linked-read sequencing data. Bioinformatics 2018, 34, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Criscione, S.W.; Zhang, Y.; Thompson, W.; Sedivy, J.M.; Neretti, N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genom. 2014, 15, 583. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.R.; Ardeljan, D.; Pacyna, C.N.; Payer, L.M.; Burns, K.H. SQuIRE reveals locus-specific regulation of interspersed repeat expression. Nucleic Acids Res. 2019, 47, e27. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Carreira, P.E.; Sanchez-Luque, F.J.; Schauer, S.N.; Fagg, A.C.; Richardson, S.R.; Davies, C.M.; Jesuadian, J.S.; Kempen, M.-J.H.; Troskie, R.-L.; et al. L1 Retrotransposon Heterogeneity in Ovarian Tumor Cell Evolution. Cell Rep. 2018, 23, 3730–3740. [Google Scholar] [CrossRef] [PubMed]
- Schauer, S.N.; Carreira, P.E.; Shukla, R.; Gerhardt, D.J.; Gerdes, P.; Sanchez-Luque, F.J.; Nicoli, P.; Kindlova, M.; Ghisletti, S.; Dos Santos, A.; et al. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res. 2018, 28, 639–653. [Google Scholar] [CrossRef]
- Jung, H.; Choi, J.K.; Lee, E.A. Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers. Genome Res. 2018, 28, 1136–1146. [Google Scholar] [CrossRef]
- Sultana, T.; van Essen, D.; Siol, O.; Bailly-Bechet, M.; Philippe, C.; El Aabidine, A.Z.; Pioger, L.; Nigumann, P.; Saccani, S.; Andrau, J.-C.; et al. The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-insertion Sequence Biases and Post-insertion Selection. Mol. Cell 2019, 74, 555–570.e7. [Google Scholar] [CrossRef]
- Rodriguez-Martin, B.; Alvarez, E.G.; Baez-Ortega, A.; Zamora, J.; Supek, F.; Demeulemeester, J.; Santamarina, M.; Ju, Y.S.; Temes, J.; Garcia-Souto, D.; et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020, 52, 306–319. [Google Scholar] [CrossRef]
- Guler, G.D.; Tindell, C.A.; Pitti, R.; Wilson, C.; Nichols, K.; Cheung, T.K.; Kim, H.-J.; Wongchenko, M.; Yan, Y.; Haley, B.; et al. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell 2017, 32, 221–237.e13. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Rose, C.M.; Cass, A.A.; Williams, A.G.; Darwish, M.; Lianoglou, S.; Haverty, P.M.; Tong, A.-J.; Blanchette, C.; Albert, M.L.; et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 2019, 10, 5228. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Li, H.; Torabi, S.J.; Yarbrough, W.G.; Mehra, S.; Osborn, H.A.; Judson, B. Association of Human Papillomavirus Status at Head and Neck Carcinoma Subsites With Overall Survival. JAMA Otolaryngol Head Neck Surg. 2018, 144, 519–525. [Google Scholar] [CrossRef]
- Ishak, C.A.; Classon, M.; De Carvalho, D.D. Deregulation of Retroelements as an Emerging Therapeutic Opportunity in Cancer. Trends Cancer 2018, 4, 583–597. [Google Scholar] [CrossRef]
- Roers, A.; Hiller, B.; Hornung, V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 2016, 44, 739–754. [Google Scholar] [CrossRef]
- Shen, Y.J.; Le Bert, N.; Chitre, A.A.; Koo, C.X.; Nga, X.H.; Ho, S.S.W.; Khatoo, M.; Tan, N.Y.; Ishii, K.J.; Gasser, S. Genome-Derived Cytosolic DNA Mediates Type I Interferon-Dependent Rejection of B Cell Lymphoma Cells. Cell Rep. 2015, 11, 460–473. [Google Scholar] [CrossRef]
- Stok, E.J.; Oosenbrug, T.; ter Haar, L.R.; Gravekamp, D.; Bromley, C.P.; Zelenay, S.; e Sousa, C.R.; van der Veen, A.G. RNA sensing via the RIG-I-like receptor LGP2 is essential for the induction of a type I IFN response in ADAR1 deficiency. EMBO J. 2022, 41, e109760. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Lübben, W.; Slomka, H.; Gebler, J.; Konert, M.; Cai, C.; Neubrandt, L.; da Costa, O.P.; Paul, S.; Dehnert, S.; et al. Nucleic Acid-Sensing Toll-like Receptors Are Essential for the Control of Endogenous Retrovirus Viremia and ERV-Induced Tumors. Immunity 2012, 37, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Pratt, G.A.; Sundararaman, B.; Townsend, M.J.; Chaivorapol, C.; Bhangale, T.; Graham, R.R.; Ortmann, W.; Criswell, L.A.; Yeo, G.W.; et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 2015, 350, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Luqman-Fatah, A.; Watanabe, Y.; Uno, K.; Ishikawa, F.; Moran, J.V.; Miyoshi, T. The interferon stimulated gene-encoded protein HELZ2 inhibits human LINE-1 retrotransposition and LINE-1 RNA-mediated type I interferon induction. Nat. Commun. 2023, 14, 203. [Google Scholar] [CrossRef]
- Irwin, R.E.; Scullion, C.; Thursby, S.J.; Sun, M.; Thakur, A.; Hilman, L.; Callaghan, B.; Thompson, P.D.; McKenna, D.J.; Rothbart, S.B.; et al. The UHRF1 protein is a key regulator of retrotransposable elements and innate immune response to viral RNA in human cells. Epigenetics. 2023, 18, 2216005. [Google Scholar] [CrossRef]
- Ahmad, S.; Mu, X.; Yang, F.; Greenwald, E.; Park, J.W.; Jacob, E.; Zhang, C.Z.; Hur, S. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell. 2018, 172, 797–810. [Google Scholar] [CrossRef]
- Neulinger-Muñoz, M.; Schaack, D.; Grekova, S.P.; Bauer, A.S.; Giese, T.; Salg, G.A.; Espinet, E.; Leuchs, B.; Heller, A.; Nüesch, J.P.F.; et al. Human Retrotransposons and the Global Shutdown of Homeostatic Innate Immunity by Oncolytic Parvovirus H-1PV in Pancreatic Cancer. Viruses 2021, 13, 1019. [Google Scholar] [CrossRef]
- Cuellar, T.L.; Herzner, A.M.; Zhang, X.; Goyal, Y.; Watanabe, C.; Friedman, B.A.; Janakiraman, V.; Durinck, S.; Stinson, J.; Arnott, D.; et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 2017, 216, 3535–3549. [Google Scholar] [CrossRef]
- Yan, J.; Zhao, Y.; Du, J.; Wang, Y.; Wang, S.; Wang, Q.; Zhao, X.; Xu, W.; Zhao, K. RNA sensor MDA5 suppresses LINE-1 retrotransposition by regulating the promoter activity of LINE-1 5’-UTR. Mob DNA 2022, 13, 10. [Google Scholar] [CrossRef]
- Volkmann, B.; Wittmann, S.; Lagisquet, J.; Deutschmann, J.; Eissmann, K.; Ross, J.J.; Biesinger, B.; Gramberg, T. Human TRIM5α senses and restricts LINE-1 elements. Proc. Natl. Acad. Sci. USA 2020, 117, 17965–17976. [Google Scholar] [CrossRef]
- Schmidt, N.; Domingues, P.; Golebiowski, F.; Patzina, C.; Tatham, M.H.; Hay, R.T.; Hale, B.G. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17399–17408. [Google Scholar] [CrossRef]
- Novototskaya-Vlasova, K.A.; Neznanov, N.S.; Molodtsov, I.; Hall, B.M.; Commane, M.; Gleiberman, A.S.; Murray, J.; Haber, M.; Norris, M.D.; Leonova, K.I.; et al. Inflammatory response to retrotransposons drives tumor drug resistance that can be prevented by reverse transcriptase inhibitors. Proc. Natl. Acad. Sci. USA 2022, 119, e2213146119. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, H.; Ørskov, A.D.; Helbo, A.S.; Gillberg, L.; Liu, M.; Zhou, W.; Ungerstedt, J.; Hellström-Lindberg, E.; Sun, W.; Liang, G.; et al. Activation of a Subset of Evolutionarily Young Transposable Elements and Innate Immunity Are Linked to Clinical Responses to 5-Azacytidine. Cancer Res. 2020, 80, 2441–2450. [Google Scholar] [CrossRef] [PubMed]
- Cañadas, I.; Thummalapalli, R.; Kim, J.W.; Kitajima, S.; Jenkins, R.W.; Christensen, C.L.; Campisi, M.; Kuang, Y.; Zhang, Y.; Gjini, E.; et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 2018, 24, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Hou, G.; Diep, J.; Ooi, Y.S.; Akopyants, N.S.; Beverley, S.M.; Carette, J.E.; Greenberg, H.B.; Ding, S. Inhibitor of growth protein 3 epigenetically silences endogenous retroviral elements and prevents innate immune activation. Nucleic Acids Res. 2021, 49, 12706–12715. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Guo, G. Immunomodulatory Function of the Tumor Suppressor p53 in Host Immune Response and the Tumor Microenvironment. Int. J. Mol. Sci. 2016, 17, 1942. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Macip, S.; Martínez-Sobrido, L.; Brown, L.; Ashour, J.; García-Sastre, A.; Lee, S.W.; Aaronson, S.A. Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 2008, 205, 1929–1938. [Google Scholar] [CrossRef]
- Leonova, K.I.; Brodsky, L.; Lipchick, B.; Pal, M.; Novototskaya, L.; Chenchik, A.A.; Sen, G.C.; Komarova, E.A.; Gudkov, A.V. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, E89–E98. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, I.; Marcel, V.; Olivier, M.; Oren, M.; Rotter, V.; Hainaut, P. Understanding wild-type and mutant p53 activities in human cancer: New landmarks on the way to targeted therapies. Cancer Gene Ther. 2011, 18, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Fung, K.Y.; Mangan, N.E.; Cumming, H.; Horvat, J.C.; Mayall, J.R.; Stifter, S.A.; De Weerd, N.; Roisman, L.C.; Rossjohn, J.; Robertson, S.A.; et al. Interferon-ε Protects the Female Reproductive Tract from Viral and Bacterial Infection. Science 2013, 339, 1088–1092. [Google Scholar] [CrossRef]
- Mason, C.E.; Shu, F.-J.; Wang, C.; Session, R.M.; Kallen, R.G.; Sidell, N.; Yu, T.; Liu, M.H.; Cheung, E.; Kallen, C.B. Location analysis for the estrogen receptor-α reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 2010, 38, 2355–2368. [Google Scholar] [CrossRef]
- Lamsal, A.; Andersen, S.B.; Johansson, I.; Vietri, M.; Bokil, A.A.; Kurganovs, N.J.; Rylander, F.; Bjørkøy, G.; Pettersen, K.; Giambelluca, M.S. Opposite and dynamic regulation of the interferon response in metastatic and non-metastatic breast cancer. Cell Commun. Signal. 2023, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- Sciamanna, I.; De Luca, C.; Spadafora, C. The Reverse Transcriptase Encoded by LINE-1 Retrotransposons in the Genesis, Progression, and Therapy of Cancer. Front. Chem. 2016, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Galindo, R.; Kaplan, M.H.; Leissner, P.; Verjat, T.; Ferlenghi, I.; Bagnoli, F.; Giusti, F.; Dosik, M.H.; Hayes, D.F.; Gitlin, S.D.; et al. Human Endogenous Retrovirus K (HML-2) Elements in the Plasma of People with Lymphoma and Breast Cancer. J. Virol. 2008, 82, 9329–9336. [Google Scholar] [CrossRef]
- De Clercq, E. HIV-1-specific RT inhibitors: Highly selective inhibitors of human immunodeficiency virus type 1 that are specifically targeted at the viral reverse transcriptase. Med. Res. Rev. 1993, 13, 229–258. [Google Scholar] [CrossRef] [PubMed]
- Mangiacasale, R.; Pittoggi, C.; Sciamanna, I.; Careddu, A.; Mattei, E.; Lorenzini, R.; Travaglini, L.; Landriscina, M.; Barone, C.; Nervi, C.; et al. Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene 2003, 22, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Landriscina, M.; Spadafora, C.; Cignarelli, M.; Barone, C. Anti-Tumor Activity of Non-Nucleosidic Reverse Transcriptase Inhibitors. Curr. Pharm. Des. 2007, 13, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Sciamanna, I.; Landriscina, M.; Pittoggi, C.; Quirino, M.; Mearelli, C.; Beraldi, R.; Mattei, E.; Serafino, A.; Cassano, A.; Sinibaldi-Vallebona, P.; et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 2005, 24, 3923–3931. [Google Scholar] [CrossRef]
- Jones, R.B.; Garrison, K.E.; Wong, J.C.; Duan, E.H.; Nixon, D.F.; Ostrowski, M.A. Nucleoside Analogue Reverse Transcriptase Inhibitors Differentially Inhibit Human LINE-1 Retrotransposition. PLoS ONE 2008, 3, e1547. [Google Scholar] [CrossRef]
- Carlini, F.; Ridolfi, B.; Molinari, A.; Parisi, C.; Bozzuto, G.; Toccacieli, L.; Formisano, G.; De Orsi, D.; Paradisi, S.; Grober, O.M.V.; et al. The Reverse Transcription Inhibitor Abacavir Shows Anticancer Activity in Prostate Cancer Cell Lines. PLoS ONE 2010, 5, e14221. [Google Scholar] [CrossRef]
- Rodić, N.; Burns, K.H. Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms? Rosenberg SM, editor. PLoS Genet. 2013, 9, e1003402. [Google Scholar] [CrossRef] [PubMed]
- Hukezalie, K.R.; Thumati, N.R.; Côté, H.C.F.; Wong, J.M.Y. In Vitro and Ex Vivo Inhibition of Human Telomerase by Anti-HIV Nucleoside Reverse Transcriptase Inhibitors (NRTIs) but Not by Non-NRTIs. PLoS ONE 2012, 7, e47505. [Google Scholar] [CrossRef]
- Saito, Y.; Nakaoka, T.; Sakai, K.; Muramatsu, T.; Toshimitsu, K.; Kimura, M.; Kanai, T.; Sato, T.; Saito, H. Inhibition of DNA Methylation Suppresses Intestinal Tumor Organoids by Inducing an Anti-Viral Response. Sci. Rep. 2016, 6, 25311. [Google Scholar] [CrossRef] [PubMed]
- Natoli, M.; Gallon, J.; Lu, H.; Amgheib, A.; Pinato, D.J.; Mauri, A.F.; Marafioti, T.; Akarca, A.U.; Ullmo, I.; Ip, J.; et al. Transcriptional analysis of multiple ovarian cancer cohorts reveals prognostic and immunomodulatory consequences of ERV expression. J. Immunother. Cancer 2021, 9, e001519. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Sajed, D.; Arora, K.S.; Solovyov, A.; Rajurkar, M.; Bledsoe, J.R.; Sil, S.; Amri, R.; Tai, E.; MacKenzie, O.C.; et al. Diverse repetitive element RNA expression defines epigenetic and immunologic features of colon cancer. J. Clin. Investig. 2017, 2, e91078. [Google Scholar] [CrossRef]
- Zhao, H.; Ning, S.; Nolley, R.; Scicinski, J.; Oronsky, B.; Knox, S.J.; Peehl, D.M. The immunomodulatory anticancer agent, RRx-001, induces an interferon response through epigenetic induction of viral mimicry. Clin. Epigenet. 2017, 9, 4. [Google Scholar] [CrossRef]
- Brocks, D.; Schmidt, C.R.; Daskalakis, M.; Jang, H.S.; Shah, N.M.; Li, D.; Li, J.; Zhang, B.; Hou, Y.; Laudato, S.; et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat. Genet. 2017, 49, 1052–1060. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; Watt, A.C.; BrinJones, H.; Sceneay, J.; Li, B.B.; Khan, N.; Ubellacker, J.M.; Xie, S.; Metzger-Filho, O.; et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017, 548, 471–475. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef]
- Kawamura, Y.; Calle, A.S.; Yamamoto, Y.; Sato, T.-A.; Ochiya, T. Extracellular vesicles mediate the horizontal transfer of an active LINE-1 retrotransposon. J. Extracell. Vesicles 2019, 8, 1643214. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).