Patient-Derived Organoids as a Promising Tool for Multimodal Management of Sarcomas
Abstract
:Simple Summary
Abstract
1. Introduction
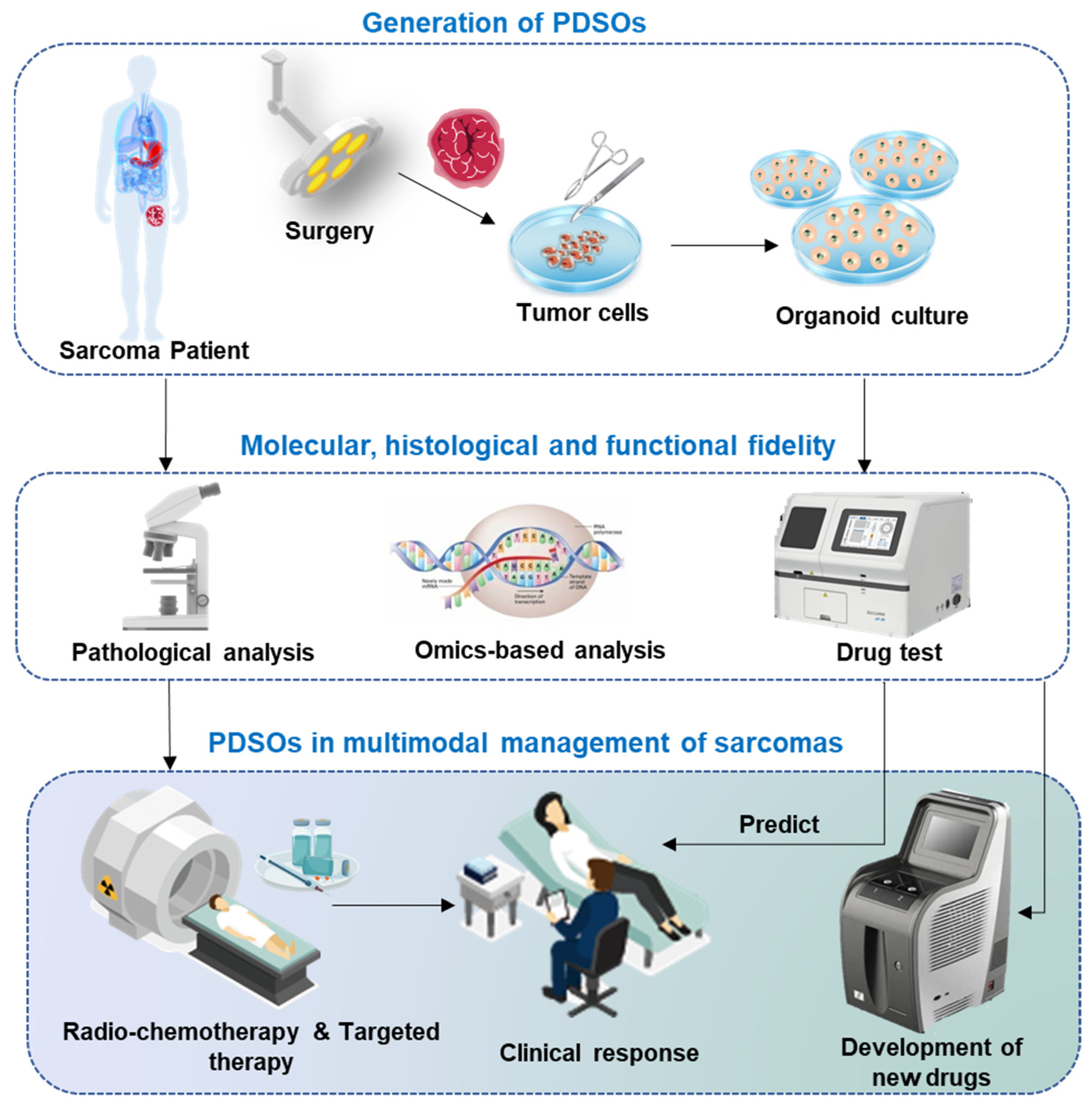
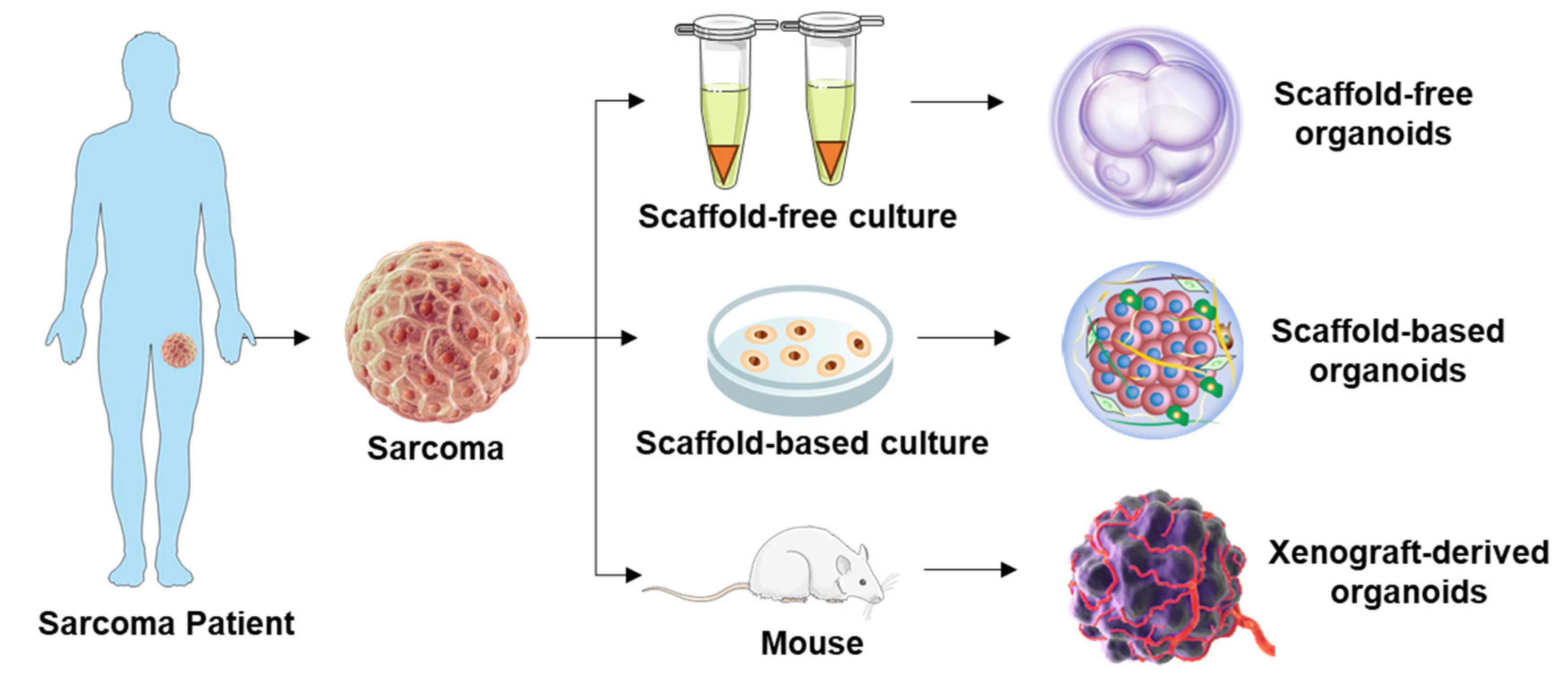
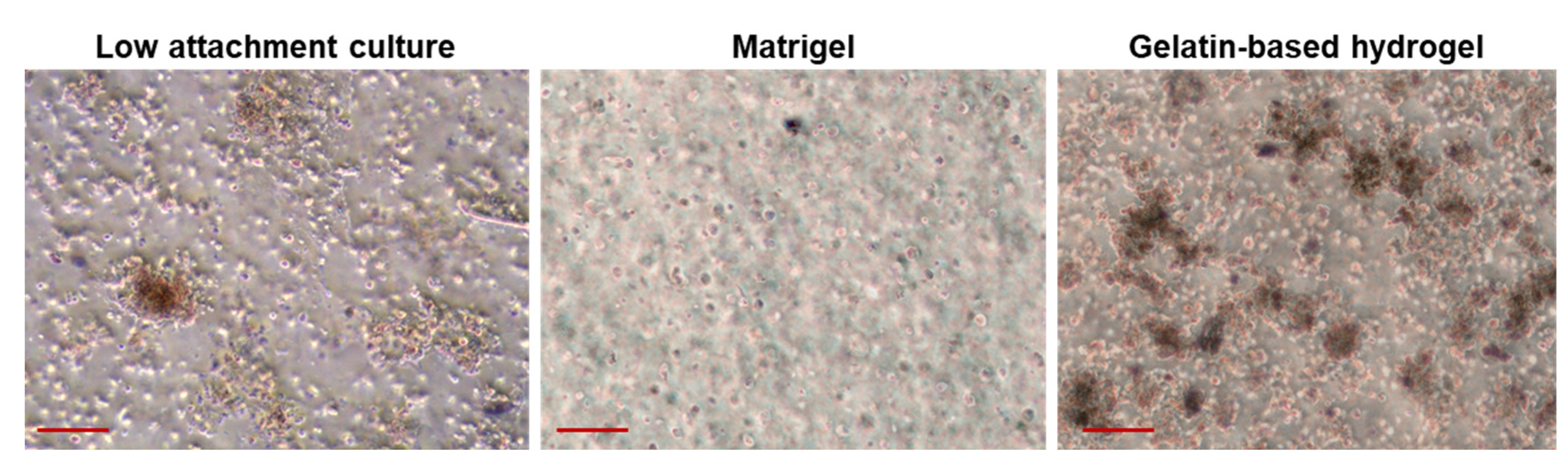
2. Establishment of PDSOs
2.1. Approaches for Obtaining Sarcoma Tissue Samples
2.2. Recapitulating Tumor Heterogeneity and Complexity in PDSOs
| Method | Organoid Establishment Methods | Sarcoma Type | Coculture Condition | Culture Media | Organoid Formation Rate and Day | Molecular results—Gene Profiling, etc. | Histological and Functional Results—Morphology, Cell Growth, etc. | Treatments | Author/Year |
|---|---|---|---|---|---|---|---|---|---|
| Scaffold-free |
|
| N/A | GM: advance DMEM supplemented with—
|
|
|
|
| Chen et al. 2023 [29] |
|
| N/A |
| N/A |
|
|
| Bangerter et al. 2023 [30] | |
| Scaffold-based |
|
| 1:3 tumor cells to immune cells (iPDSO). |
|
|
|
|
| Forsythe et al. 2022 [25] |
| 2 × ES | N/A |
| N/A |
|
|
| Wakamatsu et al. 2022 [32] | |
|
| N/A |
|
|
|
|
| Meister et al. 2022 [33] | |
| XenograftODX |
|
| N/A | N/A |
|
|
| N/A | Costa et al. 2023 [36] |
|
| N/A | GM | 30–60 days for average outgrowth time |
|
| N/A | Suzuki et al. 2023 [50] | |
|
| N/A | N/A |
|
|
|
| Xu et al. 2022 [37] |
2.3. Molecular, Histological, and Functional Fidelity of PDSOs
2.3.1. Molecular Fidelity
2.3.2. Histological Fidelity
2.3.3. Functional Fidelity
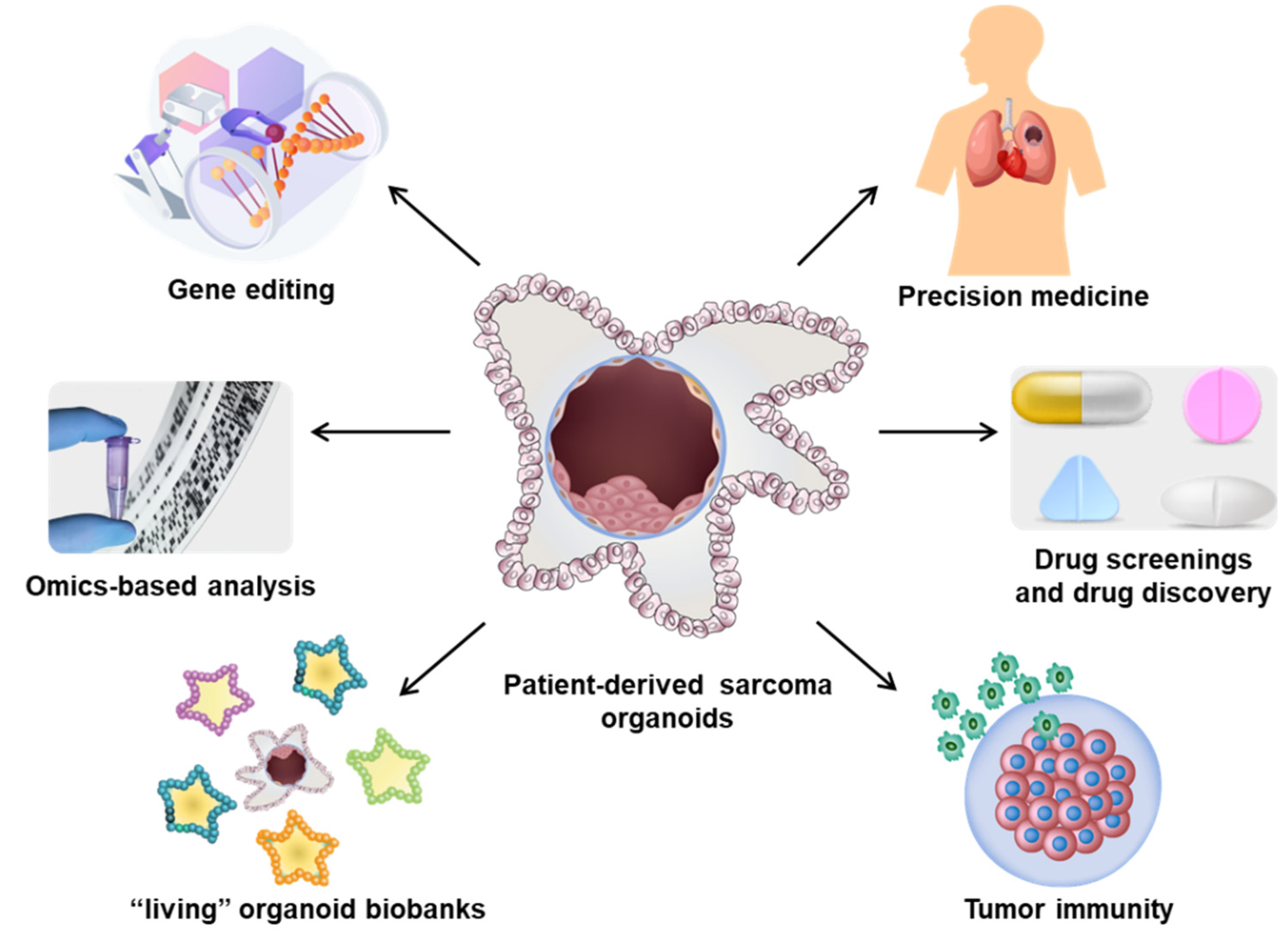
3. PDSOs in Multimodal Management of Sarcomas
3.1. Pathological and Molecular Genetic Evaluation

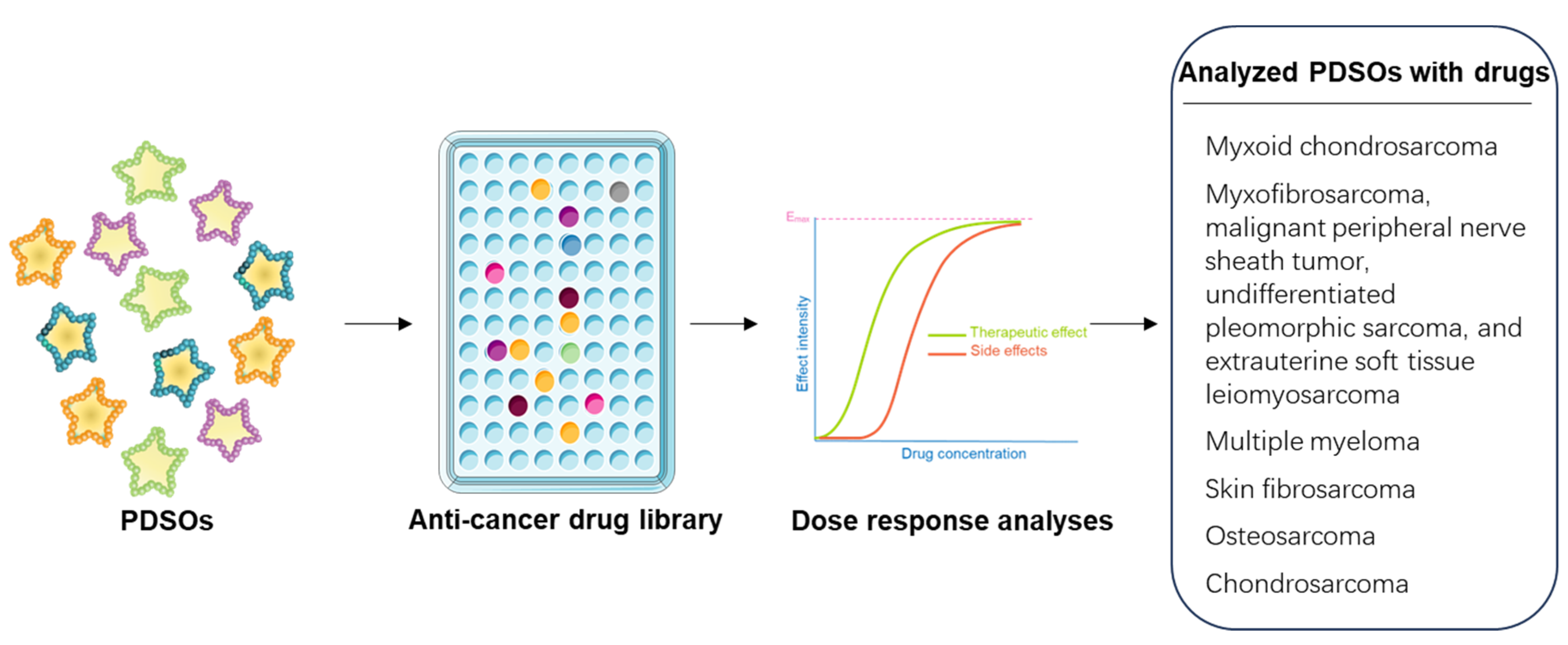
3.2. Drug Screening and and Development of Effective Treatment
3.3. Personalized Multimodal Management
3.4. Challenges and Limitations of PDSOs in Multimodality Managements
4. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviation
| Adenosine 5′-triphosphate (ATP) |
| Angiosarcoma (AS) |
| Base medium (BM) |
| BRM-associated factor (BAF) |
| BRCA1-associated RING domain 1 (BARD1) |
| BRM-associated factor (BAF) |
| Chondrosarcoma (CS) |
| Chordoma (CD) |
| Ewing sarcoma (ES) |
| Extrauterine soft tissue leiomyosarcoma (ESTL) |
| Extraskeletal myxoid chondrosarcoma (EMC) |
| Epithelioid sarcoma (ES) |
| Dermatofibrosarcoma protuberans (DFSP) |
| Gastrointestinal stromal tumor (GST) |
| Giant osteosarcoma (GOS) |
| Growth media (GM) |
| Hemangiopericytoma (HP) |
| Hyaluronic acid (HA) |
| Immunohistochemistry (IHC) |
| Immune-enhanced patient-derived sarcoma organoids (iPDSO) |
| Leiomyosarcoma (LMS) |
| Liposarcoma (LS) |
| Malignant fibrous histiocytoma (MFH) |
| malignant giant-cell tumor (MGT) |
| Malignant peripheral nerve sheath tumor (MPNST) |
| malignant schwannoma (MS) |
| Metalloproteinases (MMP9) |
| Myxofibrosarcoma (MFS) |
| Not applicable (N/A) |
| Osteosarcoma (OS) |
| Patient-derived organoids (PDO) |
| Patient-derived sarcoma organoids (PDSO) |
| Pleiomorphic abdominal sarcoma (PAS) |
| Poly [ADP-ribose] polymerase 1 (PARP1) |
| Programmed cell death protein 1 (PD1) |
| Rhabdomyosarcoma (RMS) |
| Short tandem repeat (STR) |
| SRY-box transcription factor 9 (Sox9) |
| Synovial sarcoma (SS) |
| Three-dimensional (3-D) |
| Tumor mutational burden (TMB) |
| Two-dimensional (2-D) |
| Undifferentiated pleomorphic sarcoma (UPS) |
| Vimentin (VIM) |
References
- Farid, M.; Ngeow, J. Sarcomas Associated With Genetic Cancer Predisposition Syndromes: A Review. Oncologist 2016, 21, 1002–1013. [Google Scholar] [CrossRef] [PubMed]
- Gounder, M.M.; Agaram, N.P.; Trabucco, S.E.; Robinson, V.; Ferraro, R.A.; Millis, S.Z.; Krishnan, A.; Lee, J.; Attia, S.; Abida, W.; et al. Clinical genomic profiling in the management of patients with soft tissue and bone sarcoma. Nat. Commun. 2022, 13, 3406. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, H.-S.; Han, I. Actual long-term survival after resection of stage III soft tissue sarcoma. BMC Cancer 2021, 21, 21. [Google Scholar] [CrossRef]
- Sugiura, H.; Tsukushi, S.; Yoshida, M.; Nishida, Y. What is the success of repeat surgical treatment of a local recurrence after initial wide resection of soft tissue sarcomas? Clin. Orthop. Relat. Res. 2018, 476, 1791. [Google Scholar] [CrossRef]
- Psilopatis, I.; Kokkali, S.; Palamaris, K.; Digklia, A.; Vrettou, K.; Theocharis, S. Organoids: A New Chapter in Sarcoma Diagnosis and Treatment. Int. J. Mol. Sci. 2022, 23, 11271. [Google Scholar] [CrossRef] [PubMed]
- Longhi, A.; Errani, C.; De Paolis, M.; Mercuri, M.; Bacci, G. Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat. Rev. 2006, 32, 423–436. [Google Scholar] [CrossRef]
- Fleuren, E.D.; Versleijen-Jonkers, Y.M.; Boerman, O.C.; van der Graaf, W.T. Targeting receptor tyrosine kinases in osteosarcoma and Ewing sarcoma: Current hurdles and future perspectives. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2014, 1845, 266–276. [Google Scholar] [CrossRef]
- Eyre, R.; Feltbower, R.G.; Mubwandarikwa, E.; Eden, T.O.; McNally, R.J. Epidemiology of bone tumours in children and young adults. Pediatr. Blood Cancer 2009, 53, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, X.; Lv, Y.; Li, Z. Malignant fibrous histiocytoma of the floor of mouth: A case report and review of the literature. J. Stomatol. Oral Maxillofac. Surg. 2022, 123, e106–e111. [Google Scholar] [CrossRef]
- Rosser, C.J.; Slaton, J.W.; Izawa, J.; Levy, L.B.; Dinney, C.P. Clinical presentation and outcome of high-grade urinary bladder leiomyosarcoma in adults. Urology 2003, 61, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Souhami, R.; Tobias, J.S. Cancer and Its Management; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- DeVita, V.T.; Lawrence, T.S.; Rosenberg, S.A. DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008; Volume 2. [Google Scholar]
- Kallen, M.E.; Hornick, J.L. The 2020 WHO Classification: What’s new in soft tissue tumor pathology? Am. J. Surg. Pathol. 2021, 45, e1–e23. [Google Scholar] [CrossRef]
- Wilky, B.A. Immune checkpoint inhibitors: The linchpins of modern immunotherapy. Immunol. Rev. 2019, 290, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Chouliaras, K.; Senehi, R.; Ethun, C.G.; Poultsides, G.; Tran, T.; Grignol, V.; Gamblin, T.C.; Roggin, K.K.; Tseng, J.; Fields, R.C.; et al. Recurrence patterns after resection of retroperitoneal sarcomas: An eight-institution study from the US Sarcoma Collaborative. J. Surg. Oncol. 2019, 120, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Gortzak, E.; Azzarelli, A.; Buesa, J.; Bramwell, V.; van Coevorden, F.; van Geel, A.; Ezzat, A.; Santoro, A.; Oosterhuis, J.; van Glabbeke, M.; et al. A randomised phase II study on neo-adjuvant chemotherapy for ‘high-risk’ adult soft-tissue sarcoma. Eur. J. Cancer 2001, 37, 1096–1103. [Google Scholar] [CrossRef]
- Chouliaras, K.; Senehi, R.; Ethun, C.G.; Poultsides, G.; Grignol, V.; Clarke, C.N.; Roggin, K.K.; Fields, R.C.; Schwartz, P.B.; Ronnekleiv-Kelly, S.M.; et al. Role of radiation therapy for retroperitoneal sarcomas: An eight-institution study from the US Sarcoma Collaborative. J. Surg. Oncol. 2019, 120, 1227–1234. [Google Scholar] [CrossRef]
- Gartland, A.; Pasello, M.; Lézot, F.; Lamoureux, F. Editorial: New therapies in the treatment of sarcomas. Front. Endocrinol. 2023, 14, 1137736. [Google Scholar] [CrossRef] [PubMed]
- Koelsche, C.; Schrimpf, D.; Stichel, D.; Sill, M.; Sahm, F.; Reuss, D.E.; Blattner, M.; Worst, B.; Heilig, C.E.; Beck, K.; et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 2021, 12, 498. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Barretina, J.; Maki, R.G.; Antonescu, C.R.; Singer, S.; Ladanyi, M. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer 2011, 11, 541–557. [Google Scholar] [CrossRef]
- Birdi, H.K.; Jirovec, A.; Cortés-Kaplan, S.; Werier, J.; Nessim, C.; Diallo, J.-S.; Ardolino, M. Immunotherapy for sarcomas: New frontiers and unveiled opportunities. J. Immunother. Cancer 2021, 9, e001580. [Google Scholar] [CrossRef]
- Petitprez, F.; de Reyniès, A.; Keung, E.Z.; Chen, T.W.-W.; Sun, C.-M.; Calderaro, J.; Jeng, Y.-M.; Hsiao, L.-P.; Lacroix, L.; Bougoüin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Vivarelli, S.; Candido, S.; Caruso, G.; Falzone, L.; Libra, M. Patient-Derived Tumor Organoids for Drug Repositioning in Cancer Care: A Promising Approach in the Era of Tailored Treatment. Cancers 2020, 12, 3636. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.D.; Sivakumar, H.; Erali, R.A.; Wajih, N.; Li, W.; Shen, P.; Levine, E.A.; Miller, K.E.; Skardal, A.; Votanopoulos, K.I. Patient-Specific Sarcoma Organoids for Personalized Translational Research: Unification of the Operating Room with Rare Cancer Research and Clinical Implications. Ann. Surg. Oncol. 2022, 29, 7354–7367. [Google Scholar] [CrossRef]
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. Npj Precis. Oncol. 2021, 5, 30. [Google Scholar] [CrossRef]
- Smith, A.P.; Dueber, J.C.; Allison, D.B. Seminars in Diagnostic Pathology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 187–198. [Google Scholar]
- Singer, S.; Demetri, G.D.; Baldini, E.H.; Fletcher, C.D. Management of soft-tissue sarcomas: An overview and update. Lancet Oncol. 2000, 1, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Herzog, M.; Pliego-Mendieta, A.; Bühler, M.M.; Harnisch, K.J.; Haberecker, M.; Arnold, F.; Planas-Paz, L.; Pauli, C. Addressing Modern Diagnostic Pathology for Patient-Derived Soft Tissue Sarcosphere Models in the Era of Functional Precision Oncology. Lab. Investig. 2023, 103, 100039. [Google Scholar] [CrossRef] [PubMed]
- Bangerter, J.L.; Harnisch, K.J.; Chen, Y.; Hagedorn, C.; Planas-Paz, L.; Pauli, C. Establishment, characterization and functional testing of two novel ex vivo extraskeletal myxoid chondrosarcoma (EMC) cell models. Hum. Cell 2022, 36, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Boulay, G.; Cironi, L.; Garcia, S.P.; Rengarajan, S.; Xing, Y.-H.; Lee, L.; E Awad, M.; Naigles, B.; Iyer, S.; Broye, L.C.; et al. The chromatin landscape of primary synovial sarcoma organoids is linked to specific epigenetic mechanisms and dependencies. Life Sci. Alliance 2020, 4, e202000808. [Google Scholar] [CrossRef]
- Wakamatsu, T.; Ogawa, H.; Yoshida, K.; Matsuoka, Y.; Shizuma, K.; Imura, Y.; Tamiya, H.; Nakai, S.; Yagi, T.; Nagata, S.; et al. Establishment of Organoids From Human Epithelioid Sarcoma With the Air-Liquid Interface Organoid Cultures. Front. Oncol. 2022, 12, 893592. [Google Scholar] [CrossRef] [PubMed]
- Meister, M.T.; Groot Koerkamp, M.J.; de Souza, T.; Breunis, W.B.; Frazer-Mendelewska, E.; Brok, M.; DeMartino, J.; Manders, F.; Calandrini, C.; Kerstens, H.H.; et al. Mesenchymal tumor organoid models recapitulate rhabdomyosarcoma subtypes. EMBO Mol. Med. 2022, 14, e16001. [Google Scholar] [CrossRef]
- Komatsu, A.; Matsumoto, K.; Yoshimatsu, Y.; Sin, Y.; Kubota, A.; Saito, T.; Mizumoto, A.; Ohashi, S.; Muto, M.; Noguchi, R.; et al. The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine. Cells 2021, 10, 2613. [Google Scholar] [CrossRef] [PubMed]
- Maloney, E.; Clark, C.; Sivakumar, H.; Yoo, K.; Aleman, J.; Rajan, S.A.P.; Forsythe, S.; Mazzocchi, A.; Laxton, A.W.; Tatter, S.B.; et al. Immersion Bioprinting of Tumor Organoids in Multi-Well Plates for Increasing Chemotherapy Screening Throughput. Micromachines 2020, 11, 208. [Google Scholar] [CrossRef]
- MEMd, C.; Zaidi, S.; Scoazec, J.; Droit, R.; Lim, W.C.; Marchais, A.; Salmon, J.; Cherkaoui, S.; Morscher, R.; Laurent, A.; et al. Pediatric Patient-Derived-Xenograft Development in MAPPYACTS–International Pediatric Cancer Precision Medicine Trial in Relapsed and Refractory Tumors. 2023. Available online: https://europepmc.org/article/ppr/ppr620267 (accessed on 16 August 2023).
- Xu, H.; Zheng, H.; Zhang, Q.; Song, H.; Wang, Q.; Xiao, J.; Dong, Y.; Shen, Z.; Wang, S.; Wu, S.; et al. A Multicentre Clinical Study of Sarcoma Personalised Treatment Using Patient-Derived Tumour Xenografts. Clin. Oncol. 2023, 35, e48–e59. [Google Scholar] [CrossRef] [PubMed]
- Colella, G.; Fazioli, F.; Gallo, M.; De Chiara, A.; Apice, G.; Ruosi, C.; Cimmino, A.; de Nigris, F. Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine. Int. J. Mol. Sci. 2018, 19, 615. [Google Scholar] [CrossRef]
- Nami, B.; Wang, Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Cortesi, M.; Zamagni, A.; Arienti, C.; Pignatta, S.; Tesei, A. Modeling neoplastic disease with spheroids and organoids. J. Hematol. Oncol. 2020, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Brunson, D.; Tang, Q.; Do, D.; Iftimia, N.A.; Moore, J.C.; Hayes, M.N.; Welker, A.M.; Garcia, E.G.; Dubash, T.D.; et al. Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell 2019, 177, 1903–1914.e14. [Google Scholar] [CrossRef] [PubMed]
- Bruna, A.; Rueda, O.M.; Greenwood, W.; Batra, A.S.; Callari, M.; Batra, R.N.; Pogrebniak, K.; Sandoval, J.; Cassidy, J.W.; Tufegdzic-Vidakovic, A.; et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 2016, 167, 260–274.e22. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.; Federico, S.M.; Chen, X.; Shelat, A.A.; Bradley, C.; Gordon, B.; Karlstrom, A.; Twarog, N.R.; Clay, M.R.; Bahrami, A.; et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017, 549, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Woo, X.Y.; PDXNET Consortium; Giordano, J.; Srivastava, A.; Zhao, Z.-M.; Lloyd, M.W.; de Bruijn, R.; Suh, Y.-S.; Patidar, R.; Chen, L.; et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat. Genet. 2021, 53, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Xu, K.; Mercer, K.S.; Boop, F.; Klimo, P.; DeCupyere, M.; Grenet, J.; Robinson, S.; Dunphy, P.; Baker, S.J.; et al. Patient-derived orthotopic xenografts of pediatric brain tumors: A St. Jude resource. Acta Neuropathol. 2020, 140, 209–225. [Google Scholar] [CrossRef]
- Petljak, M.; Alexandrov, L.B.; Brammeld, J.S.; Price, S.; Wedge, D.C.; Grossmann, S.; Dawson, K.J.; Ju, Y.S.; Iorio, F.; Tubio, J.M.; et al. Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell 2019, 176, 1282–1294.e20. [Google Scholar] [CrossRef]
- Pompili, L.; Porru, M.; Caruso, C.; Biroccio, A.; Leonetti, C. Patient-derived xenografts: A relevant preclinical model for drug development. J. Exp. Clin. Cancer Res. 2016, 35, 189. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef]
- Suzuki, R.; Wakamatsu, T.; Matsuoka, Y.; Takami, H.; Nakai, S.; Tamiya, H.; Kakunaga, S.; Yagi, T.; Yoshida, K.-I.; Imura, Y.; et al. Genetic characterization of a novel organoid from human malignant giant-cell tumor. J. Bone Oncol. 2023, 41, 100486. [Google Scholar] [CrossRef]
- Hinson, A.R.P.; Jones, R.; Crose, L.E.S.; Belyea, B.C.; Barr, F.G.; Linardic, C.M. Human Rhabdomyosarcoma Cell Lines for Rhabdomyosarcoma Research: Utility and Pitfalls. Front. Oncol. 2013, 3, 183. [Google Scholar] [CrossRef] [PubMed]
- Vibert, J.; Watson, S. The Molecular Biology of Soft Tissue Sarcomas: Current Knowledge and Future Perspectives. Cancers 2022, 14, 2548. [Google Scholar] [CrossRef]
- Lamhamedi-Cherradi, S.-E.; Maitituoheti, M.; Menegaz, B.A.; Krishnan, S.; Vetter, A.M.; Camacho, P.; Wu, C.-C.; Beird, H.C.; Porter, R.W.; Ingram, D.R.; et al. The androgen receptor is a therapeutic target in desmoplastic small round cell sarcoma. Nat. Commun. 2022, 13, 3057. [Google Scholar] [CrossRef]
- Fujii, M.; Sato, T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 2020, 20, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hu, H.; Kung, H.; Zou, R.; Dai, Y.; Hu, Y.; Wang, T.; Lv, T.; Yu, J.; Li, F. Organoids: The current status and biomedical applications. Medcomm 2023, 4, e274. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Fdez, A.; Sharma, A.K.; Tiriac, H.; Sicklick, J.K. Patient-Derived Sarcoma Organoids Offer a Novel Platform for Personalized Precision Medicine. Ann. Surg. Oncol. 2022, 29, 7239–7241. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef]
- Pankova, V.; Thway, K.; Jones, R.L.; Huang, P.H. The Extracellular Matrix in Soft Tissue Sarcomas: Pathobiology and Cellular Signalling. Front. Cell Dev. Biol. 2021, 9, 763640. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, M.; Kliffen, M.; Krestin, G.P.; Van Dijke, C.F. Soft tissue sarcomas at a glance: Clinical, histological, and MR imaging features of malignant extremity soft tissue tumors. Eur. Radiol. 2009, 19, 1499–1511. [Google Scholar] [CrossRef] [PubMed]
- Molina, E.R.; Chim, L.K.; Barrios, S.; Ludwig, J.A.; Mikos, A.G. Modeling the Tumor Microenvironment and Pathogenic Signaling in Bone Sarcoma. Tissue Eng. Part B Rev. 2020, 26, 249–271. [Google Scholar] [CrossRef]
- Rae, C.; Amato, F.; Braconi, C. Patient-Derived Organoids as a Model for Cancer Drug Discovery. Int. J. Mol. Sci. 2021, 22, 3483. [Google Scholar] [CrossRef]
- Maurer, L.M.; Daley, J.D.; Mukherjee, E.; Venier, R.E.; Julian, C.M.; Bailey, N.G.; Jacobs, M.F.; Kumar-Sinha, C.; Raphael, H.; Periyapatna, N.; et al. BRCA1-Associated RING Domain-1 (BARD1) Loss and GBP1 Expression Enhance Sensitivity to DNA Damage in Ewing Sarcoma. Cancer Res. Commun. 2022, 2, 220–232. [Google Scholar] [CrossRef]
- He, A.; Huang, Y.; Cheng, W.; Zhang, D.; He, W.; Bai, Y.; Gu, C.; Ma, Z.; He, Z.; Si, G.; et al. Organoid culture system for patient-derived lung metastatic osteosarcoma. Med. Oncol. 2020, 37, 105. [Google Scholar] [CrossRef]
- Ashley, J.D.; Quinlan, C.J.; Schroeder, V.A.; Suckow, M.A.; Pizzuti, V.J.; Kiziltepe, T.; Bilgicer, B. Dual Carfilzomib and Doxorubicin–Loaded Liposomal Nanoparticles for Synergistic Efficacy in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 1452–1459. [Google Scholar] [CrossRef]
- Schroeder, M.A.; Fiala, M.A.; Huselton, E.; Cardone, M.H.; Jaeger, S.; Jean, S.R.; Shea, K.; Ghobadi, A.; Wildes, T.; Stockerl-Goldstein, K.E.; et al. A Phase I/II Trial of Carfilzomib, Pegylated Liposomal Doxorubicin, and Dexamethasone for the Treatment of Relapsed/Refractory Multiple MyelomaKDD for RRMM. Clin. Cancer Res. 2019, 25, 3776–3783. [Google Scholar] [CrossRef] [PubMed]
- Gaebler, M.; Silvestri, A.; Reichardt, P.; Wardelmann, E.; Gambara, G.; Haybaeck, J.; Stroebel, P.; Niethard, M.; Erdmann, G.; Regenbrecht, C.R. Abstract 469: Patient-derived sarcoma models: First results from the SARQMA study. Cancer Res. 2019, 79, 469. [Google Scholar] [CrossRef]
- Johansson, S. Patient-derived Organoid Culture for 3D Culture of Colorectal Cancer, Renal Cancer and Osteosarcoma. 2019. Available online: https://www.diva-portal.org/smash/get/diva2:1344438/FULLTEXT01.pdf (accessed on 16 August 2023).
- Veys, C.; Benmoussa, A.; Contentin, R.; Duchemin, A.; Brotin, E.; Lafont, J.E.; Saintigny, Y.; Poulain, L.; Denoyelle, C.; Demoor, M.; et al. Tumor Suppressive Role of miR-342-5p in Human Chondrosarcoma Cells and 3D Organoids. Int. J. Mol. Sci. 2021, 22, 5590. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Zhao, L. Recent advances and application of PD-1 blockade in sarcoma. OncoTargets Ther. 2019, 12, 6887–6896. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Mahoney, M.R.; Van Tine, B.A.; Atkins, J.; Milhem, M.M.; Jahagirdar, B.N.; Antonescu, C.R.; Horvath, E.; Tap, W.D.; Schwartz, G.K.; et al. Nivolumab with or without ipilimumab treatment for metastatic sarcoma (Alliance A091401): Two open-label, non-comparative, randomised, phase 2 trials. Lancet Oncol. 2018, 19, 416–426. [Google Scholar] [CrossRef]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef]
- Murciano-Goroff, Y.R.; Warner, A.B.; Wolchok, J.D. The future of cancer immunotherapy: Microenvironment-targeting combinations. Cell Res. 2020, 30, 507–519. [Google Scholar] [CrossRef] [PubMed]
| Cell Category | Example of Cell Type | Contribution to Cellular Heterogeneity |
|---|---|---|
| Differentiated cells |
|
|
| Cancer stem cells |
| |
| Stromal cells |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Tan, S.; Guo, L. Patient-Derived Organoids as a Promising Tool for Multimodal Management of Sarcomas. Cancers 2023, 15, 4339. https://doi.org/10.3390/cancers15174339
Xu S, Tan S, Guo L. Patient-Derived Organoids as a Promising Tool for Multimodal Management of Sarcomas. Cancers. 2023; 15(17):4339. https://doi.org/10.3390/cancers15174339
Chicago/Turabian StyleXu, Songfeng, ShihJye Tan, and Ling Guo. 2023. "Patient-Derived Organoids as a Promising Tool for Multimodal Management of Sarcomas" Cancers 15, no. 17: 4339. https://doi.org/10.3390/cancers15174339
APA StyleXu, S., Tan, S., & Guo, L. (2023). Patient-Derived Organoids as a Promising Tool for Multimodal Management of Sarcomas. Cancers, 15(17), 4339. https://doi.org/10.3390/cancers15174339