Zinc Ions Modulate YY1 Activity: Relevance in Carcinogenesis
Abstract
Simple Summary
Abstract
1. Introduction
2. Zinc Homeostasis
3. Cellular Zinc Level—What Is the True Meaning?
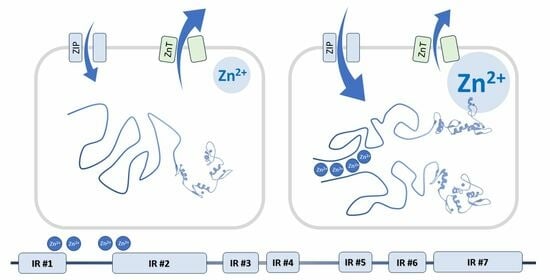
4. Zinc Ions Affect the Structure and Function of Yin Yang 1
5. YY1—A Protein of Numerous Activities and Binding Partners
6. Regions of YY1 Molecule Responsible for Interactions with Its Molecular Partners
| Partner Protein | Partner Fragment | YY1 Fragment | Additional Partner | Method | Reference |
|---|---|---|---|---|---|
| AMRP (α-2-macroglobulin receptor-associated protein) | FL | 1–414 | CP | [145] | |
| AAMDC (Mth938 domain-containing protein) | FL | 1–414 | CP | [145] | |
| AGO2 (Argonaute 2) | FL | 1–414 | coIP | [146] | |
| AKT (PKB) | 1–108 | 201–226 | CP, coIP | [113,132] | |
| ALOXE3 | FL | 1–414 | 2H | [147] | |
| ALR | FL | 1–414 | CP, coIP | [148] | |
| AP2 | 166–437 | 1–330 | CP, coIP | [149,150] | |
| AR (androgen receptor) | 556–919 | 331–414 | CP | [142] | |
| ARB1 (β-arrestin) | FL | 1–414 | coIP | [151] | |
| ATF2 | FL | 1–414 | CP | [136] | |
| ATF6 | 273–373 | 261–333 | CP | [152,153] | |
| ATFa1 | FL | 1–414 | CP | [136] | |
| ATFa2 | 334–399 | 224–330 | CP | [136] | |
| ATFa3 | FL | 1–414 | CP | [136] | |
| ATXN2L | FL | 1–414 | CP | [145] | |
| Aurora A (AURKA) | FL | 1–414 | CP | [154] | |
| BAP1 | 642–686 | 331–414 | HCF-1 | CP, coIP | [143] |
| BAX | FL | 1–414 | CP | [155] | |
| BCCIP | 1–258 | 213–270 | CP, coIP | [156] | |
| BCL6 | FL | 1–414 | CP | [157] | |
| BMI1 | FL | 1–414 | CP | [158,159,160] | |
| BRD1 | FL | 1–414 | CP | [161] | |
| BRD2 | FL | 1–414 | CP | [161] | |
| BRD4 | FL | 1–414 | CP | [162] | |
| CAND-1 | FL | 1–414 | coIP | [163] | |
| CAPB | FL | 1–414 | CP | [164] | |
| CBP | 451–721 | 154–199; 296–399 | CP, coIP | [127,165] | |
| CBX4 | FL | 1–414 | CP | [145] | |
| CCNT1 | FL | 1–414 | CP | [145] | |
| CCNT2 | FL | 1–414 | CP | [145] | |
| CDK9 | FL | 1–414 | CP | [145] | |
| C/EBP β | FL | 1–414 | CP | [166] | |
| CEP76 | FL | 1–414 | 2H | [147] | |
| CHD8 | FL | 1–414 | CP | [164] | |
| CIC | FL | 1–414 | CP | [167] | |
| CKIδ | FL | 1–414 | CP | [145] | |
| cortactin | FL | 1–414 | CP | [145] | |
| CP2 | 308–368 | 294–320 | HDAC1 | CP | [138,168,169] |
| CPSF1 | FL | 1–414 | CP | [145] | |
| CPSF6 | FL | 1–414 | CP | [145] | |
| CPSF7 | FL | 1–414 | CP | [145] | |
| CREB (ATF) | FL | 282–414 | CP, 2H | [136,170] | |
| CRKL | FL | 1–414 | 2H | [171] | |
| CTCF | 1–583 | 313–414 | CP, coIP | [128] | |
| CUL3 (cullin 3) | FL | 1–414 | coIP | [172] | |
| Cyclophilin A | FL | 1–414 | 2H | [173] | |
| CYSRT1 | FL | 1–414 | 2H | [147] | |
| DCAF13 | FL | 1–414 | CP | [145] | |
| DDX3X | FL | 1–414 | coIP | [174] | |
| DDX5 | FL | 1–414 | coIP | [174] | |
| DDX6 | FL | 1–414 | CP | [145] | |
| DDX42 | FL | 1–414 | CP | [145] | |
| DDX56 | FL | 1–414 | CP | [145] | |
| Dot1L | FL | 1–414 | coIP | [175] | |
| DNAPK | FL | 1–414 | CP | [112] | |
| DNMT3L | FL | 1–414 | PA | [176] | |
| DRBP76 | FL | 261–333 | CP, coIP | [177] | |
| E1A | 15–35; 140–188 | 54–260; 332–414 | p300 | CP, CS, FWB | [125,136,178,179,180,181] |
| EED | 502–535 | 250–414 | 2H, CP, coIP | [4] | |
| EIF5A | FL | 1–414 | CP | [145] | |
| ESM1 | FL | 1–414 | 2H | [147] | |
| EVI1 | FL | 1–414 | CP | [182] | |
| EZH2 | 493–519 | 201–226 | SPR, CP, coIP | [4,113,183,184] | |
| FAM76A | FL | 1–414 | CP | [145] | |
| FAM67B | FL | 1–414 | CP | [145] | |
| FAM98A | FL | 1–414 | CP | [145] | |
| FBW7 | FL | 1–414 | coIP | [185] | |
| FHL2 | FL | 1–414 | 2H | [147] | |
| FIP1 | FL | 1–414 | CP | [145] | |
| FKBP12 | FL | 1–414 | 2H | [173] | |
| FKBP25 | 1–90 | 300–333 | CP, coIP | [139] | |
| c-Fos | FL | 1–414 | coIP | [186] | |
| FOX-B1 | FL | 1–414 | CP | [187] | |
| FOX-J2 | FL | 1–414 | CP | [187] | |
| FOX-L1 | FL | 1–414 | CP | [187] | |
| FOX-N1 | FL | 1–414 | CP | [187] | |
| GMCL1 | FL | 1–414 | 2H | [147] | |
| GON4L | 611–1364 | 1–414 | CP, coIP | [188,189] | |
| granulin (GRN) | FL | 1–414 | 2H | [171] | |
| H4FA | FL | 142–260 | CP | [190] | |
| HCF-1 | FL | 142–260 | BAP1 | CP | [143] |
| HCVGP1 HCV core fusion protein | FL | 1–414 | CP, coIP | [130] | |
| HDAC1 | FL | 170–200; 261–333 | BA, CP, coIP | [126,191,192,193] | |
| HDAC2 (RPD3) | FL | 170–200; 261–333 | DNA | BA, CP, coIP | [126,191,192,194,195,196,197] |
| HDAC3 | 373–428 | 170–200; 261–333 | p300 | BA, CP, coIP | [126,181,191,198,199] |
| HDAC3a | FL | 170–200; 261–333 | CP | [191] | |
| HDAC4 | FL | 1–414 | CP | [200,201,202] | |
| HDAC5 | FL | 1–414 | coIP | [203,204] | |
| HEXIM1 | FL | 1–414 | CP | [145] | |
| HMGB1B | FL | 1–414 | 2H | [205] | |
| HOXA11 | 229–314 | 205–226 | HDAC2 | CP, coIP | [135] |
| HSPA4 | FL | 1–414 | CP | [112] | |
| IL-10 | FL | 1–414 | 2H | [147] | |
| INO80 (KIAA1259) | 273–521 | 1–414 | CP, coIP | [112,145,164,206,207] | |
| INO80B | FL | 1–414 | CP | [206] | |
| INO80C | FL | 1–414 | CP | [164,206] | |
| INO80D | FL | 1–414 | CP | [206] | |
| INO80E | FL | 1–414 | CP | [145,206] | |
| INO80F | FL | 1–414 | CP | [145,206] | |
| INO80G (NFRκB) | FL | 1–414 | CP | [145,164,206] | |
| INO80H (RUVBL1/TIP49A) | FL | 1–414 | CP, coIP | [112,145,164,206] | |
| INO80J (RUVBL2/TIP49B) | FL | 1–414 | CP, coIP | [110,112,164,174,206] | |
| INO80K (ACTL6A) | FL | 1–414 | CP, coIP | [112,164,206] | |
| INO80M (ACTR5) | FL | 1–414 | CP | [112,155,164,206] | |
| INO80N (ACTR8) | FL | 1–414 | CP, coIP | [112,145,164,174,206] | |
| INO80Q (MCRS1) | FL | 1–414 | CP | [145,206] | |
| INO80R (UCHL5) | FL | 1–414 | CP | [145,164,206] | |
| ITFG-1 | FL | 1–414 | CP | [208] | |
| c-Jun | FL | 1–414 | coIP | [209] | |
| JunB | FL | 1–414 | coIP | [210] | |
| JunD | FL | 1–414 | coIP | [210] | |
| KP1-3 | FL | 1–414 | 2H | [147] | |
| KAP1-5 | FL | 1–414 | 2H | [147] | |
| KAP2-3 | FL | 1–414 | 2H | [147] | |
| KAP2-4 | FL | 1–414 | 2H | [147] | |
| KAP4-2 | FL | 1–414 | 2H | [147] | |
| KAP4-5 | FL | 1–414 | 2H | [147] | |
| KAP5-6 | FL | 1–414 | 2H | [147] | |
| KAP9-3 | FL | 1–414 | 2H | [147] | |
| KAP9-8 | FL | 1–414 | 2H | [147] | |
| KAP10-5 | FL | 1–414 | 2H | [147] | |
| KAP10-8 | FL | 1–414 | 2H | [147] | |
| KAP10-9 | FL | 1–414 | 2H | [147] | |
| KAP12-2 | FL | 1–414 | 2H | [147] | |
| KAP12-3 | FL | 1–414 | 2H | [147] | |
| KAP17-1 | FL | 1–414 | 2H | [147] | |
| Ki-67 | FL | 1–414 | CP | [211] | |
| Ku70 | FL | 1–414 | coIP | [212] | |
| Ku80 | FL | 1–414 | coIP | [212] | |
| L3MBTL2 | 170–625 | 199–228 | SPR | [131] | |
| LHX3 | FL | 1–414 | 2H | [147] | |
| LHX4 | FL | 1–414 | 2H | [147] | |
| LYAR | FL | 1–414 | CP | [145] | |
| MAX | FL | 1–414 | coIP | [165] | |
| MBTD1 | 130–566 | 199–228 | SPR, CC | [131] | |
| MDFI | FL | 1–414 | 2H | [147] | |
| MDM2 (HDM2) | 150–290 | 200–295 | p53 | CP, coIP | [5,113] |
| MEPCE | FL | 1–414 | CP | [145] | |
| MeCP2 | 202–255 | 1–414 | CP, coIP | [213] | |
| MED20 | FL | 1–414 | 2H | [147] | |
| MEN1 (menin) | FL | 1–414 | coIP | [175] | |
| METTL17 | FL | 1–414 | CP | [145] | |
| MFAP1 | FL | 1–414 | CP | [145] | |
| MLL5 | FL | 1–414 | CP | [214] | |
| MMTAG2 | FL | 1–414 | CP | [145] | |
| MSL2 | FL | 1–414 | CP | [164] | |
| MTA2 | FL | 1–414 | CP | [215] | |
| mTOR | FL | 1–414 | Raptor | coIP | [134] |
| c-Myc | 262–439 | 154–199; 296–399 | 2H, CP | [129,216] | |
| n-Myc | FL | 1–414 | coIP | [217] | |
| NCAP (SARS-CoV-2) | FL | 1–414 | CP | [218] | |
| NEDD4 | FL | 1–414 | BA, coIP | [219] | |
| NEDD4L | FL | 1–414 | BA, coIP | [219] | |
| NFκB | FL | 1–414 | 2H | [171] | |
| NIRF | FL | 1–414 | coIP | [220] | |
| Notch1 | 1821–2095 | 295–414 | CP, coIP | [221] | |
| NR1H2 (nuclear receptor 1H2) | FL | 1–414 | 2H | [171] | |
| NRF2 | FL | 1–414 | DNA | coIP | [222] |
| NSRP1 | FL | 1–414 | CP | [145] | |
| Nucleophosmin | 127–144 | 155–198 | CP, coIP | [130] | |
| NUDT21 | FL | 1–414 | CP | [145] | |
| NUFP2 | FL | 1–414 | CP | [145] | |
| p14ARF (CDKN2A, INK4) | FL | 116–224 | CP, coIP | [5] | |
| p27 (CDKN1B, KIP1) | FL | 1–414 | CP | [223] | |
| p53 | 290–393 | 142–224; 331–414 | CP | [5,144,223,224,225] | |
| p300 | 1572–2370 | 170–200; 397–414 | HDAC3, c-Myc, Max | BA, 2H, CP, coIP | [125,126,170,181,199] |
| PARP1 (ADPRT) | 337–573 | 1–414 | BA, CP, coIP | [226,227,228] | |
| PCAF | FL | 170–200; 261–333 | BA | [126] | |
| PCGF2 (rnf110) | FL | 1–414 | CP | [160] | |
| PGC-1α | 400–797 | 350–380 | CP, coIP | [134] | |
| PIASγ | 100–202 | 331–414 | CP, coIP | [144] | |
| PIRH2 | FL | 1–414 | CP | [229] | |
| PIPK | FL | 1–414 | CP | [145] | |
| PKHF2 | FL | 1–414 | 2H | [147] | |
| PLEKH4 | FL | 1–414 | CP | [230] | |
| POGZ | FL | 1–414 | CP | [164] | |
| POP1 | FL | 1–414 | CP | [145] | |
| PPIL4 | FL | 1–414 | CP | [145] | |
| PPP1R10 | FL | 1–414 | CP | [145] | |
| PR38A | FL | 1–414 | CP | [145] | |
| PR40A | FL | 1–414 | CP | [145] | |
| PRMT1 | FL | 1–414 | coIP | [153,177] | |
| PRP4 | FL | 1–414 | CP | [145] | |
| PSP1 | FL | 1–414 | CP | [145] | |
| Raf-1 | FL | 1–414 | 2H | [171] | |
| Raptor | FL | 203–235 | coIP | [134] | |
| RB1 | FL | 1–414 | coIP | [3,231] | |
| RBM15B | FL | 1–414 | CP | [145] | |
| RBM25 | FL | 1–414 | CP | [145] | |
| RCL1 | FL | 1–414 | CP | [145] | |
| Rel-B | FL | 1–414 | coIP | [232] | |
| RhoGAP | FL | 1–414 | CP | [233] | |
| RhoGEF | FL | 1–414 | CP | [233] | |
| Ring1 | FL | 1–200; 343–414 | CP | [160] | |
| RNAP II (large subunit) | FL | 1–414 | CP | [170] | |
| RNF2 | FL | 1–414 | CP | [158,160] | |
| RNF144 | FL | 1–414 | CP | [234] | |
| RPL23A | FL | 1–414 | CP | [145] | |
| RPS19 | FL | 1–414 | CP | [145] | |
| RPSA | FL | 1–414 | CP | [145] | |
| RYBP (YEAF1) | 42–118; 207–227 | 272–333 | GABPB1 | CP, coIP, 2H | [141,235] |
| SAP30 | 129–220 | 295–414 | HDAC1 | CP, 2H | [192] |
| SART1 | FL | 1–414 | CP | [145] | |
| SF3A2 | FL | 1–414 | 2H | [147] | |
| SF3B4 | FL | 1–414 | CP | [145] | |
| SFMBT2 | 44–447 | 199–228 | SPR | [131] | |
| SLC39A7 | FL | 1–414 | 2H | [171] | |
| Smad1 | 12–136 | 1–200 | CP, coIP | [236,237] | |
| Smad2 | 10–176 | 1–414 | CP, coIP | [236,238] | |
| Smad3 | 10–136 | 1–414 | CP, coIP | [236,238] | |
| Smad4 (Co-Smad) | 18–142 | 1–200 | CP, coIP | [236,237] | |
| Smad7 | 261–426 | 1–414 | CP, coIP | [239] | |
| SMURF2 | FL | 248–252 | CP | [240] | |
| SNIP1 | FL | 1–414 | CP | [145] | |
| SMARCAD1 | FL | 1–414 | CP | [241] | |
| Sp1 | 620–778 | 260–331 | DNA with SP1 site | CP, coIP | [140,242,243,244,245] |
| Sp3 | FL | 1–414 | coIP | [245] | |
| Sp100 | FL | 1–414 | CP | [145] | |
| SPRTN | FL | 1–414 | CP | [246] | |
| SPRY1 | FL | 1–414 | 2H | [171] | |
| SREBP-1a | 321–490 | 256–354 | CP | [243] | |
| SUZ-12 | FL | 1–414 | CP | [158] | |
| TACO1 | FL | 1–414 | CP | [145] | |
| TAF2 | FL | 1–414 | CP | [145] | |
| TAF7 (TAFII55) | 1–117 | 154–199; 296–399 | CP | [127,247] | |
| Tat (HIV-1) | FL | 1–414 | CP | [248] | |
| TBP | FL | 154–199; 296–399 | CP | [127,129,170] | |
| TCF3 | FL | 1–414 | CP | [206] | |
| TESK1 | FL | 1–414 | 2H | [171] | |
| TF2B | FL | 154–199; 296–414 | CP | [127,129,249] | |
| TF2I | FL | 1–414 | CP | [145] | |
| TOP1 | FL | 1–414 | CP | [145] | |
| TOX4 | FL | 1–414 | CP | [145] | |
| TRABID | FL | 1–414 | CP | [145] | |
| TRF-1 | FL | 1–414 | CP | [250] | |
| TRF-2 | FL | 1–414 | CP | [250] | |
| TRIM42 | FL | 1–414 | 2H | [147] | |
| TRIM67 | FL | 1–414 | coIP | [251] | |
| TRIP12 | FL | 1–414 | coIP | [163] | |
| TWIST-1 | FL | 1–414 | CP | [225] | |
| Ubiquitin B (UBB) | FL | 1–414 | coIP | [163] | |
| Ubc9 | FL | 1–414 | BA, CP, coIP | [144] | |
| USP7 | FL | 1–414 | CP | [252] | |
| USP8 | FL | 1–414 | coIP | [163] | |
| VWC2 | FL | 1–414 | 2H | [147] | |
| WDR82 | FL | 1–414 | CP | [145] | |
| WIZ | FL | 1–414 | CP | [164] | |
| XAGE1B | FL | 1–414 | 2H | [147] | |
| XAGE1E | FL | 1–414 | 2H | [147] | |
| YAF2 | FL | 271–333 | 2H | [195] | |
| YY1 | 65–80; 201–226 | 65–80; 201–226 | SEC, BiFC, EM, SPR, CP | [6,9,110,111,113] | |
| YY1AP | 1–260; 475–608 | 1–414 | 2H, CP, coIP | [253] | |
| YY2 | FL | 1–414 | CP | [164] | |
| ZHX1 | FL | 1–414 | CP | [164] | |
| ZNF85 | FL | 1–414 | 2H | [147] | |
| ZNF232 | FL | 1–414 | 2H | [254] | |
| ZNF644 | FL | 1–414 | CP | [164] |
7. YY1 Binding of Zinc Ions May Interfere with the Interactions of Molecular Partners
7.1. EZH2
7.2. AR
7.3. MDM2/p53
7.4. YY1 Dimers in Chromatin Loops
7.5. E1A
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deng, Z.; Cao, P.; Wan, M.M.; Sui, G. Yin Yang 1: A multifaceted protein beyond a transcription factor. Transcription 2010, 1, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Figiel, M.; Gorecki, A. Physical Interaction of Human Yin Yang 1 Protein with DNA. Crit. Rev. Oncog. 2017, 22, 75–97. [Google Scholar] [CrossRef] [PubMed]
- Petkova, V.; Romanowski, M.J.; Sulijoadikusumo, I.; Rohne, D.; Kang, P.; Shenk, T.; Usheva, A. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J. Biol. Chem. 2001, 276, 7932–7936. [Google Scholar] [CrossRef] [PubMed]
- Satijn, D.P.; Hamer, K.M.; den Blaauwen, J.; Otte, A.P. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 2001, 21, 1360–1369. [Google Scholar] [CrossRef]
- Sui, G.; Affarel, B.; Shi, Y.; Brignone, C.; Wall, N.R.; Yin, P.; Donohoe, M.; Luke, M.P.; Calvo, D.; Grossman, S.R.; et al. Yin Yang 1 is a negative regulator of p53. Cell 2004, 117, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588.e1528. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, M.E.; Zhang, X.; McGinnis, L.; Biggers, J.; Li, E.; Shi, Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999, 19, 7237–7244. [Google Scholar] [CrossRef]
- Gabriele, M.; Vulto-van Silfhout, A.T.; Germain, P.L.; Vitriolo, A.; Kumar, R.; Douglas, E.; Haan, E.; Kosaki, K.; Takenouchi, T.; Rauch, A.; et al. YY1 Haploinsufficiency Causes an Intellectual Disability Syndrome Featuring Transcriptional and Chromatin Dysfunction. Am. J. Hum. Genet. 2017, 100, 907–925. [Google Scholar] [CrossRef]
- Figiel, M.; Szubert, F.; Luchinat, E.; Bonarek, P.; Baranowska, A.; Wajda-Nikiel, K.; Wilamowski, M.; Milek, P.; Dziedzicka-Wasylewska, M.; Banci, L.; et al. Zinc controls operator affinity of human transcription factor YY1 by mediating dimerization via its N-terminal region. Biochim. Biophys. Acta Gene Regul. Mech. 2023, 1866, 194905. [Google Scholar] [CrossRef]
- Houbaviy, H.B.; Usheva, A.; Shenk, T.; Burley, S.K. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. USA 1996, 93, 13577–13582. [Google Scholar] [CrossRef]
- Gorecki, A.; Bonarek, P.; Gorka, A.K.; Figiel, M.; Wilamowski, M.; Dziedzicka-Wasylewska, M. Intrinsic disorder of human Yin Yang 1 protein. Proteins 2015, 83, 1284–1296. [Google Scholar] [CrossRef]
- Lioumi, M.; Ferguson, C.A.; Sharpe, P.T.; Freeman, T.; Marenholz, I.; Mischke, D.; Heizmann, C.; Ragoussis, J. Isolation and characterization of human and mouse ZIRTL, a member of the IRT1 family of transporters, mapping within the epidermal differentiation complex. Genomics 1999, 62, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Milon, B.; Dhermy, D.; Pountney, D.; Bourgeois, M.; Beaumont, C. Differential subcellular localization of hZip1 in adherent and non-adherent cells. FEBS Lett. 2001, 507, 241–246. [Google Scholar] [CrossRef]
- Franklin, R.B.; Feng, P.; Milon, B.; Desouki, M.M.; Singh, K.K.; Kajdacsy-Balla, A.; Bagasra, O.; Costello, L.C. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol. Cancer 2005, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Desouki, M.M.; Geradts, J.; Milon, B.; Franklin, R.B.; Costello, L.C. hZip2 and hZip3 zinc transporters are down regulated in human prostate adenocarcinomatous glands. Mol. Cancer 2007, 6, 37. [Google Scholar] [CrossRef]
- Dong, X.; Kong, C.; Zhang, Z.; Liu, X.; Zhan, B.; Chen, Z.; Shi, D. hZIP1 that is down-regulated in clear cell renal cell carcinoma is negatively associated with the malignant potential of the tumor. Urol. Oncol. 2014, 32, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Huo, R.; Zhi, Q.; Zhan, M.; Chen, X.; Hua, Z.C. Increased expression of zinc transporter ZIP4, ZIP11, ZnT1, and ZnT6 predicts poor prognosis in pancreatic cancer. J. Trace Elem. Med. Biol. 2021, 65, 126734. [Google Scholar] [CrossRef] [PubMed]
- Gaither, L.A.; Eide, D.J. Functional expression of the human hZIP2 zinc transporter. J. Biol. Chem. 2000, 275, 5560–5564. [Google Scholar] [CrossRef]
- Dufner-Beattie, J.; Huang, Z.L.; Geiser, J.; Xu, W.; Andrews, G.K. Generation and characterization of mice lacking the zinc uptake transporter ZIP3. Mol. Cell. Biol. 2005, 25, 5607–5615. [Google Scholar] [CrossRef]
- Kelleher, S.L.; Lopez, V.; Lonnerdal, B.; Dufner-Beattie, J.; Andrews, G.K. Zip3 (Slc39a3) functions in zinc reuptake from the alveolar lumen in lactating mammary gland. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R194–R201. [Google Scholar] [CrossRef]
- Costello, L.C.; Zou, J.; Desouki, M.M.; Franklin, R.B. Evidence for changes in RREB-1, ZIP3, and Zinc in the early development of pancreatic adenocarcinoma. J. Gastrointest. Cancer 2012, 43, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Kury, S.; Dreno, B.; Bezieau, S.; Giraudet, S.; Kharfi, M.; Kamoun, R.; Moisan, J.P. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat. Genet. 2002, 31, 239–240. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, Y.; Liu, Z.; Bharadwaj, U.; Wang, H.; Wang, X.; Zhang, S.; Liuzzi, J.P.; Chang, S.M.; Cousins, R.J.; et al. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc. Natl. Acad. Sci. USA 2007, 104, 18636–18641. [Google Scholar] [CrossRef] [PubMed]
- Kambe, T.; Andrews, G.K. Novel proteolytic processing of the ectodomain of the zinc transporter ZIP4 (SLC39A4) during zinc deficiency is inhibited by acrodermatitis enteropathica mutations. Mol. Cell. Biol. 2009, 29, 129–139. [Google Scholar] [CrossRef]
- Wu, D.M.; Liu, T.; Deng, S.H.; Han, R.; Xu, Y. SLC39A4 expression is associated with enhanced cell migration, cisplatin resistance, and poor survival in non-small cell lung cancer. Sci. Rep. 2017, 7, 7211. [Google Scholar] [CrossRef]
- Fan, Q.; Cai, Q.; Li, P.; Wang, W.; Wang, J.; Gerry, E.; Wang, T.L.; Shih, I.M.; Nephew, K.P.; Xu, Y. The novel ZIP4 regulation and its role in ovarian cancer. Oncotarget 2017, 8, 90090–90107. [Google Scholar] [CrossRef]
- Gartmann, L.; Wex, T.; Grungreiff, K.; Reinhold, D.; Kalinski, T.; Malfertheiner, P.; Schutte, K. Expression of zinc transporters ZIP4, ZIP14 and ZnT9 in hepatic carcinogenesis-An immunohistochemical study. J. Trace Elem. Med. Biol. 2018, 49, 35–42. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, Y.M.; Liu, J.; Han, J.; Guo, J.X.; Lu, S.; Huang, X.M.; Yi, P.; Lang, J.Y.; Zhang, P.; et al. Inhibition of ZIP4 reverses epithelial-to-mesenchymal transition and enhances the radiosensitivity in human nasopharyngeal carcinoma cells. Cell Death Dis. 2019, 10, 588. [Google Scholar] [CrossRef]
- Zhang, C.; Sui, D.; Zhang, T.; Hu, J. Molecular Basis of Zinc-Dependent Endocytosis of Human ZIP4 Transceptor. Cell Rep. 2020, 31, 107582. [Google Scholar] [CrossRef]
- Zhu, B.; Yang, C.; Sun, L.; Li, Z.; Li, J.; Hua, Z.C. Expression pattern and prognostic implication of zinc homeostasis-related genes in acute myeloid leukemia. Metallomics 2023, 15, mfad022. [Google Scholar] [CrossRef]
- Wang, F.; Kim, B.E.; Petris, M.J.; Eide, D.J. The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J. Biol. Chem. 2004, 279, 51433–51441. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Morgan, H.E.; Johnson, A.; Hadley, L.J.; Nicholson, R.I. Structure-function analysis of LIV-1, the breast cancer-associated protein that belongs to a new subfamily of zinc transporters. Biochem. J. 2003, 375, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Morikawa, H.; Kamon, H.; Iguchi, M.; Hojyo, S.; Fukada, T.; Yamashita, S.; Kaisho, T.; Akira, S.; Murakami, M.; et al. Toll-like receptor-mediated regulation of zinc homeostasis influences dendritic cell function. Nat. Immunol. 2006, 7, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Hogstrand, C.; Kille, P.; Ackland, M.L.; Hiscox, S.; Taylor, K.M. A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3). Biochem. J. 2013, 455, 229–237. [Google Scholar] [CrossRef]
- Cheng, X.; Wei, L.; Huang, X.; Zheng, J.; Shao, M.; Feng, T.; Li, J.; Han, Y.; Tan, W.; Tan, W.; et al. Solute Carrier Family 39 Member 6 Gene Promotes Aggressiveness of Esophageal Carcinoma Cells by Increasing Intracellular Levels of Zinc, Activating Phosphatidylinositol 3-Kinase Signaling, and Up-regulating Genes That Regulate Metastasis. Gastroenterology 2017, 152, 1985–1997.e12. [Google Scholar] [CrossRef]
- Taylor, K.M.; Morgan, H.E.; Johnson, A.; Nicholson, R.I. Structure-function analysis of HKE4, a member of the new LIV-1 subfamily of zinc transporters. Biochem. J. 2004, 377, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Kirschke, C.P.; Zhang, Y.; Yu, Y.Y. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J. Biol. Chem. 2005, 280, 15456–15463. [Google Scholar] [CrossRef]
- Taylor, K.M.; Vichova, P.; Jordan, N.; Hiscox, S.; Hendley, R.; Nicholson, R.I. ZIP7-mediated intracellular zinc transport contributes to aberrant growth factor signaling in antihormone-resistant breast cancer Cells. Endocrinology 2008, 149, 4912–4920. [Google Scholar] [CrossRef]
- Wei, Y.; Dong, J.; Li, F.; Wei, Z.; Tian, Y. Knockdown of SLC39A7 suppresses cell proliferation, migration and invasion in cervical cancer. EXCLI J. 2017, 16, 1165–1176. [Google Scholar] [CrossRef]
- Begum, N.A.; Kobayashi, M.; Moriwaki, Y.; Matsumoto, M.; Toyoshima, K.; Seya, T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: Identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics 2002, 80, 630–645. [Google Scholar] [CrossRef]
- Liu, M.J.; Bao, S.; Galvez-Peralta, M.; Pyle, C.J.; Rudawsky, A.C.; Pavlovicz, R.E.; Killilea, D.W.; Li, C.; Nebert, D.W.; Wewers, M.D.; et al. ZIP8 regulates host defense through zinc-mediated inhibition of NF-kappaB. Cell Rep. 2013, 3, 386–400. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, W.; Yamazaki, T.; Yamaguchi-Iwai, Y.; Masuda, S.; Nagao, M.; Andrews, G.K.; Kambe, T. SLC39A9 (ZIP9) regulates zinc homeostasis in the secretory pathway: Characterization of the ZIP subfamily I protein in vertebrate cells. Biosci. Biotechnol. Biochem. 2009, 73, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Pang, Y.; Dong, J.; Berg, A.H. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology 2014, 155, 4250–4265. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Converse, A.; Berg, H.A. ZIP9, a novel membrane androgen receptor and zinc transporter protein. Gen. Comp. Endocrinol. 2018, 257, 130–136. [Google Scholar] [CrossRef]
- Deng, H.; Qiao, X.; Xie, T.; Fu, W.; Li, H.; Zhao, Y.; Guo, M.; Feng, Y.; Chen, L.; Zhao, Y.; et al. SLC-30A9 is required for Zn(2+) homeostasis, Zn(2+) mobilization, and mitochondrial health. Proc. Natl. Acad. Sci. USA 2021, 118, e2023909118. [Google Scholar] [CrossRef]
- Ma, T.; Zhao, L.; Zhang, J.; Tang, R.; Wang, X.; Liu, N.; Zhang, Q.; Wang, F.; Li, M.; Shan, Q.; et al. A pair of transporters controls mitochondrial Zn(2+) levels to maintain mitochondrial homeostasis. Protein Cell 2022, 13, 180–202. [Google Scholar] [CrossRef] [PubMed]
- Kagara, N.; Tanaka, N.; Noguchi, S.; Hirano, T. Zinc and its transporter ZIP10 are involved in invasive behavior of breast cancer cells. Cancer Sci. 2007, 98, 692–697. [Google Scholar] [CrossRef]
- Miyai, T.; Hojyo, S.; Ikawa, T.; Kawamura, M.; Irie, T.; Ogura, H.; Hijikata, A.; Bin, B.H.; Yasuda, T.; Kitamura, H.; et al. Zinc transporter SLC39A10/ZIP10 facilitates antiapoptotic signaling during early B-cell development. Proc. Natl. Acad. Sci. USA 2014, 111, 11780–11785. [Google Scholar] [CrossRef]
- Taylor, K.M.; Muraina, I.A.; Brethour, D.; Schmitt-Ulms, G.; Nimmanon, T.; Ziliotto, S.; Kille, P.; Hogstrand, C. Zinc transporter ZIP10 forms a heteromer with ZIP6 which regulates embryonic development and cell migration. Biochem. J. 2016, 473, 2531–2544. [Google Scholar] [CrossRef]
- Landry, G.M.; Furrow, E.; Holmes, H.L.; Hirata, T.; Kato, A.; Williams, P.; Strohmaier, K.; Gallo, C.J.R.; Chang, M.; Pandey, M.K.; et al. Cloning, function, and localization of human, canine, and Drosophila ZIP10 (SLC39A10), a Zn(2+) transporter. Am. J. Physiol. Renal Physiol. 2019, 316, F263–F273. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, A.; Zhang, Z.; Yan, G.; Zhang, F.; Zhang, L.; Shen, X.; Hu, R.; Zhang, Y.; Zhang, K.; et al. Characterization of the GufA subfamily member SLC39A11/Zip11 as a zinc transporter. J. Nutr. Biochem. 2013, 24, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chaffee, K.G.; Parker, A.S.; Sicotte, H.; Petersen, G.M. Zinc transporter genes and urological cancers: Integrated analysis suggests a role for ZIP11 in bladder cancer. Tumour Biol. 2015, 36, 7431–7437. [Google Scholar] [CrossRef] [PubMed]
- Olea-Flores, M.; Kan, J.; Carlson, A.; Syed, S.A.; McCann, C.; Mondal, V.; Szady, C.; Ricker, H.M.; McQueen, A.; Navea, J.G.; et al. ZIP11 Regulates Nuclear Zinc Homeostasis in HeLa Cells and Is Required for Proliferation and Establishment of the Carcinogenic Phenotype. Front. Cell Dev. Biol. 2022, 10, 895433. [Google Scholar] [CrossRef] [PubMed]
- Chowanadisai, W.; Graham, D.M.; Keen, C.L.; Rucker, R.B.; Messerli, M.A. Neurulation and neurite extension require the zinc transporter ZIP12 (slc39a12). Proc. Natl. Acad. Sci. USA 2013, 110, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Oliver, E.; Maratou, K.; Atanur, S.S.; Dubois, O.D.; Cotroneo, E.; Chen, C.N.; Wang, L.; Arce, C.; Chabosseau, P.L.; et al. The zinc transporter ZIP12 regulates the pulmonary vascular response to chronic hypoxia. Nature 2015, 524, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.N.; Strong, M.D.; Chambers, E.; Hart, M.D.; Bettaieb, A.; Clarke, S.L.; Smith, B.J.; Stoecker, B.J.; Lucas, E.A.; Lin, D.; et al. A role for zinc transporter gene SLC39A12 in the nervous system and beyond. Gene 2021, 799, 145824. [Google Scholar] [CrossRef]
- Fukada, T.; Civic, N.; Furuichi, T.; Shimoda, S.; Mishima, K.; Higashiyama, H.; Idaira, Y.; Asada, Y.; Kitamura, H.; Yamasaki, S.; et al. The zinc transporter SLC39A13/ZIP13 is required for connective tissue development; its involvement in BMP/TGF-beta signaling pathways. PLoS ONE 2008, 3, e3642. [Google Scholar] [CrossRef]
- Bin, B.H.; Fukada, T.; Hosaka, T.; Yamasaki, S.; Ohashi, W.; Hojyo, S.; Miyai, T.; Nishida, K.; Yokoyama, S.; Hirano, T. Biochemical characterization of human ZIP13 protein: A homo-dimerized zinc transporter involved in the spondylocheiro dysplastic Ehlers-Danlos syndrome. J. Biol. Chem. 2011, 286, 40255–40265. [Google Scholar] [CrossRef]
- Lee, M.G.; Bin, B.H. Different Actions of Intracellular Zinc Transporters ZIP7 and ZIP13 Are Essential for Dermal Development. Int. J. Mol. Sci. 2019, 20, 3941. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, X.; Zhao, H.; Yang, Q.; Xu, Z. Downregulation of the zinc transporter SLC39A13 (ZIP13) is responsible for the activation of CaMKII at reperfusion and leads to myocardial ischemia/reperfusion injury in mouse hearts. J. Mol. Cell. Cardiol. 2021, 152, 69–79. [Google Scholar] [CrossRef]
- Taylor, K.M.; Morgan, H.E.; Johnson, A.; Nicholson, R.I. Structure-function analysis of a novel member of the LIV-1 subfamily of zinc transporters, ZIP14. FEBS Lett. 2005, 579, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, T.B.; Troche, C.; Kim, M.H.; Cousins, R.J. Hepatic ZIP14-mediated Zinc Transport Contributes to Endosomal Insulin Receptor Trafficking and Glucose Metabolism. J. Biol. Chem. 2016, 291, 23939–23951. [Google Scholar] [CrossRef] [PubMed]
- Lopez, V.; Foolad, F.; Kelleher, S.L. ZnT2-overexpression represses the cytotoxic effects of zinc hyper-accumulation in malignant metallothionein-null T47D breast tumor cells. Cancer Lett. 2011, 304, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.B.; Wenzel, H.J.; Kafer, K.E.; Schwartzkroin, P.A.; Palmiter, R.D. Elimination of zinc from synaptic vesicles in the intact mouse brain by disruption of the ZnT3 gene. Proc. Natl. Acad. Sci. USA 1999, 96, 1716–1721. [Google Scholar] [CrossRef]
- Henshall, S.M.; Afar, D.E.; Rasiah, K.K.; Horvath, L.G.; Gish, K.; Caras, I.; Ramakrishnan, V.; Wong, M.; Jeffry, U.; Kench, J.G.; et al. Expression of the zinc transporter ZnT4 is decreased in the progression from early prostate disease to invasive prostate cancer. Oncogene 2003, 22, 6005–6012. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.P.; Zhang, Y.; Hiscox, S.; Guo, G.L.; Apte, U.; Taylor, K.M.; Sheline, C.T.; Wang, L.; Andrews, G.K. Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PLoS ONE 2010, 5, e13158. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, Y.; Wang, Y.; Yang, J.; Zhu, V.F.; Liu, Y.; Cui, X.; Chen, L.; Yan, W.; Jiang, T.; et al. ZIP4 is a novel molecular marker for glioma. Neuro Oncol. 2013, 15, 1008–1016. [Google Scholar] [CrossRef]
- Kambe, T.; Narita, H.; Yamaguchi-Iwai, Y.; Hirose, J.; Amano, T.; Sugiura, N.; Sasaki, R.; Mori, K.; Iwanaga, T.; Nagao, M. Cloning and characterization of a novel mammalian zinc transporter, zinc transporter 5, abundantly expressed in pancreatic beta cells. J. Biol. Chem. 2002, 277, 19049–19055. [Google Scholar] [CrossRef]
- Cragg, R.A.; Christie, G.R.; Phillips, S.R.; Russi, R.M.; Kury, S.; Mathers, J.C.; Taylor, P.M.; Ford, D. A novel zinc-regulated human zinc transporter, hZTL1, is localized to the enterocyte apical membrane. J. Biol. Chem. 2002, 277, 22789–22797. [Google Scholar] [CrossRef]
- Inoue, K.; Matsuda, K.; Itoh, M.; Kawaguchi, H.; Tomoike, H.; Aoyagi, T.; Nagai, R.; Hori, M.; Nakamura, Y.; Tanaka, T. Osteopenia and male-specific sudden cardiac death in mice lacking a zinc transporter gene, Znt5. Hum. Mol. Genet. 2002, 11, 1775–1784. [Google Scholar] [CrossRef]
- Devergnas, S.; Chimienti, F.; Naud, N.; Pennequin, A.; Coquerel, Y.; Chantegrel, J.; Favier, A.; Seve, M. Differential regulation of zinc efflux transporters ZnT-1, ZnT-5 and ZnT-7 gene expression by zinc levels: A real-time RT-PCR study. Biochem. Pharmacol. 2004, 68, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Yamazaki, T.; Ishida, Y.; Suzuki, T.; Oda, K.; Nagao, M.; Yamaguchi-Iwai, Y.; Kambe, T. Zinc transport complexes contribute to the homeostatic maintenance of secretory pathway function in vertebrate cells. J. Biol. Chem. 2006, 281, 17743–17750. [Google Scholar] [CrossRef] [PubMed]
- Ogo, O.A.; Tyson, J.; Cockell, S.J.; Howard, A.; Valentine, R.A.; Ford, D. The zinc finger protein ZNF658 regulates the transcription of genes involved in zinc homeostasis and affects ribosome biogenesis through the zinc transcriptional regulatory element. Mol. Cell. Biol. 2015, 35, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Barresi, V.; Valenti, G.; Spampinato, G.; Musso, N.; Castorina, S.; Rizzarelli, E.; Condorelli, D.F. Transcriptome analysis reveals an altered expression profile of zinc transporters in colorectal cancer. J. Cell. Biochem. 2018, 119, 9707–9719. [Google Scholar] [CrossRef]
- Huang, L.; Kirschke, C.P.; Gitschier, J. Functional characterization of a novel mammalian zinc transporter, ZnT6. J. Biol. Chem. 2002, 277, 26389–26395. [Google Scholar] [CrossRef]
- Seve, M.; Chimienti, F.; Devergnas, S.; Favier, A. In silico identification and expression of SLC30 family genes: An expressed sequence tag data mining strategy for the characterization of zinc transporters’ tissue expression. BMC Genomics 2004, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Chimienti, F.; Favier, A.; Seve, M. ZnT-8, a pancreatic beta-cell-specific zinc transporter. Biometals 2005, 18, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Smidt, K.; Pedersen, S.B.; Brock, B.; Schmitz, O.; Fisker, S.; Bendix, J.; Wogensen, L.; Rungby, J. Zinc-transporter genes in human visceral and subcutaneous adipocytes: Lean versus obese. Mol. Cell Endocrinol. 2007, 264, 68–73. [Google Scholar] [CrossRef]
- Overbeck, S.; Uciechowski, P.; Ackland, M.L.; Ford, D.; Rink, L. Intracellular zinc homeostasis in leukocyte subsets is regulated by different expression of zinc exporters ZnT-1 to ZnT-9. J. Leukoc. Biol. 2008, 83, 368–380. [Google Scholar] [CrossRef]
- Perez, Y.; Shorer, Z.; Liani-Leibson, K.; Chabosseau, P.; Kadir, R.; Volodarsky, M.; Halperin, D.; Barber-Zucker, S.; Shalev, H.; Schreiber, R.; et al. SLC30A9 mutation affecting intracellular zinc homeostasis causes a novel cerebro-renal syndrome. Brain 2017, 140, 928–939. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Gbadamosi, O.; Kolor, K.; Sosa, J.; Andrzejczuk, L.; Gibson, G.; St Croix, C.; Chikina, M.; Aizenman, E.; Clark, N.; et al. Evolutionary rate covariation identifies SLC30A9 (ZnT9) as a mitochondrial zinc transporter. Biochem. J. 2021, 478, 3205–3220. [Google Scholar] [CrossRef] [PubMed]
- Quadri, M.; Federico, A.; Zhao, T.; Breedveld, G.J.; Battisti, C.; Delnooz, C.; Severijnen, L.A.; Di Toro Mammarella, L.; Mignarri, A.; Monti, L.; et al. Mutations in SLC30A10 cause parkinsonism and dystonia with hypermanganesemia, polycythemia, and chronic liver disease. Am. J. Hum. Genet. 2012, 90, 467–477. [Google Scholar] [CrossRef]
- Patrushev, N.; Seidel-Rogol, B.; Salazar, G. Angiotensin II requires zinc and downregulation of the zinc transporters ZnT3 and ZnT10 to induce senescence of vascular smooth muscle cells. PLoS ONE 2012, 7, e33211. [Google Scholar] [CrossRef] [PubMed]
- Bosomworth, H.J.; Thornton, J.K.; Coneyworth, L.J.; Ford, D.; Valentine, R.A. Efflux function, tissue-specific expression and intracellular trafficking of the Zn transporter ZnT10 indicate roles in adult Zn homeostasis. Metallomics 2012, 4, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Cherian, M.G.; Jayasurya, A.; Bay, B.H. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat. Res. 2003, 533, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Yap, X.; Tan, H.Y.; Huang, J.; Lai, Y.; Yip, G.W.; Tan, P.H.; Bay, B.H. Over-expression of metallothionein predicts chemoresistance in breast cancer. J. Pathol. 2009, 217, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Yang, C.; Liu, D.; Zhi, Q.; Hua, Z.C. Zinc depletion induces JNK/p38 phosphorylation and suppresses Akt/mTOR expression in acute promyelocytic NB4 cells. J. Trace Elem. Med. Biol. 2023, 79, 127264. [Google Scholar] [CrossRef]
- Dalto, D.B.; Audet, I.; Roy, C.; Novais, A.K.; Deschene, K.; Goulet, K.; Matte, J.J.; Lapointe, J. Effects of dietary zinc oxide levels on the metabolism of zinc and copper in weaned pigs. J. Anim. Sci. 2023, 101, skad055. [Google Scholar] [CrossRef]
- Lakha, R.; Hachicho, C.; Mehlenbacher, M.R.; Wilcox, D.E.; Austin, R.N.; Vizcarra, C.L. Metallothionein-3 attenuates the effect of Cu(2+) ions on actin filaments. J. Inorg. Biochem. 2023, 242, 112157. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, M.; Nordberg, G.F. Toxicological aspects of metallothionein. Cell Mol. Biol. (Noisy-Le-Grand) 2000, 46, 451–463. [Google Scholar]
- Meloni, G.; Zovo, K.; Kazantseva, J.; Palumaa, P.; Vasak, M. Organization and assembly of metal-thiolate clusters in epithelium-specific metallothionein-4. J. Biol. Chem. 2006, 281, 14588–14595. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, H.; Xu, Z.; Cheng, X. Zinc dysregulation in cancers and its potential as a therapeutic target. Cancer Biol. Med. 2020, 17, 612–625. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Yoshigai, E.; Ohashi, T.; Fukada, T. Zinc transporters as potential therapeutic targets: An updated review. J. Pharmacol. Sci. 2022, 148, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Stovall, D.B.; Wang, W.; Sui, G. Advances of Zinc Signaling Studies in Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 667. [Google Scholar] [CrossRef]
- Laity, J.H.; Andrews, G.K. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1). Arch. Biochem. Biophys. 2007, 463, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Peck, E.J., Jr.; Ray, W.J., Jr. Metal complexes of phosphoglucomutase in vivo. Alterations induced by insulin. J. Biol. Chem. 1971, 246, 1160–1167. [Google Scholar] [CrossRef]
- Zalewski, P.D.; Forbes, I.J.; Betts, W.H. Correlation of apoptosis with change in intracellular labile Zn(II) using zinquin [(2-methyl-8-p-toluenesulphonamido-6-quinolyloxy)acetic acid], a new specific fluorescent probe for Zn(II). Biochem. J. 1993, 296 Pt 2, 403–408. [Google Scholar] [CrossRef]
- Kikuchi, K.; Komatsu, K.; Nagano, T. Zinc sensing for cellular application. Curr. Opin. Chem. Biol. 2004, 8, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Devinney, M.J., 2nd; Reynolds, I.J.; Dineley, K.E. Simultaneous detection of intracellular free calcium and zinc using fura-2FF and FluoZin-3. Cell Calcium 2005, 37, 225–232. [Google Scholar] [CrossRef]
- Krezel, A.; Maret, W. Zinc-buffering capacity of a eukaryotic cell at physiological pZn. J. Biol. Inorg. Chem. 2006, 11, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Coyle, P.; Zalewski, P.D.; Philcox, J.C.; Forbes, I.J.; Ward, A.D.; Lincoln, S.F.; Mahadevan, I.; Rofe, A.M. Measurement of zinc in hepatocytes by using a fluorescent probe, zinquin: Relationship to metallothionein and intracellular zinc. Biochem. J. 1994, 303 Pt 3, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Colvin, R.A.; Holmes, W.R.; Fontaine, C.P.; Maret, W. Cytosolic zinc buffering and muffling: Their role in intracellular zinc homeostasis. Metallomics 2010, 2, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Krezel, A.; Wojcik, J.; Maciejczyk, M.; Bal, W. May GSH and L-His contribute to intracellular binding of zinc? Thermodynamic and solution structural study of a ternary complex. Chem. Commun. 2003, 6, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Franklin, R.B. A comprehensive review of the role of zinc in normal prostate function and metabolism; and its implications in prostate cancer. Arch. Biochem. Biophys. 2016, 611, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Maret, W. Metals on the move: Zinc ions in cellular regulation and in the coordination dynamics of zinc proteins. Biometals 2011, 24, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Potter, B.M.; Feng, L.S.; Parasuram, P.; Matskevich, V.A.; Wilson, J.A.; Andrews, G.K.; Laity, J.H. The six zinc fingers of metal-responsive element binding transcription factor-1 form stable and quasi-ordered structures with relatively small differences in zinc affinities. J. Biol. Chem. 2005, 280, 28529–28540. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Chen, H.; Qi, M.; Dou, Y.; Wang, Q. Balance between metallothionein and metal response element binding transcription factor 1 is mediated by zinc ions (review). Mol. Med. Rep. 2015, 11, 1582–1586. [Google Scholar] [CrossRef]
- Li, L.; Williams, P.; Ren, W.; Wang, M.Y.; Gao, Z.; Miao, W.; Huang, M.; Song, J.; Wang, Y. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol. 2021, 17, 161–168. [Google Scholar] [CrossRef]
- Lopez-Perrote, A.; Alatwi, H.E.; Torreira, E.; Ismail, A.; Ayora, S.; Downs, J.A.; Llorca, O. Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases. J. Biol. Chem. 2014, 289, 22614–22629. [Google Scholar] [CrossRef]
- Belak, Z.R.; Nair, M.; Ovsenek, N. Parameters for effective in vitro production of zinc finger nucleic acid-binding proteins. Biotechnol. Appl. Biochem. 2011, 58, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Shi, Y.; Mulligan, P.; Gay, F.; Landry, J.; Liu, H.; Lu, J.; Qi, H.H.; Wang, W.; Nickoloff, J.A.; et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 2007, 14, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Wang, W.; Yi, C.; Xu, Q.; Wang, W.; Shi, J.; Stovall, D.B.; Li, D.; Sui, G. YY1 Oligomerization Is Regulated by Its OPB Domain and Competes with Its Regulation of Oncoproteins. Cancers 2022, 14, 1611. [Google Scholar] [CrossRef] [PubMed]
- Passerini, A.; Andreini, C.; Menchetti, S.; Rosato, A.; Frasconi, P. Predicting zinc binding at the proteome level. BMC Bioinform. 2007, 8, 39. [Google Scholar] [CrossRef]
- Wang, Y.; Lorenzi, I.; Georgiev, O.; Schaffner, W. Metal-responsive transcription factor-1 (MTF-1) selects different types of metal response elements at low vs. high zinc concentration. Biol. Chem. 2004, 385, 623–632. [Google Scholar] [CrossRef]
- Klar, M. It is not necessarily YY1—The frequently forgotten Yin-Yang-2 transcription factor. Proc. Natl. Acad. Sci. USA 2010, 107, E190, author reply E191. [Google Scholar] [CrossRef]
- Kaufhold, S.; Aziz, N.; Bonavida, B. The Forgotten YY2 in Reported YY1 Expression Levels in Human Cancers. Crit. Rev. Oncog. 2017, 22, 63–73. [Google Scholar] [CrossRef]
- Arai, M.; Sugase, K.; Dyson, H.J.; Wright, P.E. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc. Natl. Acad. Sci. USA 2015, 112, 9614–9619. [Google Scholar] [CrossRef]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J. Proteome Res. 2006, 5, 888–898. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7-13. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Galvin, K.M.; See, R.H.; Eckner, R.; Livingston, D.; Moran, E.; Shi, Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes. Dev. 1995, 9, 1188–1198. [Google Scholar] [CrossRef]
- Yao, Y.L.; Yang, W.M.; Seto, E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001, 21, 5979–5991. [Google Scholar] [CrossRef]
- Austen, M.; Luscher, B.; Luscher-Firzlaff, J.M. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 1997, 272, 1709–1717. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, M.E.; Zhang, L.F.; Xu, N.; Shi, Y.; Lee, J.T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell 2007, 25, 43–56. [Google Scholar] [CrossRef]
- Austen, M.; Cerni, C.; Luscher-Firzlaff, J.M.; Luscher, B. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene 1998, 17, 511–520. [Google Scholar] [CrossRef]
- Mai, R.T.; Yeh, T.S.; Kao, C.F.; Sun, S.K.; Huang, H.H.; Wu Lee, Y.H. Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene 2006, 25, 448–462. [Google Scholar] [CrossRef]
- Alfieri, C.; Gambetta, M.C.; Matos, R.; Glatt, S.; Sehr, P.; Fraterman, S.; Wilm, M.; Muller, J.; Muller, C.W. Structural basis for targeting the chromatin repressor Sfmbt to Polycomb response elements. Genes. Dev. 2013, 27, 2367–2379. [Google Scholar] [CrossRef]
- Zhang, Q.; Wan, M.; Shi, J.; Horita, D.A.; Miller, L.D.; Kute, T.E.; Kridel, S.J.; Kulik, G.; Sui, G. Yin Yang 1 promotes mTORC2-mediated AKT phosphorylation. J. Mol. Cell. Biol. 2016, 8, 232–243. [Google Scholar] [CrossRef]
- Wilkinson, F.H.; Park, K.; Atchison, M.L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA 2006, 103, 19296–19301. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, J.T.; Rodgers, J.T.; Arlow, D.H.; Vazquez, F.; Mootha, V.K.; Puigserver, P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007, 450, 736–740. [Google Scholar] [CrossRef]
- Luke, M.P.; Sui, G.; Liu, H.; Shi, Y. Yin Yang 1 physically interacts with Hoxa11 and represses Hoxa11-dependent transcription. J. Biol. Chem. 2006, 281, 33226–33232. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Gedrich, R.W.; Engel, D.A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J. Virol. 1995, 69, 4323–4330. [Google Scholar] [CrossRef] [PubMed]
- Wai, D.C.; Shihab, M.; Low, J.K.; Mackay, J.P. The zinc fingers of YY1 bind single-stranded RNA with low sequence specificity. Nucleic Acids Res. 2016, 44, 9153–9165. [Google Scholar] [CrossRef]
- Coull, J.J.; Romerio, F.; Sun, J.M.; Volker, J.L.; Galvin, K.M.; Davie, J.R.; Shi, Y.; Hansen, U.; Margolis, D.M. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 2000, 74, 6790–6799. [Google Scholar] [CrossRef]
- Yang, W.M.; Yao, Y.L.; Seto, E. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J. 2001, 20, 4814–4825. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Lewis, B.; Shenk, T. Interaction between transcription factors Sp1 and YY1. Nature 1993, 365, 462–464. [Google Scholar] [CrossRef]
- Garcia, E.; Marcos-Gutierrez, C.; del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418. [Google Scholar] [CrossRef]
- Deng, Z.; Wan, M.; Cao, P.; Rao, A.; Cramer, S.D.; Sui, G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene 2009, 28, 3746–3757. [Google Scholar] [CrossRef]
- Yu, H.; Mashtalir, N.; Daou, S.; Hammond-Martel, I.; Ross, J.; Sui, G.; Hart, G.W.; Rauscher, F.J., 3rd; Drobetsky, E.; Milot, E.; et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 2010, 30, 5071–5085. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Wan, M.; Sui, G. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol. Cell. Biol. 2007, 27, 3780–3792. [Google Scholar] [CrossRef]
- Marcon, E.; Ni, Z.; Pu, S.; Turinsky, A.L.; Trimble, S.S.; Olsen, J.B.; Silverman-Gavrila, R.; Silverman-Gavrila, L.; Phanse, S.; Guo, H.; et al. Human-chromatin-related protein interactions identify a demethylase complex required for chromosome segregation. Cell Rep. 2014, 8, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.Y.; Ko, C.Y.; Wang, S.M.; Lin, P.I.; Wang, H.Y.; Lin, W.C.; Wu, D.Y.; Wang, L.H.; Wang, J.M. Bortezomib-induced miRNAs direct epigenetic silencing of locus genes and trigger apoptosis in leukemia. Cell Death Dis. 2017, 8, e3167. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Huang, J.; Xie, P.; Dong, Y.; An, W. Inhibition of Drp1 SUMOylation by ALR protects the liver from ischemia-reperfusion injury. Cell Death Differ. 2021, 28, 1174–1192. [Google Scholar] [CrossRef]
- Begon, D.Y.; Delacroix, L.; Vernimmen, D.; Jackers, P.; Winkler, R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J. Biol. Chem. 2005, 280, 24428–24434. [Google Scholar] [CrossRef]
- Wu, F.; Lee, A.S. YY1 as a regulator of replication-dependent hamster histone H3.2 promoter and an interactive partner of AP-2. J. Biol. Chem. 2001, 276, 28–34. [Google Scholar] [CrossRef]
- Yue, R.; Kang, J.; Zhao, C.; Hu, W.; Tang, Y.; Liu, X.; Pei, G. Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression. Cell 2009, 139, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Baumeister, P.; Roy, B.; Phan, T.; Foti, D.; Luo, S.; Lee, A.S. ATF6 as a transcription activator of the endoplasmic reticulum stress element: Thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 2000, 20, 5096–5106. [Google Scholar] [CrossRef]
- Baumeister, P.; Luo, S.; Skarnes, W.C.; Sui, G.; Seto, E.; Shi, Y.; Lee, A.S. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: Activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell. Biol. 2005, 25, 4529–4540. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.; Long, Z.J.; Xu, D.; Lv, S.S.; Liu, B.; Wang, C.L.; Xu, J.; Lam, E.W.; Liu, Q. Aurora kinase A regulates Survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis 2017, 6, e298. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Li, F.; Wu, T.; Ding, J.; Lu, Z.; Wang, L.; Yang, Y.; Wang, F.; Zhao, L.; Zhu, H.; et al. BCCIP binds to and activates its promoter in a YY1-dependent fashion in HCT116 cells. FEBS J. 2018, 285, 3026–3040. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.R.; Crockett, D.K.; Lim, M.S.; Elenitoba-Johnson, K.S. Analysis of BCL6-interacting proteins by tandem mass spectrometry. Mol. Cell Proteomics 2005, 4, 1898–1909. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, X.; Zhao, M.; Yang, R.; Malik, R.; Qiao, Y.; Poliakov, A.; Yocum, A.K.; Li, Y.; Chen, W.; et al. The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 2014, 5, 3127. [Google Scholar] [CrossRef]
- Freire-Beneitez, V.; Pomella, N.; Millner, T.O.; Dumas, A.A.; Niklison-Chirou, M.V.; Maniati, E.; Wang, J.; Rajeeve, V.; Cutillas, P.; Marino, S. Elucidation of the BMI1 interactome identifies novel regulatory roles in glioblastoma. NAR Cancer 2021, 3, zcab009. [Google Scholar] [CrossRef]
- Lorente, M.; Perez, C.; Sanchez, C.; Donohoe, M.; Shi, Y.; Vidal, M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech. Dev. 2006, 123, 312–320. [Google Scholar] [CrossRef]
- Kim, J.J.; Lee, S.Y.; Gong, F.; Battenhouse, A.M.; Boutz, D.R.; Bashyal, A.; Refvik, S.T.; Chiang, C.M.; Xhemalce, B.; Paull, T.T.; et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019, 33, 1751–1774. [Google Scholar] [CrossRef]
- Shu, S.; Wu, H.J.; Ge, J.Y.; Zeid, R.; Harris, I.S.; Jovanovic, B.; Murphy, K.; Wang, B.; Qiu, X.; Endress, J.E.; et al. Synthetic Lethal and Resistance Interactions with BET Bromodomain Inhibitors in Triple-Negative Breast Cancer. Mol. Cell 2020, 78, 1096–1113. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, D.; Niu, X.; Wu, H.; Yang, J.; Zhang, Y.; Song, S.; Lv, D.; Chai, Y.; Lu, H.; et al. Mild iron overload induces TRIP12-mediated degradation of YY1 to trigger hepatic inflammation. Free Radic. Biol. Med. 2020, 161, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.H.; Cheveralls, K.C.; Brunner, A.D.; Kim, K.; Michaelis, A.C.; Raghavan, P.; Kobayashi, H.; Savy, L.; Li, J.Y.; Canaj, H.; et al. OpenCell: Endogenous tagging for the cartography of human cellular organization. Science 2022, 375, eabi6983. [Google Scholar] [CrossRef] [PubMed]
- de Nigris, F.; Botti, C.; Rossiello, R.; Crimi, E.; Sica, V.; Napoli, C. Cooperation between Myc and YY1 provides novel silencing transcriptional targets of alpha3beta1-integrin in tumour cells. Oncogene 2007, 26, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Bauknecht, T.; See, R.H.; Shi, Y. A novel C/EBP beta-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region. J. Virol. 1996, 70, 7695–7705. [Google Scholar] [CrossRef] [PubMed]
- Weissmann, S.; Cloos, P.A.; Sidoli, S.; Jensen, O.N.; Pollard, S.; Helin, K. The Tumor Suppressor CIC Directly Regulates MAPK Pathway Genes via Histone Deacetylation. Cancer Res. 2018, 78, 4114–4125. [Google Scholar] [CrossRef]
- Romerio, F.; Gabriel, M.N.; Margolis, D.M. Repression of human immunodeficiency virus type 1 through the novel cooperation of human factors YY1 and LSF. J. Virol. 1997, 71, 9375–9382. [Google Scholar] [CrossRef]
- Kang, H.C.; Chung, B.M.; Chae, J.H.; Yang, S.-I.; Kim, C.G.; Kim, C.G. Identification and characterization of four novel peptide motifs that recognize distinct regions of the transcription factor CP2. FEBS J. 2005, 272, 1265–1277. [Google Scholar] [CrossRef]
- Galvin, K.M.; Shi, Y. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 1997, 17, 3723–3732. [Google Scholar] [CrossRef]
- Wang, J.; Huo, K.; Ma, L.; Tang, L.; Li, D.; Huang, X.; Yuan, Y.; Li, C.; Wang, W.; Guan, W.; et al. Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 2011, 7, 536. [Google Scholar] [CrossRef]
- Kouranti, I.; Abdel Khalek, W.; Mazurkiewicz, S.; Loisel-Ferreira, I.; Gautreau, A.M.; Pintard, L.; Jeunemaitre, X.; Clauser, E. Cullin 3 Exon 9 Deletion in Familial Hyperkalemic Hypertension Impairs Cullin3-Ring-E3 Ligase (CRL3) Dynamic Regulation and Cycling. Int. J. Mol. Sci. 2022, 23, 5151. [Google Scholar] [CrossRef]
- Yang, W.M.; Inouye, C.J.; Seto, E. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J. Biol. Chem. 1995, 270, 15187–15193. [Google Scholar] [CrossRef] [PubMed]
- Vella, P.; Barozzi, I.; Cuomo, A.; Bonaldi, T.; Pasini, D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012, 40, 3403–3418. [Google Scholar] [CrossRef] [PubMed]
- Salvati, A.; Melone, V.; Sellitto, A.; Rizzo, F.; Tarallo, R.; Nyman, T.A.; Giurato, G.; Nassa, G.; Weisz, A. Combinatorial targeting of a chromatin complex comprising Dot1L, menin and the tyrosine kinase BAZ1B reveals a new therapeutic vulnerability of endocrine therapy-resistant breast cancer. Breast Cancer Res. 2022, 24, 52. [Google Scholar] [CrossRef] [PubMed]
- Pacaud, R.; Sery, Q.; Oliver, L.; Vallette, F.M.; Tost, J.; Cartron, P.F. DNMT3L interacts with transcription factors to target DNMT3L/DNMT3B to specific DNA sequences: Role of the DNMT3L/DNMT3B/p65-NFkappaB complex in the (de-)methylation of TRAF1. Biochimie 2014, 104, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Rezai-Zadeh, N.; Zhang, X.; Namour, F.; Fejer, G.; Wen, Y.D.; Yao, Y.L.; Gyory, I.; Wright, K.; Seto, E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003, 17, 1019–1029. [Google Scholar] [CrossRef]
- Zhou, Q.; Engel, D.A. Adenovirus E1A243 disrupts the ATF/CREB-YY1 complex at the mouse c-fos promoter. J. Virol. 1995, 69, 7402–7409. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; See, R.H.; Galvin, K.M.; Wang, J.; Shi, Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995, 23, 925–931. [Google Scholar] [CrossRef]
- Lewis, B.A.; Tullis, G.; Seto, E.; Horikoshi, N.; Weinmann, R.; Shenk, T. Adenovirus E1A proteins interact with the cellular YY1 transcription factor. J. Virol. 1995, 69, 1628–1636. [Google Scholar] [CrossRef]
- Kadeppagari, R.K.; Sankar, N.; Thimmapaya, B. Adenovirus transforming protein E1A induces c-Myc in quiescent cells by a novel mechanism. J. Virol. 2009, 83, 4810–4822. [Google Scholar] [CrossRef]
- Bard-Chapeau, E.A.; Gunaratne, J.; Kumar, P.; Chua, B.Q.; Muller, J.; Bard, F.A.; Blackstock, W.; Copeland, N.G.; Jenkins, N.A. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc. Natl. Acad. Sci. USA 2013, 110, E2885–E2894. [Google Scholar] [CrossRef]
- Caretti, G.; Di Padova, M.; Micales, B.; Lyons, G.E.; Sartorelli, V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004, 18, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Li, G.; Wang, W.; Sun, Y.; Zhang, Y.; Zhong, C.; Stovall, D.B.; Li, D.; Shi, J.; Sui, G. Disruption of YY1-EZH2 Interaction Using Synthetic Peptides Inhibits Breast Cancer Development. Cancers 2021, 13, 2402. [Google Scholar] [CrossRef]
- Xu, F.; Li, J.; Ni, M.; Cheng, J.; Zhao, H.; Wang, S.; Zhou, X.; Wu, X. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol. Cancer 2021, 20, 45. [Google Scholar] [CrossRef]
- Zhang, N.; Chan, C.W.; Sanchez-Guerrero, E.; Khachigian, L.M. Repression of PDGF-R-alpha after cellular injury involves TNF-alpha, formation of a c-Fos-YY1 complex, and negative regulation by HDAC. Am. J. Physiol. Cell Physiol. 2012, 302, C1590–C1598. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, W.; Wang, J.; Malovannaya, A.; Xi, Y.; Li, W.; Guerra, R.; Hawke, D.H.; Qin, J.; Chen, J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015, 11, 775. [Google Scholar] [CrossRef]
- Lu, P.; Hankel, I.L.; Hostager, B.S.; Swartzendruber, J.A.; Friedman, A.D.; Brenton, J.L.; Rothman, P.B.; Colgan, J.D. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J. Biol. Chem. 2011, 286, 18311–18319. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Dancik, G.M.; Goodspeed, A.; Costello, J.C.; Owens, C.; Duex, J.E.; Theodorescu, D. GON4L Drives Cancer Growth through a YY1-Androgen Receptor-CD24 Axis. Cancer Res. 2016, 76, 5175–5185. [Google Scholar] [CrossRef]
- Nakamura, K.; Saredi, G.; Becker, J.R.; Foster, B.M.; Nguyen, N.V.; Beyer, T.E.; Cesa, L.C.; Faull, P.A.; Lukauskas, S.; Frimurer, T.; et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019, 21, 311–318. [Google Scholar] [CrossRef]
- Yang, W.M.; Yao, Y.L.; Sun, J.M.; Davie, J.R.; Seto, E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997, 272, 28001–28007. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.E.; Lin, C.H.; Lin, Y.S.; Yu, W.C. Modulation of YY1 activity by SAP30. Biochem. Biophys. Res. Commun. 2003, 306, 267–275. [Google Scholar] [CrossRef]
- He, Y.; Sandoval, J.; Casaccia-Bonnefil, P. Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: The role of HDAC and YY1. Neuron Glia Biol. 2007, 3, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Glenn, D.J.; Wang, F.; Chen, S.; Nishimoto, M.; Gardner, D.G. Endothelin-stimulated human B-type natriuretic peptide gene expression is mediated by Yin Yang 1 in association with histone deacetylase 2. Hypertension 2009, 53, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Kalenik, J.L.; Chen, D.; Bradley, M.E.; Chen, S.J.; Lee, T.C. Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 1997, 25, 843–849. [Google Scholar] [CrossRef]
- Yang, W.M.; Inouye, C.; Zeng, Y.; Bearss, D.; Seto, E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 1996, 93, 12845–12850. [Google Scholar] [CrossRef]
- Sun, J.M.; Chen, H.Y.; Davie, J.R. Differential distribution of unmodified and phosphorylated histone deacetylase 2 in chromatin. J. Biol. Chem. 2007, 282, 33227–33236. [Google Scholar] [CrossRef]
- Villagra, A.; Ulloa, N.; Zhang, X.; Yuan, Z.; Sotomayor, E.; Seto, E. Histone deacetylase 3 down-regulates cholesterol synthesis through repression of lanosterol synthase gene expression. J. Biol. Chem. 2007, 282, 35457–35470. [Google Scholar] [CrossRef]
- Sankar, N.; Baluchamy, S.; Kadeppagari, R.K.; Singhal, G.; Weitzman, S.; Thimmapaya, B. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene 2008, 27, 5717–5728. [Google Scholar] [CrossRef]
- Han, S.; Lu, J.; Zhang, Y.; Cheng, C.; Han, L.; Wang, X.; Li, L.; Liu, C.; Huang, B. Recruitment of histone deacetylase 4 by transcription factors represses interleukin-5 transcription. Biochem. J. 2006, 400, 439–448. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Y.; Xu, L.; Chen, Y.; Zhang, Y.; Su, D.; Ren, G.; Lu, J.; Huang, B. YY1 restrained cell senescence through repressing the transcription of p16. Biochim. Biophys. Acta 2008, 1783, 1876–1883. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Zhang, G.; Dong, Z.; Liu, Z.; Li, L.; Feng, Y.; Su, D.; Zhang, Y.; Huang, B.; Lu, J. Recruitment of HDAC4 by transcription factor YY1 represses HOXB13 to affect cell growth in AR-negative prostate cancers. Int. J. Biochem. Cell Biol. 2009, 41, 1094–1101. [Google Scholar] [CrossRef]
- Sucharov, C.C.; Langer, S.; Bristow, M.; Leinwand, L. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am. J. Physiol. Cell Physiol. 2006, 291, C1029–C1037. [Google Scholar] [CrossRef]
- Sucharov, C.C.; Dockstader, K.; McKinsey, T.A. YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Mol. Biol. Cell 2008, 19, 4141–4153. [Google Scholar] [CrossRef] [PubMed]
- Barreiro-Alonso, A.; Camara-Quilez, M.; Salamini-Montemurri, M.; Lamas-Maceiras, M.; Vizoso-Vazquez, A.; Rodriguez-Belmonte, E.; Quindos-Varela, M.; Martinez-Iglesias, O.; Figueroa, A.; Cerdan, M.E. Characterization of HMGB1/2 Interactome in Prostate Cancer by Yeast Two Hybrid Approach: Potential Pathobiological Implications. Cancers 2019, 11, 1729. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Yao, T.; Gottschalk, A.J.; Swanson, S.K.; Wu, S.; Shi, Y.; Washburn, M.P.; Florens, L.; Conaway, R.C.; et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007, 14, 872–874. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Jin, J.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit organization of the human INO80 chromatin remodeling complex: An evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 2011, 286, 11283–11289. [Google Scholar] [CrossRef]
- Kato, M.; Chou, T.F.; Yu, C.Z.; DeModena, J.; Sternberg, P.W. LINKIN, a new transmembrane protein necessary for cell adhesion. Elife 2014, 3, e04449. [Google Scholar] [CrossRef]
- Kang, J.H.; Chang, S.Y.; Yeom, D.H.; Kim, S.A.; Um, S.J.; Hong, K.J. Weakening of the repressive YY-1 site on the thrombospondin-1 promoter via c-Jun/YY-1 interaction. Exp. Mol. Med. 2004, 36, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Tsai, M.F.; Dai, T.H.; Hong, T.M.; Chan, W.K.; Chen, J.J.; Yang, P.C. Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res. 2007, 67, 4816–4826. [Google Scholar] [CrossRef]
- Sobecki, M.; Mrouj, K.; Camasses, A.; Parisis, N.; Nicolas, E.; Lleres, D.; Gerbe, F.; Prieto, S.; Krasinska, L.; David, A.; et al. The cell proliferation antigen Ki-67 organises heterochromatin. Elife 2016, 5, e13722. [Google Scholar] [CrossRef] [PubMed]
- Sucharov, C.C.; Helmke, S.M.; Langer, S.J.; Perryman, M.B.; Bristow, M.; Leinwand, L. The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression. Mol. Cell. Biol. 2004, 24, 8705–8715. [Google Scholar] [CrossRef]
- Forlani, G.; Giarda, E.; Ala, U.; Di Cunto, F.; Salani, M.; Tupler, R.; Kilstrup-Nielsen, C.; Landsberger, N. The MeCP2/YY1 interaction regulates ANT1 expression at 4q35: Novel hints for Rett syndrome pathogenesis. Hum. Mol. Genet. 2010, 19, 3114–3123. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jiang, W.; Zhou, P.; Liu, L.; Wan, X.; Yuan, X.; Wang, X.; Chen, M.; Chen, J.; Yang, J.; et al. Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PLoS ONE 2015, 10, e0145023. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.L.; Yang, W.M. The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J. Biol. Chem. 2003, 278, 42560–42568. [Google Scholar] [CrossRef]
- Shrivastava, A.; Saleque, S.; Kalpana, G.V.; Artandi, S.; Goff, S.P.; Calame, K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 1993, 262, 1889–1892. [Google Scholar] [CrossRef]
- Wang, L.; Chen, C.; Song, Z.; Wang, H.; Ye, M.; Wang, D.; Kang, W.; Liu, H.; Qing, G. EZH2 depletion potentiates MYC degradation inhibiting neuroblastoma and small cell carcinoma tumor formation. Nat. Commun. 2022, 13, 12. [Google Scholar] [CrossRef]
- Somasekharan, S.P.; Gleave, M. SARS-CoV-2 nucleocapsid protein interacts with immunoregulators and stress granules and phase separates to form liquid droplets. FEBS Lett. 2021, 595, 2872–2896. [Google Scholar] [CrossRef]
- Persaud, A.; Alberts, P.; Amsen, E.M.; Xiong, X.; Wasmuth, J.; Saadon, Z.; Fladd, C.; Parkinson, J.; Rotin, D. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 2009, 5, 333. [Google Scholar] [CrossRef]
- Mori, T.; Ikeda, D.D.; Fukushima, T.; Takenoshita, S.; Kochi, H. NIRF constitutes a nodal point in the cell cycle network and is a candidate tumor suppressor. Cell Cycle 2011, 10, 3284–3299. [Google Scholar] [CrossRef]
- Yeh, T.S.; Lin, Y.M.; Hsieh, R.H.; Tseng, M.J. Association of transcription factor YY1 with the high molecular weight Notch complex suppresses the transactivation activity of Notch. J. Biol. Chem. 2003, 278, 41963–41969. [Google Scholar] [CrossRef]
- Rene, C.; Lopez, E.; Claustres, M.; Taulan, M.; Romey-Chatelain, M.C. NF-E2-related factor 2, a key inducer of antioxidant defenses, negatively regulates the CFTR transcription. Cell. Mol. Life Sci. 2010, 67, 2297–2309. [Google Scholar] [CrossRef]
- Wan, M.; Huang, W.; Kute, T.E.; Miller, L.D.; Zhang, Q.; Hatcher, H.; Wang, J.; Stovall, D.B.; Russell, G.B.; Cao, P.D.; et al. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 2012, 180, 2120–2133. [Google Scholar] [CrossRef]
- Gronroos, E.; Terentiev, A.A.; Punga, T.; Ericsson, J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 2004, 101, 12165–12170. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, M.; Pastrello, C.; Pivetta, F.; Lo Sardo, A.; Cumbaa, C.; Li, H.; Naranian, T.; Niu, Y.; Ding, Z.; Vafaee, F.; et al. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat. Methods 2015, 12, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Oei, S.L.; Griesenbeck, J.; Schweiger, M.; Babich, V.; Kropotov, A.; Tomilin, N. Interaction of the transcription factor YY1 with human poly(ADP-ribosyl) transferase. Biochem. Biophys. Res. Commun. 1997, 240, 108–111. [Google Scholar] [CrossRef]
- Oei, S.L.; Griesenbeck, J.; Schweiger, M.; Ziegler, M. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem. 1998, 273, 31644–31647. [Google Scholar] [CrossRef]
- Oei, S.L.; Shi, Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem. Biophys. Res. Commun. 2001, 285, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Daks, A.; Petukhov, A.; Fedorova, O.; Shuvalov, O.; Kizenko, A.; Tananykina, E.; Vasileva, E.; Semenov, O.; Bottrill, A.; Barlev, N. The RNA-binding protein HuR is a novel target of Pirh2 E3 ubiquitin ligase. Cell Death Dis. 2021, 12, 581. [Google Scholar] [CrossRef] [PubMed]
- Shami Shah, A.; Batrouni, A.G.; Kim, D.; Punyala, A.; Cao, W.; Han, C.; Goldberg, M.L.; Smolka, M.B.; Baskin, J.M. PLEKHA4/kramer Attenuates Dishevelled Ubiquitination to Modulate Wnt and Planar Cell Polarity Signaling. Cell Rep. 2019, 27, 2157–2170 e2158. [Google Scholar] [CrossRef]
- Hu, H.M.; Kanda, K.; Zhang, L.; Boxer, L.M. Activation of the c-myc p1 promoter in Burkitt’s lymphoma by the hs3 immunoglobulin heavy-chain gene enhancer. Leukemia 2007, 21, 747–753. [Google Scholar] [CrossRef]
- Sepulveda, M.A.; Emelyanov, A.V.; Birshtein, B.K. NF-kappa B and Oct-2 synergize to activate the human 3’ Igh hs4 enhancer in B cells. J. Immunol. 2004, 172, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.M.; Rademacher, J.; Bagshaw, R.D.; Wortmann, C.; Barth, C.; van Unen, J.; Alp, K.M.; Giudice, G.; Eccles, R.L.; Heinrich, L.E.; et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol. 2020, 22, 498–511. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Cao, J.L.; Zhang, Y.; Liao, L.; Deng, L.; Yang, S.Y.; Hu, S.Y.; Ning, Y.; Zhang, F.L.; Li, D.Q. RNF144A exerts tumor suppressor function in breast cancer through targeting YY1 for proteasomal degradation to downregulate GMFG expression. Med. Oncol. 2022, 39, 48. [Google Scholar] [CrossRef] [PubMed]
- Sawa, C.; Yoshikawa, T.; Matsuda-Suzuki, F.; Delehouzee, S.; Goto, M.; Watanabe, H.; Sawada, J.; Kataoka, K.; Handa, H. YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J. Biol. Chem. 2002, 277, 22484–22490. [Google Scholar] [CrossRef] [PubMed]
- Kurisaki, K.; Kurisaki, A.; Valcourt, U.; Terentiev, A.A.; Pardali, K.; Ten Dijke, P.; Heldin, C.H.; Ericsson, J.; Moustakas, A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 2003, 23, 4494–4510. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Evans, S.; Ruan, T.Y.; Lassar, A.B. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development 2004, 131, 4709–4723. [Google Scholar] [CrossRef]
- Brown, K.A.; Ham, A.J.; Clark, C.N.; Meller, N.; Law, B.K.; Chytil, A.; Cheng, N.; Pietenpol, J.A.; Moses, H.L. Identification of novel Smad2 and Smad3 associated proteins in response to TGF-beta1. J. Cell. Biochem. 2008, 105, 596–611. [Google Scholar] [CrossRef]
- Yan, X.; Pan, J.; Xiong, W.; Cheng, M.; Sun, Y.; Zhang, S.; Chen, Y. Yin Yang 1 (YY1) synergizes with Smad7 to inhibit TGF-beta signaling in the nucleus. Sci. China Life Sci. 2014, 57, 128–136. [Google Scholar] [CrossRef]
- Jeong, H.M.; Lee, S.H.; Yum, J.; Yeo, C.Y.; Lee, K.Y. Smurf2 regulates the degradation of YY1. Biochim. Biophys. Acta 2014, 1843, 2005–2011. [Google Scholar] [CrossRef]
- Rowbotham, S.P.; Barki, L.; Neves-Costa, A.; Santos, F.; Dean, W.; Hawkes, N.; Choudhary, P.; Will, W.R.; Webster, J.; Oxley, D.; et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell 2011, 42, 285–296. [Google Scholar] [CrossRef]
- Lee, J.S.; Galvin, K.M.; Shi, Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. USA 1993, 90, 6145–6149. [Google Scholar] [CrossRef]
- Bennett, M.K.; Ngo, T.T.; Athanikar, J.N.; Rosenfeld, J.M.; Osborne, T.F. Co-stimulation of promoter for low density lipoprotein receptor gene by sterol regulatory element-binding protein and Sp1 is specifically disrupted by the yin yang 1 protein. J. Biol. Chem. 1999, 274, 13025–13032. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Wang, Z.; Liu, X.; Guo, L.; Huang, L.; Gao, L.; McNutt, M.A.; Li, G. The role of transcription factors Sp1 and YY1 in proximal promoter region in initiation of transcription of the mu opioid receptor gene in human lymphocytes. J. Cell. Biochem. 2008, 104, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; May, J.M. Regulation of the human ascorbate transporter SVCT2 exon 1b gene by zinc-finger transcription factors. Free Radic. Biol. Med. 2011, 50, 1196–1209. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perry, M.; Biegert, M.; Kollala, S.S.; Mallard, H.; Su, G.; Kodavati, M.; Kreiling, N.; Holbrook, A.; Ghosal, G. USP11 mediates repair of DNA-protein cross-links by deubiquitinating SPRTN metalloprotease. J. Biol. Chem. 2021, 296, 100396. [Google Scholar] [CrossRef]
- Chiang, C.M.; Roeder, R.G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 1995, 267, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Gautier, V.W.; Gu, L.; O’Donoghue, N.; Pennington, S.; Sheehy, N.; Hall, W.W. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology 2009, 6, 47. [Google Scholar] [CrossRef]
- Usheva, A.; Shenk, T. YY1 transcriptional initiator: Protein interactions and association with a DNA site containing unpaired strands. Proc. Natl. Acad. Sci. USA 1996, 93, 13571–13576. [Google Scholar] [CrossRef]
- Giannone, R.J.; McDonald, H.W.; Hurst, G.B.; Shen, R.F.; Wang, Y.; Liu, Y. The protein network surrounding the human telomere repeat binding factors TRF1, TRF2, and POT1. PLoS ONE 2010, 5, e12407. [Google Scholar] [CrossRef]
- Demirdizen, E.; Al-Ali, R.; Narayanan, A.; Sun, X.; Varga, J.P.; Steffl, B.; Brom, M.; Krunic, D.; Schmidt, C.; Schmidt, G.; et al. TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing. Neuro Oncol. 2023, 25, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Sarkar, S.; Mohanta, P.S.; Kumar, K.; Chakrabarti, S.; Basu, M.; Ghosh, M.K. USP7 targets XIAP for cancer progression: Establishment of a p53-independent therapeutic avenue for glioma. Oncogene 2022, 41, 5061–5075. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Liang, Y.J.; Lin, Y.S.; Shih, H.M.; Jou, Y.S.; Yu, W.C. YY1AP, a novel co-activator of YY1. J. Biol. Chem. 2004, 279, 17750–17755. [Google Scholar] [CrossRef]
- Ravasi, T.; Suzuki, H.; Cannistraci, C.V.; Katayama, S.; Bajic, V.B.; Tan, K.; Akalin, A.; Schmeier, S.; Kanamori-Katayama, M.; Bertin, N.; et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 2010, 140, 744–752. [Google Scholar] [CrossRef]
- Rulisek, L.; Vondrasek, J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J. Inorg. Biochem. 1998, 71, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.A.; Yu, J. EZH2, an epigenetic driver of prostate cancer. Protein Cell 2013, 4, 331–341. [Google Scholar] [CrossRef]
- Honda, R.; Tanaka, H.; Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997, 420, 25–27. [Google Scholar] [CrossRef]
- Wang, W.; Qiao, S.; Li, G.; Cheng, J.; Yang, C.; Zhong, C.; Stovall, D.B.; Shi, J.; Teng, C.; Li, D.; et al. A histidine cluster determines YY1-compartmentalized coactivators and chromatin elements in phase-separated enhancer clusters. Nucleic Acids Res. 2022, 50, 4917–4937. [Google Scholar] [CrossRef]
- Singh, V.; Xu, L.; Boyko, S.; Surewicz, K.; Surewicz, W.K. Zinc promotes liquid-liquid phase separation of tau protein. J. Biol. Chem. 2020, 295, 5850–5856. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Zhong, T.; Wang, L.Q.; Zhang, N.; Zeng, Y.; Hu, J.Y.; Dang, H.B.; Chen, J.; Liang, Y. Zinc enhances liquid-liquid phase separation of Tau protein and aggravates mitochondrial damages in cells. Int. J. Biol. Macromol. 2022, 209, 703–715. [Google Scholar] [CrossRef]
- Das, B.; Roychowdhury, S.; Mohanty, P.; Rizuan, A.; Chakraborty, J.; Mittal, J.; Chattopadhyay, K. A Zn-dependent structural transition of SOD1 modulates its ability to undergo phase separation. EMBO J. 2023, 42, e111185. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.Y.; Fang, Y.L.; Shankar, S.; Lee, S.P.; Hu, H.T.; Chen, H.; Wang, T.F.; Hsia, K.C.; Hsueh, Y.P. Phase separation and zinc-induced transition modulate synaptic distribution and association of autism-linked CTTNBP2 and SHANK3. Nat. Commun. 2022, 13, 2664. [Google Scholar] [CrossRef] [PubMed]
- Rhie, S.K.; Perez, A.A.; Lay, F.D.; Schreiner, S.; Shi, J.; Polin, J.; Farnham, P.J. A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome. Nat. Commun. 2019, 10, 4154. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.Q.; Lind, S.E. Metal ionophores—An emerging class of anticancer drugs. IUBMB Life 2009, 61, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- To, P.K.; Do, M.H.; Cho, Y.S.; Kwon, S.Y.; Kim, M.S.; Jung, C. Zinc Inhibits Expression of Androgen Receptor to Suppress Growth of Prostate Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3062. [Google Scholar] [CrossRef]
- Qi, Y.; Yan, T.; Chen, L.; Zhang, Q.; Wang, W.; Han, X.; Li, D.; Shi, J.; Sui, G. Characterization of YY1 OPB Peptide for its Anticancer Activity. Curr. Cancer Drug Targets 2019, 19, 504–511. [Google Scholar] [CrossRef]


| Family | Protein | Flux | Expression | Reference |
|---|---|---|---|---|
| ZIP | ZIP1 | EC → Cyt ER → Cyt | Ubiquitously expressed. In enterocytes and prostate epithelium located in basolateral membrane. Downregulated in prostate cancer and clear cell renal cell carcinoma. Expression inversely correlates with prognosis; enhances proliferation and invasion of ACHN cells. Upregulated in pancreatic cancer. | [12,13,14,15,16,17] |
| ZIP2 | EC → Cyt | Expressed in the apical membrane of prostate and uterine epithelial cells. Downregulated in prostate cancer | [15,18] | |
| ZIP3 | EC → Cyt | Expressed in ovary, testis, pancreas, mammary gland and in the apical membrane of prostate epithelium. Takes part in zinc reuptake in the lactating mammary gland. Downregulated in prostate cancer and pancreatic cancer. Upregulated in pancreatic cancer | [15,17,19,20,21] | |
| ZIP4 | EC → Cyt | Expressed in kidney, stomach, intestine, and colon. In enterocytes, located in apical membrane. Overexpressed in pancreatic, ovarian cancer and pancreatic cancer. Promotes proliferation of pancreatic cancer cells. Promoting invasiveness of hepatocellular carcinoma, nasopharyngeal carcinoma and non-small cell lung cancer. Overexpression predicts poor survival in acute myeloid leukemia and pancreatic cancer. Associated with cisplatin resistance, radioresistance and enhanced cell migration. Mutations cause acrodermatitis enteropathica | [17,22,23,24,25,26,27,28,29,30] | |
| ZIP5 | EC → Cyt | Expressed in liver, kidney, pancreas, small intestine, colon, spleen. In enterocytes, located in basolateral membrane. Downregulated in pancreatic cancer | [17,31] | |
| ZIP6 | EC → Cyt | Expressed in breast, prostate, placenta, kidney, pituitary gland. Triggers progression of breast cancer. Overexpressed in esophageal carcinoma and pancreatic and colorectal cancer. Overexpression promotes epithelial-to-mesenchymal transition, cell mobility and metastasis. Involved in antigen presentation to T cells | [17,32,33,34,35] | |
| ZIP7 | ER → Cyt Golgi → Cyt | Ubiquitously expressed. Overexpressed and involved in progression of cancers of breast and cervix. Upregulated in pancreatic and colorectal cancer. Overexpression promotes cell proliferation, migration and invasion in breast and cervix cancer | [17,36,37,38,39] | |
| ZIP8 | EC → Cyt | Ubiquitously expressed. Involved in inflammation. | [40,41] | |
| ZIP9 | EC → Cyt Golgi → Cyt M → Cyt | Highest expression in pancreas, testis and pituitary gland. Upregulated in pancreatic and colorectal cancer. Acts as membrane androgen receptor. Involved in testosterone-induced apoptosis of breast and prostate cancer cells | [17,42,43,44,45,46] | |
| ZIP10 | EC → Cyt | Highest expression in kidney. Involved in anti-apoptotic signaling during B cell development. Overexpressed and involved in progression of metastatic breast cancer and renal cell carcinoma. Upregulated in pancreatic and colorectal cancer. Promotes cell migration | [17,47,48,49,50] | |
| ZIP11 | Golgi → Cyt N → Cyt | Expressed in testes, stomach, intestine; Involved in progression of pancreatic, ovarian, and cervical cancer. Variants associated with increased risks of bladder cancer, and renal cell carcinoma. Promotes proliferation of Capan-1 cells. Promotes proliferation, migration, invasiveness and mitochondrial potential of HeLa cells. Overexpression correlates with poor prognosis in cervical cancer | [17,51,52,53] | |
| ZIP12 | EC → Cyt | Highest expression in brain, eye and pulmonary vasculature. | [54,55,56] | |
| ZIP13 | ER → Cyt Golgi → Cyt | Highest expression in heart, dermis and connective tissue. Upregulated in pancreatic cancer | [17,57,58,59,60] | |
| ZIP14 | EC → Cyt M → Cyt | Zinc uptake to liver in response to inflammation. Regulates glucose homeostasis in hepatocytes. Overexpressed in hepatocellular carcinoma. Downregulated in pancreatic cancer | [17,27,54,61,62] | |
| ZnT | ZnT1 | Cyt. → N Cyt. → EC | In enterocytes, located in basolateral membrane. Upregulated in pancreatic cancer. Promotes proliferation of Capan-1 cells. Underexpression predicts poor survival in acute myeloid leukemia | [17,24] |
| ZnT2 | Cyt. → ER Cyt. → E | Overexpressed in breast cancer. Serves for vesicular sequestration of the accumulated zinc. Protecting the cells from zinc cytotoxicity. Downregulated in pancreatic cancer | [17,63] | |
| ZnT3 | Cyt. → SV Cyt. → E | Transports zinc into synaptic vesicles of glutamatergic neurons | [64] | |
| ZnT4 | Cyt. → Golgi Cyt. → SV Cyt. → E | Higher expression in prostate than in other organs. Expression decreases upon the progression of prostate cancer. Overexpressed in liver and pancreas cancer and glioma. Repressing apoptosis. Enhancing proliferation and migration. Higher expression predicted poorer survival in acute myeloid leukemia | [23,30,65,66,67] | |
| ZnT5 | Cyt. → Golgi Cyt. → IG | Highly expressed in pancreas, liver, kidney and insulin-containing beta cells. Undetectable in other endocrine cell types. Decreased expression predicts poor survival in acute myeloid leukemia. Expression is regulated by zinc ions. Upregulated by endoplasmic reticulum stress. Lower expression predicted poorer survival in acute myeloid leukemia. Upregulated in pancreatic and colorectal cancers | [17,30,68,69,70,71,72,73,74] | |
| ZnT6 | Cyt. → ER Cyt. → Golgi | Expressed in brain, B-cells, colon, eye, and lung. Lower expression in bone, brain, cervix, ear, heart, kidney, muscle, nerve, pancreas, prostate, skin, stomach, and testis. Upregulated in pancreatic and colorectal cancers. Promotes proliferation of Capan-1 cells. Underexpression predicts poor survival in acute myeloid leukemia | [17,74,75,76] | |
| ZnT7 | Cyt. → ER Cyt. → Golgi | Upregulated in pancreatic and colorectal cancers. Lower expression predicted poorer survival in acute myeloid leukemia | [17,30,74] | |
| ZnT8 | Cyt. → IG | Expressed in pancreatic beta-cell-specific zinc transporter. Precent in fat tissue and T-cells and B-cells | [77,78,79] | |
| ZnT9 | Cyt. → N | Ubiquitously expressed in fetal and adult tissues and cancer cell lines. Mitochondrial zinc exporter. Upregulated in pancreatic cancer | [17,80,81] | |
| ZnT10 | Cyt. → Golgi Cyt. → E Cyt. → EC | Highest levels of expression in small intestine, liver and brain tissues. Down-regulated by zinc and angiotensin-2 | [82,83,84] | |
| MT | MT1 | Cytoplasm sequestration | Ubiquitously expressed. Overexpressed in cancers of the breast, colon, kidney, liver, skin (melanoma), lung, nasopharynx, ovary, prostate, mouth, salivary gland, testes, thyroid and urinary bladder. Underexpressed in hepatocellular carcinoma, liver adenocarcinoma and pancreatic cancer | [17,85] |
| MT2a | Ubiquitously expressed. Overexpression correlates with cancer chemoresistance and acute myeloid leukemia | [86,87] | ||
| MT3 | Expressed mainly in central nervous system. Present in kidney and jejunum. Abundant in a subset of astrocytes in the normal human brain | [88,89] | ||
| MT4 | Specifically expressed in stratified epithelia | [90,91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figiel, M.; Górka, A.K.; Górecki, A. Zinc Ions Modulate YY1 Activity: Relevance in Carcinogenesis. Cancers 2023, 15, 4338. https://doi.org/10.3390/cancers15174338
Figiel M, Górka AK, Górecki A. Zinc Ions Modulate YY1 Activity: Relevance in Carcinogenesis. Cancers. 2023; 15(17):4338. https://doi.org/10.3390/cancers15174338
Chicago/Turabian StyleFigiel, Małgorzata, Adam Kazimierz Górka, and Andrzej Górecki. 2023. "Zinc Ions Modulate YY1 Activity: Relevance in Carcinogenesis" Cancers 15, no. 17: 4338. https://doi.org/10.3390/cancers15174338
APA StyleFigiel, M., Górka, A. K., & Górecki, A. (2023). Zinc Ions Modulate YY1 Activity: Relevance in Carcinogenesis. Cancers, 15(17), 4338. https://doi.org/10.3390/cancers15174338