Lynch Syndrome Biopathology and Treatment: The Potential Role of microRNAs in Clinical Practice
Abstract
:Simple Summary
Abstract
1. Introduction
2. Lynch Syndrome
2.1. Genetic and Molecular Background
2.2. Clinical and Diagnostic Overview
2.3. LS and Breast Cancer: What Is the Real Link?
3. miRNAs
3.1. Biogenesis and Mechanism of Action
3.2. Colorectal Cancer and miRNAs
- Early diagnosis and detection: miRNAs represent promising non-invasive biomarkers for the diagnosis of CRC. They can be detected in blood or other body fluids, enabling early diagnosis of the disease. Some miRNAs, such as miR-21, miR-92a, and miR-135b, were found to be upregulated in CRC patients [112,113,114,115].
- Prognosis and risk stratification: aberrant expression of specific miRNAs was associated with the prognosis and survival of CRC patients. Among them, miR-21 and miR-29a were correlated with a poor prognosis and shorter overall survival in CRC [116,117]. This information may be useful for identifying any patients who may need more aggressive treatment or closer monitoring.
- Prediction of treatment response: miRNAs can help to predict a patient’s response to a specific treatment. For example, some miRNAs, such as miR-21 or miR-17, have been related to resistance or sensitivity to chemotherapeutic agents commonly used in the treatment of CRC, such as 5-fluorouracil (5-FU) or oxaliplatin [118,119].
- Monitoring of treatment response and relapse: changes in miRNA expression levels during treatment can be monitored to evaluate treatment response and detect potential relapse. The monitoring of specific miRNAs, e.g., miR-21 or miR-155, can provide data on treatment efficacy and help in the process of personalizing approaches [118].
3.3. Lynch Syndrome and miRNAs
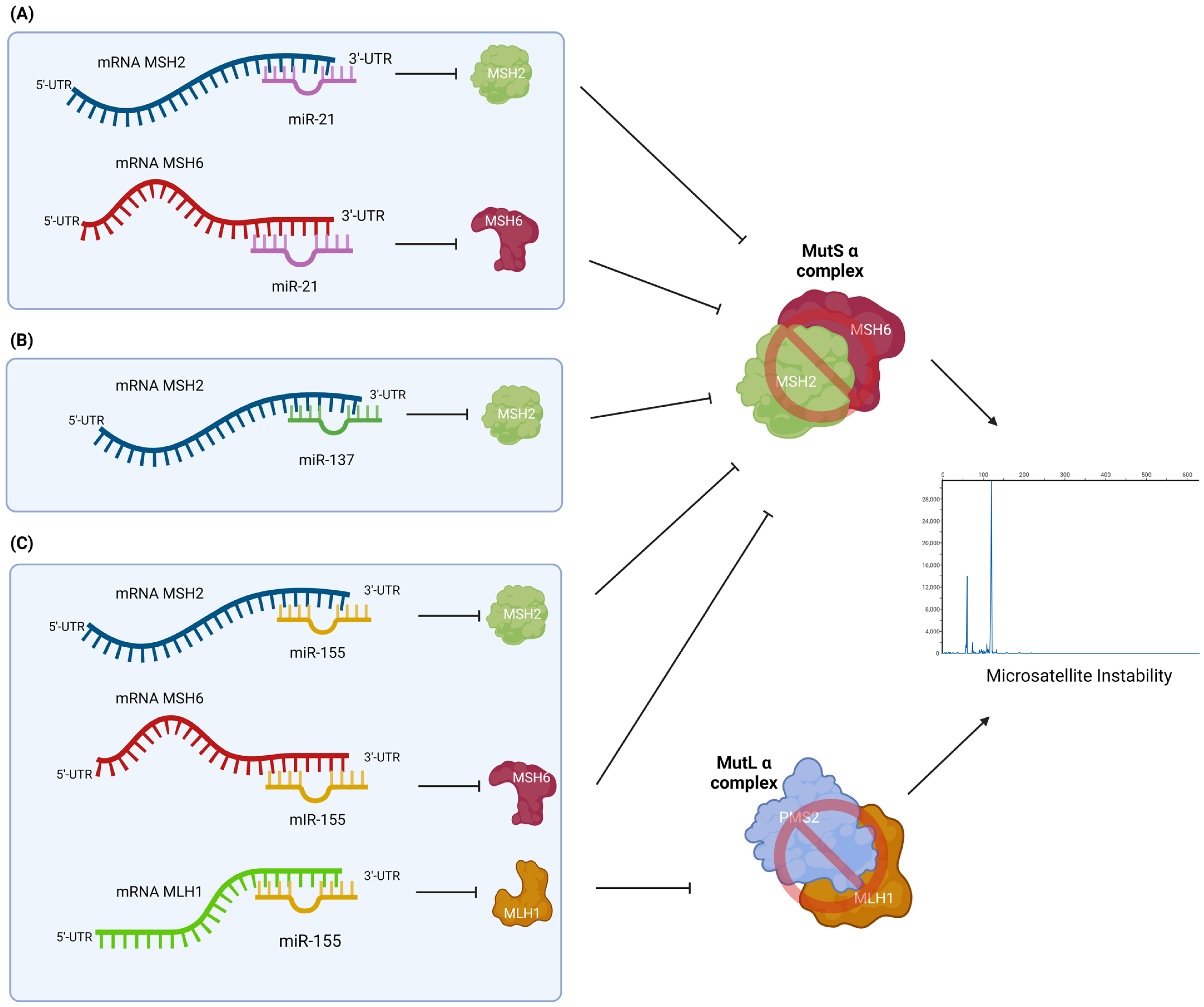
3.3.1. miR-21
3.3.2. miR-137
3.3.3. miR-155
3.3.4. Other miRNAs Player in Lynch Syndrome
3.4. miRNAs Correlated with CRC and MSI Status
3.4.1. miRNAs and MSI-L Status
miR-92
let-7
miR-145
3.4.2. miRNAs and MSI-H Status
miR-26b
miR-31
miR-223
3.4.3. miRNAs and MSS Status
miR-196a
4. Current Clinical Scenario and Future Perspectives
- Adenomatous polyp may initially develop with classic biallelic loss of APC and subsequently cause dMMR, leading to the accumulation of mutations fundamental to tumor growth [53].
- Adenomatous polyps may lose MMR early and subsequently acquire frameshift mutations in the single nucleotide repeats of the RNF43 gene, inducing malignant transformation [178].
- The traditional adenomatous polyp stage could be completely bypassed [55].
4.1. Overview of Treatment in the Era of Precision Medicine
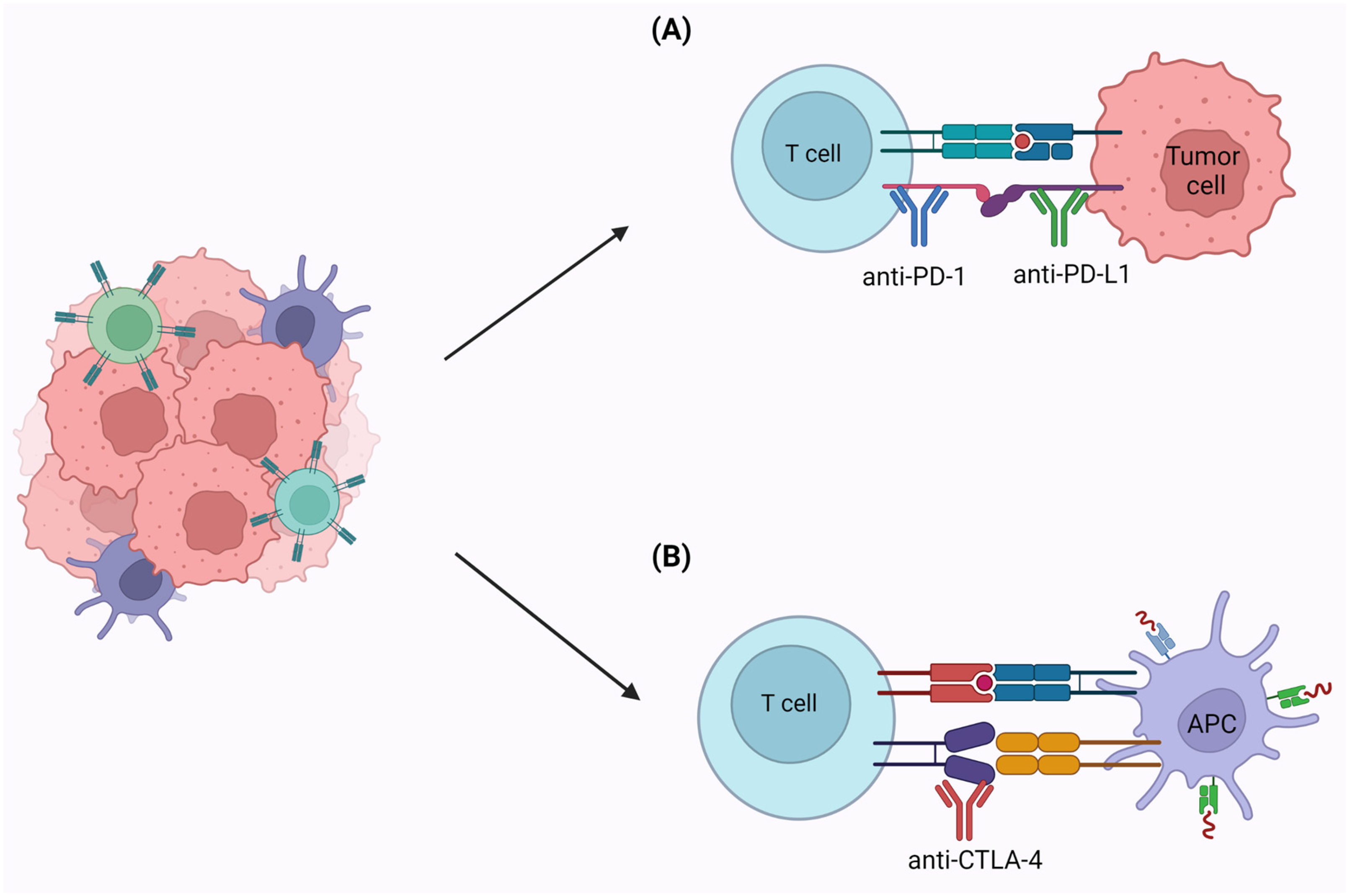
4.2. Immunotherapy
4.3. Potential Use of miRNAs as Biomarkers or Therapeutic Targets
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lynch, H.T.; de la Chapelle, A. Hereditary colorectal cancer. N. Engl. J. Med. 2003, 348, 919–932. [Google Scholar] [CrossRef]
- Lynch, H.T.; Snyder, C.L.; Shaw, T.G.; Heinen, C.D.; Hitchins, M.P. Milestones of Lynch syndrome: 1895–2015. Nat. Rev. Cancer 2015, 15, 181–194. [Google Scholar] [CrossRef]
- Burt, R.W.; DiSario, J.A.; Cannon-Albright, L. Genetics of colon cancer: Impact of inheritance on colon cancer risk. Annu. Rev. Med. 1995, 46, 371–379. [Google Scholar] [CrossRef]
- Syngal, S.; Brand, R.E.; Church, J.M.; Giardiello, F.M.; Hampel, H.L.; Burt, R.W.; American College of Gastroenterology. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am. J. Gastroenterol. 2015, 110, 223–262; quiz 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, S.A.; Pritchard, C.C.; Jarvik, G.P. Lynch Syndrome: From Screening to Diagnosis to Treatment in the Era of Modern Molecular Oncology. Annu. Rev. Genom. Hum. Genet. 2019, 20, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Senter, L. Genetic testing by cancer site: Colon (nonpolyposis syndromes). Cancer J. 2012, 18, 334–337. [Google Scholar] [CrossRef]
- Haraldsdottir, S.; Hampel, H.; Wei, L.; Wu, C.; Frankel, W.; Bekaii-Saab, T.; de la Chapelle, A.; Goldberg, R.M. Prostate cancer incidence in males with Lynch syndrome. Genet. Med. 2014, 16, 553–557. [Google Scholar] [CrossRef] [Green Version]
- Davies, H.; Morganella, S.; Purdie, C.A.; Jang, S.J.; Borgen, E.; Russnes, H.; Glodzik, D.; Zou, X.; Viari, A.; Richardson, A.L.; et al. Whole-Genome Sequencing Reveals Breast Cancers with Mismatch Repair Deficiency. Cancer Res. 2017, 77, 4755–4762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Win, A.K.; Lindor, N.M.; Jenkins, M.A. Risk of breast cancer in Lynch syndrome: A systematic review. Breast Cancer Res. 2013, 15, R27. [Google Scholar] [CrossRef]
- Harkness, E.F.; Barrow, E.; Newton, K.; Green, K.; Clancy, T.; Lalloo, F.; Hill, J.; Evans, D.G. Lynch syndrome caused by MLH1 mutations is associated with an increased risk of breast cancer: A cohort study. J. Med. Genet. 2015, 52, 553–556. [Google Scholar] [CrossRef]
- Gruchalla, R.S.; Sampson, H.A.; Liu, A.H.; Shreffler, W.; Wallace, P.K.; Togias, A.; David, G.; Calatroni, A.; LeBeau, P.; Inner-City Asthma Consortium. Effects of omalizumab on T lymphocyte function in inner-city children with asthma. Pediatr. Allergy Immunol. 2016, 27, 328–331. [Google Scholar] [CrossRef] [Green Version]
- Kanth, P.; Grimmett, J.; Champine, M.; Burt, R.; Samadder, N.J. Hereditary Colorectal Polyposis and Cancer Syndromes: A Primer on Diagnosis and Management. Am. J. Gastroenterol. 2017, 112, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.; Vasen, H.F.A.; Mecklin, J.P.; Bernstein, I.; Aarnio, M.; Jarvinen, H.J.; Myrhoj, T.; Sunde, L.; Wijnen, J.T.; Lynch, H.T. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int. J. Cancer 2008, 123, 444–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinicrope, F.A. Lynch Syndrome-Associated Colorectal Cancer. N. Engl. J. Med. 2018, 379, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Styk, J.; Pos, Z.; Pos, O.; Radvanszky, J.; Turnova, E.H.; Buglyo, G.; Klimova, D.; Budis, J.; Repiska, V.; Nagy, B.; et al. Microsatellite instability assessment is instrumental for Predictive, Preventive and Personalised Medicine: Status quo and outlook. EPMA J. 2023, 14, 143–165. [Google Scholar] [CrossRef]
- Biller, L.H.; Syngal, S.; Yurgelun, M.B. Recent advances in Lynch syndrome. Fam. Cancer 2019, 18, 211–219. [Google Scholar] [CrossRef]
- Rubenstein, J.H.; Enns, R.; Heidelbaugh, J.; Barkun, A.; Clinical Guidelines Committee. American Gastroenterological Association Institute Guideline on the Diagnosis and Management of Lynch Syndrome. Gastroenterology 2015, 149, 777–782; quiz e16-7. [Google Scholar] [CrossRef] [Green Version]
- Haraldsdottir, S.; Rafnar, T.; Frankel, W.L.; Einarsdottir, S.; Sigurdsson, A.; Hampel, H.; Snaebjornsson, P.; Masson, G.; Weng, D.; Arngrimsson, R.; et al. Comprehensive population-wide analysis of Lynch syndrome in Iceland reveals founder mutations in MSH6 and PMS2. Nat. Commun. 2017, 8, 14755. [Google Scholar] [CrossRef] [Green Version]
- Thibodeau, S.N.; Bren, G.; Schaid, D. Microsatellite instability in cancer of the proximal colon. Science 1993, 260, 816–819. [Google Scholar] [CrossRef]
- Aaltonen, L.A.; Peltomaki, P.; Leach, F.S.; Sistonen, P.; Pylkkanen, L.; Mecklin, J.P.; Jarvinen, H.; Powell, S.M.; Jen, J.; Hamilton, S.R.; et al. Clues to the pathogenesis of familial colorectal cancer. Science 1993, 260, 812–816. [Google Scholar] [CrossRef]
- Gallo Cantafio, M.E.; Grillone, K.; Caracciolo, D.; Scionti, F.; Arbitrio, M.; Barbieri, V.; Pensabene, L.; Guzzi, P.H.; Di Martino, M.T. From Single Level Analysis to Multi-Omics Integrative Approaches: A Powerful Strategy towards the Precision Oncology. High Throughput 2018, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 117. [Google Scholar] [CrossRef]
- Caracciolo, D.; Riillo, C.; Juli, G.; Scionti, F.; Todoerti, K.; Polera, N.; Grillone, K.; Fiorillo, L.; Arbitrio, M.; Di Martino, M.T.; et al. miR-22 Modulates Lenalidomide Activity by Counteracting MYC Addiction in Multiple Myeloma. Cancers 2021, 13, 4365. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Arbitrio, M.; Caracciolo, D.; Cordua, A.; Cuomo, O.; Grillone, K.; Riillo, C.; Carida, G.; Scionti, F.; Labanca, C.; et al. miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: A systematic review. Mol. Ther. Nucleic Acids 2022, 27, 1191–1224. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Tufekci, K.U.; Oner, M.G.; Meuwissen, R.L.; Genc, S. The role of microRNAs in human diseases. Methods Mol. Biol. 2014, 1107, 33–50. [Google Scholar] [PubMed]
- Detassis, S.; Grasso, M.; Del Vescovo, V.; Denti, M.A. microRNAs Make the Call in Cancer Personalized Medicine. Front. Cell Dev. Biol. 2017, 5, 86. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Fu, L.L.; Wen, X.; Liu, B.; Huang, J.; Wang, J.H.; Wei, Y.Q. Oncogenic and tumor suppressive roles of microRNAs in apoptosis and autophagy. Apoptosis 2014, 19, 1177–1189. [Google Scholar] [CrossRef]
- Lanza, G.; Ferracin, M.; Gafa, R.; Veronese, A.; Spizzo, R.; Pichiorri, F.; Liu, C.G.; Calin, G.A.; Croce, C.M.; Negrini, M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer 2007, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Xicola, R.M.; Bontu, S.; Doyle, B.J.; Rawson, J.; Garre, P.; Lee, E.; de la Hoya, M.; Bessa, X.; Clofent, J.; Bujanda, L.; et al. Association of a let-7 miRNA binding region of TGFBR1 with hereditary mismatch repair proficient colorectal cancer (MSS HNPCC). Carcinogenesis 2016, 37, 751–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
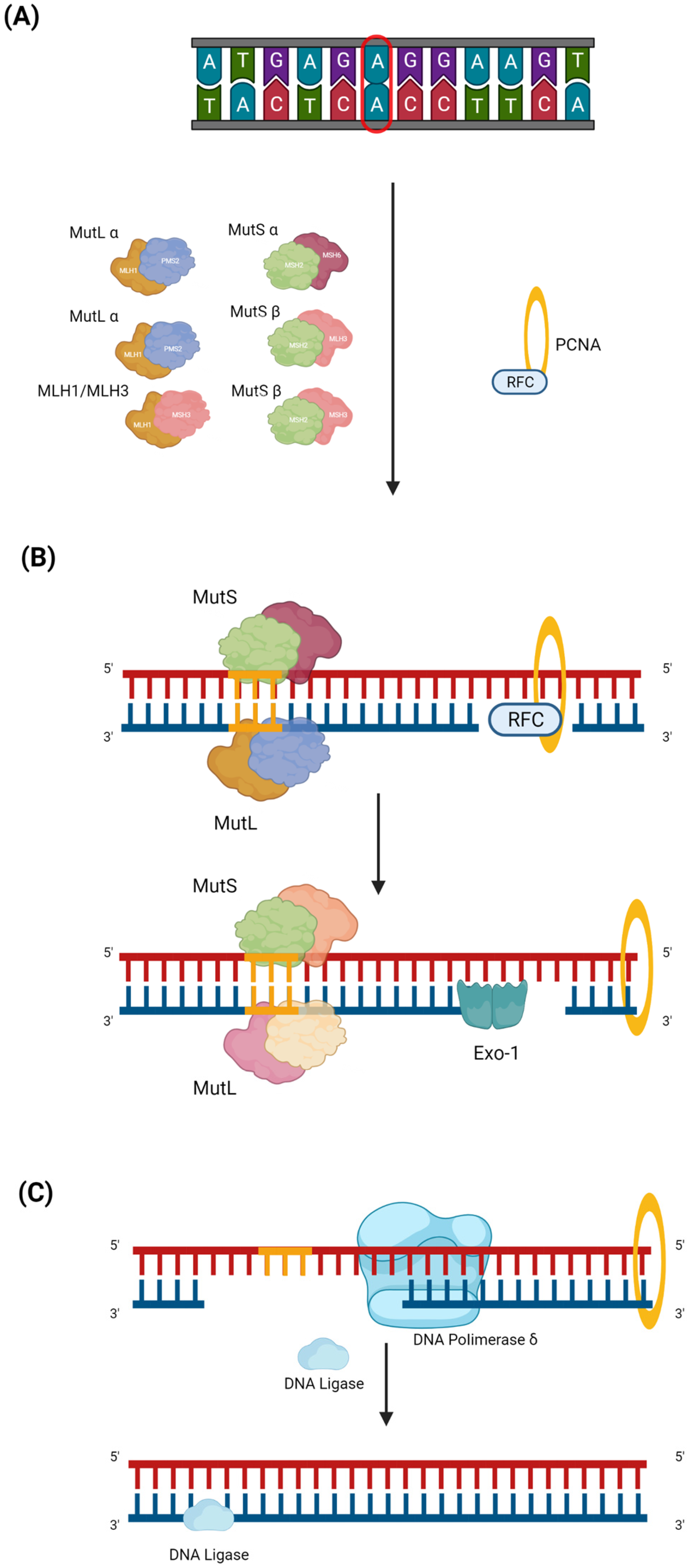
- Kunkel, T.A.; Erie, D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005, 74, 681–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rustgi, A.K. The genetics of hereditary colon cancer. Genes. Dev. 2007, 21, 2525–2538. [Google Scholar] [CrossRef] [Green Version]
- De’Angelis, G.L.; Bottarelli, L.; Azzoni, C.; De’Angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018, 89, 97–101. [Google Scholar]
- Liu, D.; Keijzers, G.; Rasmussen, L.J. DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. Rev. Mutat. Res. 2017, 773, 174–187. [Google Scholar] [CrossRef]
- Edelbrock, M.A.; Kaliyaperumal, S.; Williams, K.J. DNA mismatch repair efficiency and fidelity are elevated during DNA synthesis in human cells. Mutat. Res. 2009, 662, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Chen, L.; Li, G.M. DNA mismatch repair in trinucleotide repeat instability. Sci. China Life Sci. 2017, 60, 1087–1092. [Google Scholar] [CrossRef]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef]
- Dowty, J.G.; Win, A.K.; Buchanan, D.D.; Lindor, N.M.; Macrae, F.A.; Clendenning, M.; Antill, Y.C.; Thibodeau, S.N.; Casey, G.; Gallinger, S.; et al. Cancer risks for MLH1 and MSH2 mutation carriers. Hum. Mutat. 2013, 34, 490–497. [Google Scholar] [CrossRef] [Green Version]
- Palomaki, G.E.; McClain, M.R.; Melillo, S.; Hampel, H.L.; Thibodeau, S.N. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet. Med. 2009, 11, 42–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempers, M.J.; Kuiper, R.P.; Ockeloen, C.W.; Chappuis, P.O.; Hutter, P.; Rahner, N.; Schackert, H.K.; Steinke, V.; Holinski-Feder, E.; Morak, M.; et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: A cohort study. Lancet Oncol. 2011, 12, 49–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissman, S.M.; Burt, R.; Church, J.; Erdman, S.; Hampel, H.; Holter, S.; Jasperson, K.; Kalady, M.F.; Haidle, J.L.; Lynch, H.T.; et al. Identification of individuals at risk for Lynch syndrome using targeted evaluations and genetic testing: National Society of Genetic Counselors and the Collaborative Group of the Americas on Inherited Colorectal Cancer joint practice guideline. J. Genet. Couns. 2012, 21, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531. [Google Scholar] [CrossRef] [Green Version]
- Seth, S.; Ager, A.; Arends, M.J.; Frayling, I.M. Lynch syndrome—Cancer pathways, heterogeneity and immune escape. J. Pathol. 2018, 246, 129–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonadona, V.; Bonaiti, B.; Olschwang, S.; Grandjouan, S.; Huiart, L.; Longy, M.; Guimbaud, R.; Buecher, B.; Bignon, Y.J.; Caron, O.; et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011, 305, 2304–2310. [Google Scholar] [CrossRef] [Green Version]
- Senter, L.; Clendenning, M.; Sotamaa, K.; Hampel, H.; Green, J.; Potter, J.D.; Lindblom, A.; Lagerstedt, K.; Thibodeau, S.N.; Lindor, N.M.; et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 2008, 135, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Lynch, H.T.; Riegert-Johnson, D.L.; Snyder, C.; Lynch, J.F.; Hagenkord, J.; Boland, C.R.; Rhees, J.; Thibodeau, S.N.; Boardman, L.A.; Davies, J.; et al. Lynch syndrome-associated extracolonic tumors are rare in two extended families with the same EPCAM deletion. Am. J. Gastroenterol. 2011, 106, 1829–1836. [Google Scholar] [CrossRef] [Green Version]
- Grandval, P.; Baert-Desurmont, S.; Bonnet, F.; Bronner, M.; Buisine, M.P.; Colas, C.; Noguchi, T.; North, M.O.; Rey, J.M.; Tinat, J.; et al. Colon-specific phenotype in Lynch syndrome associated with EPCAM deletion. Clin. Genet. 2012, 82, 97–99. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, D.M.; Laudanna, C.; Migliozzi, S.; Zoppoli, P.; Santamaria, G.; Grillone, K.; Elia, L.; Mignogna, C.; Biamonte, F.; Sacco, R.; et al. Identification of different mutational profiles in cancers arising in specific colon segments by next generation sequencing. Oncotarget 2018, 9, 23960–23974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurgelun, M.B.; Goel, A.; Hornick, J.L.; Sen, A.; Turgeon, D.K.; Ruffin, M.T.t.; Marcon, N.E.; Baron, J.A.; Bresalier, R.S.; Syngal, S.; et al. Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev. Res. 2012, 5, 574–582. [Google Scholar] [CrossRef] [Green Version]
- Ahadova, A.; Gallon, R.; Gebert, J.; Ballhausen, A.; Endris, V.; Kirchner, M.; Stenzinger, A.; Burn, J.; von Knebel Doeberitz, M.; Blaker, H.; et al. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int. J. Cancer 2018, 143, 139–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloor, M.; Huth, C.; Voigt, A.Y.; Benner, A.; Schirmacher, P.; von Knebel Doeberitz, M.; Blaker, H. Prevalence of mismatch repair-deficient crypt foci in Lynch syndrome: A pathological study. Lancet Oncol. 2012, 13, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Ahadova, A.; von Knebel Doeberitz, M.; Blaker, H.; Kloor, M. CTNNB1-mutant colorectal carcinomas with immediate invasive growth: A model of interval cancers in Lynch syndrome. Fam. Cancer 2016, 15, 579–586. [Google Scholar] [CrossRef]
- Moller, P.; Seppala, T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Evans, D.G.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.; et al. Incidence of and survival after subsequent cancers in carriers of pathogenic MMR variants with previous cancer: A report from the prospective Lynch syndrome database. Gut 2017, 66, 1657–1664. [Google Scholar] [CrossRef] [Green Version]
- Schwitalle, Y.; Kloor, M.; Eiermann, S.; Linnebacher, M.; Kienle, P.; Knaebel, H.P.; Tariverdian, M.; Benner, A.; von Knebel Doeberitz, M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology 2008, 134, 988–997. [Google Scholar] [CrossRef]
- Barrow, E.; Hill, J.; Evans, D.G. Cancer risk in Lynch Syndrome. Fam. Cancer 2013, 12, 229–240. [Google Scholar] [CrossRef]
- Win, A.K.; Lindor, N.M.; Young, J.P.; Macrae, F.A.; Young, G.P.; Williamson, E.; Parry, S.; Goldblatt, J.; Lipton, L.; Winship, I.; et al. Risks of primary extracolonic cancers following colorectal cancer in lynch syndrome. J. Natl. Cancer Inst. 2012, 104, 1363–1372. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.E.; Jackson, S.A.; Susswein, L.R.; Zeinomar, N.; Ma, X.; Marshall, M.L.; Stettner, A.R.; Milewski, B.; Xu, Z.; Solomon, B.D.; et al. MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet. Med. 2018, 20, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Vasen, H.F. Clinical description of the Lynch syndrome [hereditary nonpolyposis colorectal cancer (HNPCC)]. Fam. Cancer 2005, 4, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Kalady, M.F.; McGannon, E.; Vogel, J.D.; Manilich, E.; Fazio, V.W.; Church, J.M. Risk of colorectal adenoma and carcinoma after colectomy for colorectal cancer in patients meeting Amsterdam criteria. Ann. Surg. 2010, 252, 507–511; discussion 11-3. [Google Scholar] [CrossRef] [Green Version]
- Ten Broeke, S.W.; van der Klift, H.M.; Tops, C.M.J.; Aretz, S.; Bernstein, I.; Buchanan, D.D.; de la Chapelle, A.; Capella, G.; Clendenning, M.; Engel, C.; et al. Cancer Risks for PMS2-Associated Lynch Syndrome. J. Clin. Oncol. 2018, 36, 2961–2968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurgelun, M.B.; Hampel, H. Recent Advances in Lynch Syndrome: Diagnosis, Treatment, and Cancer Prevention. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 101–109. [Google Scholar] [CrossRef]
- Vasen, H.F.; Mecklin, J.P.; Khan, P.M.; Lynch, H.T. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis. Colon Rectum 1991, 34, 424–425. [Google Scholar] [CrossRef]
- Vasen, H.F.; Watson, P.; Mecklin, J.P.; Lynch, H.T. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999, 116, 1453–1456. [Google Scholar] [CrossRef]
- Rodriguez-Bigas, M.A.; Boland, C.R.; Hamilton, S.R.; Henson, D.E.; Jass, J.R.; Khan, P.M.; Lynch, H.; Perucho, M.; Smyrk, T.; Sobin, L.; et al. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: Meeting highlights and Bethesda guidelines. J. Natl. Cancer Inst. 1997, 89, 1758–1762. [Google Scholar] [CrossRef] [Green Version]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Ruschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef]
- Kastrinos, F.; Steyerberg, E.W.; Balmana, J.; Mercado, R.; Gallinger, S.; Haile, R.; Casey, G.; Hopper, J.L.; LeMarchand, L.; Lindor, N.M.; et al. Comparison of the clinical prediction model PREMM(1,2,6) and molecular testing for the systematic identification of Lynch syndrome in colorectal cancer. Gut 2013, 62, 272–279. [Google Scholar] [CrossRef]
- Kastrinos, F.; Steyerberg, E.W.; Mercado, R.; Balmana, J.; Holter, S.; Gallinger, S.; Siegmund, K.D.; Church, J.M.; Jenkins, M.A.; Lindor, N.M.; et al. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology 2011, 140, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Wang, W.; Lee, S.; Nafa, K.; Lee, J.; Romans, K.; Watson, P.; Gruber, S.B.; Euhus, D.; Kinzler, K.W.; et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 2006, 296, 1479–1487. [Google Scholar] [CrossRef] [Green Version]
- Green, R.C.; Parfrey, P.S.; Woods, M.O.; Younghusband, H.B. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J. Natl. Cancer Inst. 2009, 101, 331–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastrinos, F.; Uno, H.; Ukaegbu, C.; Alvero, C.; McFarland, A.; Yurgelun, M.B.; Kulke, M.H.; Schrag, D.; Meyerhardt, J.A.; Fuchs, C.S.; et al. Development and Validation of the PREMM(5) Model for Comprehensive Risk Assessment of Lynch Syndrome. J. Clin. Oncol. 2017, 35, 2165–2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastrinos, F.; Ojha, R.P.; Leenen, C.; Alvero, C.; Mercado, R.C.; Balmana, J.; Valenzuela, I.; Balaguer, F.; Green, R.; Lindor, N.M.; et al. Comparison of Prediction Models for Lynch Syndrome Among Individuals With Colorectal Cancer. J. Natl. Cancer Inst. 2016, 108, djv308. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, Y.M.; de Jong, A.E.; Morreau, H.; Tops, C.M.; Vasen, H.F.; Wijnen, J.T.; Breuning, M.H.; Brocker-Vriends, A.H. Diagnostic approach and management of Lynch syndrome (hereditary nonpolyposis colorectal carcinoma): A guide for clinicians. CA Cancer J. Clin. 2006, 56, 213–225. [Google Scholar] [CrossRef]
- Guastadisegni, C.; Colafranceschi, M.; Ottini, L.; Dogliotti, E. Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur. J. Cancer 2010, 46, 2788–2798. [Google Scholar] [CrossRef]
- Tsibel, B.N.; Bochkareva, A.K. Functional morphology of adenohypophysis, thymus, and adrenal cortex in sudden infant death syndrome. Arkh. Patol. 1998, 60, 23–27. [Google Scholar]
- Adar, T.; Rodgers, L.H.; Shannon, K.M.; Yoshida, M.; Ma, T.; Mattia, A.; Lauwers, G.Y.; Iafrate, A.J.; Chung, D.C. A tailored approach to BRAF and MLH1 methylation testing in a universal screening program for Lynch syndrome. Mod. Pathol. 2017, 30, 440–447. [Google Scholar] [CrossRef] [Green Version]
- Hampel, H.; Pearlman, R.; Beightol, M.; Zhao, W.; Jones, D.; Frankel, W.L.; Goodfellow, P.J.; Yilmaz, A.; Miller, K.; Bacher, J.; et al. Assessment of Tumor Sequencing as a Replacement for Lynch Syndrome Screening and Current Molecular Tests for Patients With Colorectal Cancer. JAMA Oncol. 2018, 4, 806–813. [Google Scholar] [CrossRef]
- Haraldsdottir, S.; Hampel, H.; Tomsic, J.; Frankel, W.L.; Pearlman, R.; de la Chapelle, A.; Pritchard, C.C. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology 2014, 147, 1308–1316.e1. [Google Scholar] [CrossRef] [Green Version]
- Boland, P.M.; Yurgelun, M.B.; Boland, C.R. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 2018, 68, 217–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, J.A.; Yurgelun, M.B.; Bruce, J.L.; Rojas-Rudilla, V.; Hall, D.L.; Shivdasani, P.; Garcia, E.P.; Agoston, A.T.; Srivastava, A.; Ogino, S.; et al. Detection of Mismatch Repair Deficiency and Microsatellite Instability in Colorectal Adenocarcinoma by Targeted Next-Generation Sequencing. J. Mol. Diagn. 2017, 19, 84–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, J.M. Is breast cancer a part of Lynch syndrome? Breast Cancer Res. 2012, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Buerki, N.; Gautier, L.; Kovac, M.; Marra, G.; Buser, M.; Mueller, H.; Heinimann, K. Evidence for breast cancer as an integral part of Lynch syndrome. Genes. Chromosomes Cancer 2012, 51, 83–91. [Google Scholar] [CrossRef]
- Schwartz, C.J.; da Silva, E.M.; Marra, A.; Gazzo, A.M.; Selenica, P.; Rai, V.K.; Mandelker, D.; Pareja, F.; Misyura, M.; D’Alfonso, T.M.; et al. Morphologic and Genomic Characteristics of Breast Cancers Occurring in Individuals with Lynch Syndrome. Clin. Cancer Res. 2022, 28, 404–413. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [Green Version]
- Sajjadi, E.; Venetis, K.; Piciotti, R.; Invernizzi, M.; Guerini-Rocco, E.; Haricharan, S.; Fusco, N. Mismatch repair-deficient hormone receptor-positive breast cancers: Biology and pathological characterization. Cancer Cell Int. 2021, 21, 266. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [Green Version]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [Green Version]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Vergani-Junior, C.A.; Tonon-da-Silva, G.; Inan, M.D.; Mori, M.A. DICER: Structure, function, and regulation. Biophys. Rev. 2021, 13, 1081–1090. [Google Scholar] [CrossRef]
- Hock, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef]
- Jo, M.H.; Song, J.J.; Hohng, S. Single-molecule fluorescence measurements reveal the reaction mechanisms of the core RISC, composed of human Argonaute 2 and a guide RNA. BMB Rep. 2015, 48, 643–644. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; San Lucas, A.; Wang, Z.; Liu, Y. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014, 15 (Suppl. 7), S4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhou, W.; Liu, Y.; Liu, T.; Li, C.; Wang, L. Oncogenic role of microRNA-532-5p in human colorectal cancer via targeting of the 5′UTR of RUNX3. Oncol. Lett. 2018, 15, 7215–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Li, L.; Wang, D.; Zhang, C.Y.; Zen, K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014, 289, 10270–10275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottini, S.; Hamouda-Tekaya, N.; Mategot, R.; Zaragosi, L.E.; Audebert, S.; Pisano, S.; Grandjean, V.; Mauduit, C.; Benahmed, M.; Barbry, P.; et al. Post-transcriptional gene silencing mediated by microRNAs is controlled by nucleoplasmic Sfpq. Nat. Commun. 2017, 8, 1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allo, M.; Agirre, E.; Bessonov, S.; Bertucci, P.; Gomez Acuna, L.; Buggiano, V.; Bellora, N.; Singh, B.; Petrillo, E.; Blaustein, M.; et al. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 15622–15629. [Google Scholar] [CrossRef]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef] [Green Version]
- Weitz, J.; Koch, M.; Debus, J.; Hohler, T.; Galle, P.R.; Buchler, M.W. Colorectal cancer. Lancet 2005, 365, 153–165. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Zhang, N.; Hu, X.; Du, Y.; Du, J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed. Pharmacother. 2021, 134, 111099. [Google Scholar] [CrossRef] [PubMed]
- Bastaminejad, S.; Taherikalani, M.; Ghanbari, R.; Akbari, A.; Shabab, N.; Saidijam, M. Investigation of MicroRNA-21 Expression Levels in Serum and Stool as a Potential Non-Invasive Biomarker for Diagnosis of Colorectal Cancer. Iran. Biomed. J. 2017, 21, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.O.; Senda, T. Circulating microRNA-92a-3p in colorectal cancer: A review. Med. Mol. Morphol. 2021, 54, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Faltejskova, P.; Bocanek, O.; Sachlova, M.; Svoboda, M.; Kiss, I.; Vyzula, R.; Slaby, O. Circulating miR-17-3p, miR-29a, miR-92a and miR-135b in serum: Evidence against their usage as biomarkers in colorectal cancer. Cancer Biomark. 2012, 12, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Fellizar, A.; Refuerzo, V.; Ramos, J.D.; Albano, P.M. Expression of specific microRNAs in tissue and plasma in colorectal cancer. J. Pathol. Transl. Med. 2023, 57, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.P.; Ma, A. The ratio of miR-21/miR-24 as a promising diagnostic and poor prognosis biomarker in colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8649–8656. [Google Scholar]
- Zdralevic, M.; Raonic, J.; Popovic, N.; Vuckovic, L.; Rovcanin Dragovic, I.; Vukcevic, B.; Todorovic, V.; Vukmirovic, F.; Marzano, F.; Tullo, A.; et al. The role of miRNA in colorectal cancer diagnosis: A pilot study. Oncol. Lett. 2023, 25, 267. [Google Scholar] [CrossRef]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef]
- Jung, E.; Choi, J.; Kim, J.S.; Han, T.S. MicroRNA-Based Therapeutics for Drug-Resistant Colorectal Cancer. Pharmaceuticals 2021, 14, 136. [Google Scholar] [CrossRef]
- Hampel, H.; Frankel, W.L.; Martin, E.; Arnold, M.; Khanduja, K.; Kuebler, P.; Nakagawa, H.; Sotamaa, K.; Prior, T.W.; Westman, J.; et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N. Engl. J. Med. 2005, 352, 1851–1860. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Adachi, Y.; Taniguchi, H.; Kunimoto, H.; Nosho, K.; Suzuki, H.; Shinomura, Y. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J. Gastroenterol. 2012, 18, 2745–2755. [Google Scholar] [CrossRef]
- Hrasovec, S.; Glavac, D. MicroRNAs as Novel Biomarkers in Colorectal Cancer. Front. Genet. 2012, 3, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaguer, F.; Moreira, L.; Lozano, J.J.; Link, A.; Ramirez, G.; Shen, Y.; Cuatrecasas, M.; Arnold, M.; Meltzer, S.J.; Syngal, S.; et al. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin. Cancer Res. 2011, 17, 6239–6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sanchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velazquez, I.A.; Gonzalez-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manriquez, R.; Castro-Hernandez, C.; Fragoso-Ontiveros, V.; Alvarez-Gomez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids. 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Valeri, N.; Gasparini, P.; Braconi, C.; Paone, A.; Lovat, F.; Fabbri, M.; Sumani, K.M.; Alder, H.; Amadori, D.; Patel, T.; et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. USA 2010, 107, 21098–21103. [Google Scholar] [CrossRef]
- Sun, G.; Ye, P.; Murai, K.; Lang, M.F.; Li, S.; Zhang, H.; Li, W.; Fu, C.; Yin, J.; Wang, A.; et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2011, 2, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandres, E.; Agirre, X.; Bitarte, N.; Ramirez, N.; Zarate, R.; Roman-Gomez, J.; Prosper, F.; Garcia-Foncillas, J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer. 2009, 125, 2737–2743. [Google Scholar] [CrossRef]
- Balaguer, F.; Link, A.; Lozano, J.J.; Cuatrecasas, M.; Nagasaka, T.; Boland, C.R.; Goel, A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010, 70, 6609–6618. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Li, J.; Li, J.; Wan, Y.; Li, T.; Ma, P.; Wang, Y.; Sang, H. Hsa-miR-137, hsa-miR-520e and hsa-miR-590-3p perform crucial roles in Lynch syndrome. Oncol. Lett. 2016, 12, 2011–2017. [Google Scholar] [CrossRef] [Green Version]
- Liccardo, R.; Nolano, A.; Lambiase, M.; Della Ragione, C.; De Rosa, M.; Izzo, P.; Duraturo, F. MSH2 Overexpression Due to an Unclassified Variant in 3′-Untranslated Region in a Patient with Colon Cancer. Biomedicines 2020, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Liccardo, R.; Sessa, R.; Trombetti, S.; De Rosa, M.; Izzo, P.; Grosso, M.; Duraturo, F. MiR-137 Targets the 3′ Untranslated Region of MSH2: Potential Implications in Lynch Syndrome-Related Colorectal Cancer. Cancers 2021, 13, 4662. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta 2009, 1792, 497–505. [Google Scholar] [CrossRef]
- Amodio, N.; Gallo Cantafio, M.E.; Botta, C.; Agosti, V.; Federico, C.; Caracciolo, D.; Ronchetti, D.; Rossi, M.; Driessen, C.; Neri, A.; et al. Replacement of miR-155 Elicits Tumor Suppressive Activity and Antagonizes Bortezomib Resistance in Multiple Myeloma. Cancers 2019, 11, 236. [Google Scholar] [CrossRef] [Green Version]
- Valeri, N.; Gasparini, P.; Fabbri, M.; Braconi, C.; Veronese, A.; Lovat, F.; Adair, B.; Vannini, I.; Fanini, F.; Bottoni, A.; et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA 2010, 107, 6982–6987. [Google Scholar] [CrossRef]
- Kaur, S.; Lotsari, J.E.; Al-Sohaily, S.; Warusavitarne, J.; Kohonen-Corish, M.R.; Peltomaki, P. Identification of subgroup-specific miRNA patterns by epigenetic profiling of sporadic and Lynch syndrome-associated colorectal and endometrial carcinoma. Clin. Epigenet. 2015, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.B.; Luo, H.P.; Shi, Q.; Hao, Z.N.; Ding, Y.; Wang, Q.S.; Li, S.B.; Xiao, G.C.; Tong, S.L. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J. Gastroenterol. 2014, 20, 6515–6522. [Google Scholar] [CrossRef] [PubMed]
- Mokutani, Y.; Uemura, M.; Munakata, K.; Okuzaki, D.; Haraguchi, N.; Takahashi, H.; Nishimura, J.; Hata, T.; Murata, K.; Takemasa, I.; et al. Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer. Ann. Surg. Oncol. 2016, 23 (Suppl. 5), 599–608. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Xiang, G.; Duan, X.; Wang, H.; He, K.; Xiao, J. MiR-132-3p inhibits proliferation, invasion and migration of colorectal cancer cells via down-regulating FOXP2 expression. Acta Biochim. Pol. 2022, 69, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.T.; Wang, J.L.; Du, W.; Hong, J.; Zhao, S.L.; Wang, Y.C.; Xiong, H.; Chen, H.M.; Fang, J.Y. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis 2011, 32, 1207–1215. [Google Scholar] [CrossRef] [Green Version]
- Rezayi Soufiani, A.; Dolatkhah, R.; Raeisi, M.; Chavoshi, H.; Mohammadi, P.; Mehdinavaz Aghdam, A. Hypermethylation of MIR129-2 Regulates SOX4 Transcription and Associates with Metastasis in Patients with Colorectal Cancer. J. Gastrointest. Cancer 2022, 53, 718–724. [Google Scholar] [CrossRef]
- Earle, J.S.; Luthra, R.; Romans, A.; Abraham, R.; Ensor, J.; Yao, H.; Hamilton, S.R. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J. Mol. Diagn. 2010, 12, 433–440. [Google Scholar] [CrossRef]
- Brinzan, C.; Aschie, M.; Cozaru, G.; Dumitru, E.; Mitroi, A. The diagnostic value of miR-92a, -143, and -145 expression levels in patients with colorectal adenocarcinoma from Romania. Medicine 2020, 99, e21895. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.K.; Chong, W.W.; Jin, H.; Lam, E.K.; Shin, V.Y.; Yu, J.; Poon, T.C.; Ng, S.S.; Sung, J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Moridikia, A.; Mirzaei, H.; Sahebkar, A.; Salimian, J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J. Cell Physiol. 2018, 233, 901–913. [Google Scholar] [CrossRef]
- Han, H.B.; Gu, J.; Zuo, H.J.; Chen, Z.G.; Zhao, W.; Li, M.; Ji, D.B.; Lu, Y.Y.; Zhang, Z.Q. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 2012, 226, 544–555. [Google Scholar] [CrossRef]
- Khodadadi Kohlan, A.; Saidijam, M.; Amini, R.; Samadi, P.; Najafi, R. Induction of let-7e gene expression attenuates oncogenic phenotype in HCT-116 colorectal cancer cells through targeting of DCLK1 regulation. Life Sci. 2019, 228, 221–227. [Google Scholar] [CrossRef]
- Shan, Y.; Liu, Y.; Zhao, L.; Liu, B.; Li, Y.; Jia, L. MicroRNA-33a and let-7e inhibit human colorectal cancer progression by targeting ST8SIA1. Int. J. Biochem. Cell Biol. 2017, 90, 48–58. [Google Scholar] [CrossRef]
- Qin, J.; Wang, F.; Jiang, H.; Xu, J.; Jiang, Y.; Wang, Z. MicroRNA-145 suppresses cell migration and invasion by targeting paxillin in human colorectal cancer cells. Int. J. Clin. Exp. Pathol. 2015, 8, 1328–1340. [Google Scholar] [PubMed]
- Arndt, G.M.; Dossey, L.; Cullen, L.M.; Lai, A.; Druker, R.; Eisbacher, M.; Zhang, C.; Tran, N.; Fan, H.; Retzlaff, K.; et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 2009, 9, 374. [Google Scholar] [CrossRef] [Green Version]
- Fan, D.; Lin, X.; Zhang, F.; Zhong, W.; Hu, J.; Chen, Y.; Cai, Z.; Zou, Y.; He, X.; Chen, X.; et al. MicroRNA 26b promotes colorectal cancer metastasis by downregulating phosphatase and tensin homolog and wingless-type MMTV integration site family member 5A. Cancer Sci. 2018, 109, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Farouk, S.; El-Shenawy, R.; Khairy, A.M.; Bader El-Din, N.G. Overexpression of miRNA 26a and 26b with MMP-9 are valuable diagnostic biomarkers for colorectal cancer patients. Biomark. Med. 2023, 17, 159–169. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Z.; Liu, B.; Shan, Y.; Zhao, L.; Jia, L. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis. 2017, 8, e2892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Robertis, M.; Loiacono, L.; Fusilli, C.; Poeta, M.L.; Mazza, T.; Sanchez, M.; Marchionni, L.; Signori, E.; Lamorte, G.; Vescovi, A.L.; et al. Dysregulation of EGFR Pathway in EphA2 Cell Subpopulation Significantly Associates with Poor Prognosis in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 159–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Burwinkel, B.; Tao, S.; Brenner, H. MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer Epidemiol. Biomark. Prev. 2011, 20, 1272–1286. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Ma, P.; Wu, D.; Shu, Y.; Gao, W. Functions and mechanisms of microRNA-31 in human cancers. Biomed. Pharmacother. 2018, 108, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Moloudizargari, M.; Rahmani, J.; Asghari, M.H.; Goel, A. The prognostic role of miR-31 in colorectal cancer: The results of a meta-analysis of 4720 patients. Epigenomics 2022, 14, 101–112. [Google Scholar] [CrossRef]
- Zhang, W.W.; Ming, X.L.; Rong, Y.; Huang, C.Q.; Weng, H.; Chen, H.; Bian, J.M.; Wang, F.B. Diagnostic Value Investigation and Bioinformatics Analysis of miR-31 in Patients with Lymph Node Metastasis of Colorectal Cancer. Anal. Cell Pathol. 2019, 2019, 9740475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Huang, S.K.; Zhao, M.; Yang, M.; Zhong, J.L.; Gu, Y.Y.; Peng, H.; Che, Y.Q.; Huang, C.Z. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS ONE 2014, 9, e87451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Luo, X.; Li, H.; Yue, X.; Deng, L.; Cui, Y.; Lu, Y. MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol. Rep. 2014, 32, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.W.; Yang, Y.M.; Du, L.T.; Dong, Z.; Wang, L.L.; Zhang, X.; Zhou, X.J.; Zheng, G.X.; Qu, A.L.; Wang, C.X. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med. Oncol. 2014, 31, 256. [Google Scholar] [CrossRef]
- Ding, J.; Zhao, Z.; Song, J.; Luo, B.; Huang, L. MiR-223 promotes the doxorubicin resistance of colorectal cancer cells via regulating epithelial-mesenchymal transition by targeting FBXW7. Acta Biochim. Biophys. Sin. 2018, 50, 597–604. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.Y.; Chen, C.C.; Chang, Y.S.; Tsai, W.S.; You, J.F.; Lin, G.P.; Chen, T.W.; Chen, J.S.; Chan, E.C. MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget 2016, 7, 10663–10675. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, H.A.; El Amin, H.A.; Ahmed, E.S.M.; Kenawy, A.G.; El-Ebidi, A.M.; ElNakeeb, I.; Kholef, E.F.M.; Elsewify, W.A.E. Role of MicroRNA-223 and MicroRNA-182 as Novel Biomarkers in Early Detection of Colorectal Cancer. Int. J. Gen. Med. 2022, 15, 3281–3291. [Google Scholar] [CrossRef]
- Ren, D.; Lin, B.; Zhang, X.; Peng, Y.; Ye, Z.; Ma, Y.; Liang, Y.; Cao, L.; Li, X.; Li, R.; et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 2017, 8, 49807–49823. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Gu, L. The diagnostic effect of serum miR-196b as biomarker in colorectal cancer. Biomed. Rep. 2017, 6, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvinen, H.J.; Aarnio, M.; Mustonen, H.; Aktan-Collan, K.; Aaltonen, L.A.; Peltomaki, P.; De La Chapelle, A.; Mecklin, J.P. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000, 118, 829–834. [Google Scholar] [CrossRef]
- Vasen, H.F.; Blanco, I.; Aktan-Collan, K.; Gopie, J.P.; Alonso, A.; Aretz, S.; Bernstein, I.; Bertario, L.; Burn, J.; Capella, G.; et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): Recommendations by a group of European experts. Gut 2013, 62, 812–823. [Google Scholar] [CrossRef] [PubMed]
- Perrod, G.; Rahmi, G.; Cellier, C. Colorectal cancer screening in Lynch syndrome: Indication, techniques and future perspectives. Dig. Endosc. 2021, 33, 520–528. [Google Scholar] [CrossRef]
- Stoffel, E.M.; Mangu, P.B.; Gruber, S.B.; Hamilton, S.R.; Kalady, M.F.; Lau, M.W.; Lu, K.H.; Roach, N.; Limburg, P.J.; American Society of Clinical Oncology; et al. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J. Clin. Oncol. 2015, 33, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Moller, P.; Seppala, T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Evans, D.G.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.; et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: First report from the prospective Lynch syndrome database. Gut 2017, 66, 464–472. [Google Scholar] [CrossRef]
- Giardiello, F.M.; Allen, J.I.; Axilbund, J.E.; Boland, C.R.; Burke, C.A.; Burt, R.W.; Church, J.M.; Dominitz, J.A.; Johnson, D.A.; Kaltenbach, T.; et al. Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology 2014, 147, 502–526. [Google Scholar] [CrossRef] [Green Version]
- Sekine, S.; Mori, T.; Ogawa, R.; Tanaka, M.; Yoshida, H.; Taniguchi, H.; Nakajima, T.; Sugano, K.; Yoshida, T.; Kato, M.; et al. Mismatch repair deficiency commonly precedes adenoma formation in Lynch Syndrome-Associated colorectal tumorigenesis. Mod. Pathol. 2017, 30, 1144–1151. [Google Scholar] [CrossRef] [Green Version]
- Auranen, A.; Joutsiniemi, T. A systematic review of gynecological cancer surveillance in women belonging to hereditary nonpolyposis colorectal cancer (Lynch syndrome) families. Acta Obstet. Gynecol. Scand. 2011, 90, 437–444. [Google Scholar] [CrossRef]
- Kastrinos, F.; Mukherjee, B.; Tayob, N.; Wang, F.; Sparr, J.; Raymond, V.M.; Bandipalliam, P.; Stoffel, E.M.; Gruber, S.B.; Syngal, S. Risk of pancreatic cancer in families with Lynch syndrome. JAMA 2009, 302, 1790–1795. [Google Scholar] [CrossRef]
- Canto, M.I.; Harinck, F.; Hruban, R.H.; Offerhaus, G.J.; Poley, J.W.; Kamel, I.; Nio, Y.; Schulick, R.S.; Bassi, C.; Kluijt, I.; et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 2013, 62, 339–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haanstra, J.F.; Al-Toma, A.; Dekker, E.; Vanhoutvin, S.A.; Nagengast, F.M.; Mathus-Vliegen, E.M.; van Leerdam, M.E.; de Vos tot Nederveen Cappel, W.H.; Sanduleanu, S.; Veenendaal, R.A.; et al. Prevalence of small-bowel neoplasia in Lynch syndrome assessed by video capsule endoscopy. Gut 2015, 64, 1578–1583. [Google Scholar] [CrossRef]
- Sankila, R.; Aaltonen, L.A.; Jarvinen, H.J.; Mecklin, J.P. Better survival rates in patients with MLH1-associated hereditary colorectal cancer. Gastroenterology 1996, 110, 682–687. [Google Scholar] [CrossRef]
- Roudko, V.; Cimen Bozkus, C.; Greenbaum, B.; Lucas, A.; Samstein, R.; Bhardwaj, N. Lynch Syndrome and MSI-H Cancers: From Mechanisms to “Off-The-Shelf” Cancer Vaccines. Front. Immunol. 2021, 12, 757804. [Google Scholar] [CrossRef] [PubMed]
- Laghi, L.; Malesci, A. Microsatellite instability and therapeutic consequences in colorectal cancer. Dig. Dis. 2012, 30, 304–309. [Google Scholar] [CrossRef]
- Diao, Z.; Han, Y.; Chen, Y.; Zhang, R.; Li, J. The clinical utility of microsatellite instability in colorectal cancer. Crit. Rev. Oncol. Hematol. 2021, 157, 103171. [Google Scholar] [CrossRef] [PubMed]
- Hewish, M.; Lord, C.J.; Martin, S.A.; Cunningham, D.; Ashworth, A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat. Rev. Clin. Oncol. 2010, 7, 197–208. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Martin, S.T.; Winter, D.C. Segmental vs extended colectomy in the management of hereditary nonpolyposis colorectal cancer: A systematic review and meta-analysis. Color. Dis. 2015, 17, 382–389. [Google Scholar] [CrossRef]
- Mathers, J.C.; Movahedi, M.; Macrae, F.; Mecklin, J.P.; Moeslein, G.; Olschwang, S.; Eccles, D.; Evans, G.; Maher, E.R.; Bertario, L.; et al. Long-term effect of resistant starch on cancer risk in carriers of hereditary colorectal cancer: An analysis from the CAPP2 randomised controlled trial. Lancet Oncol. 2012, 13, 1242–1249. [Google Scholar] [CrossRef]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef] [Green Version]
- Benson, A.B., 3rd; Venook, A.P.; Cederquist, L.; Chan, E.; Chen, Y.J.; Cooper, H.S.; Deming, D.; Engstrom, P.F.; Enzinger, P.C.; Fichera, A.; et al. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2017, 15, 370–398. [Google Scholar] [CrossRef]
- Zaanan, A.; Shi, Q.; Taieb, J.; Alberts, S.R.; Meyers, J.P.; Smyrk, T.C.; Julie, C.; Zawadi, A.; Tabernero, J.; Mini, E.; et al. Role of Deficient DNA Mismatch Repair Status in Patients With Stage III Colon Cancer Treated With FOLFOX Adjuvant Chemotherapy: A Pooled Analysis From 2 Randomized Clinical Trials. JAMA Oncol. 2018, 4, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Andre, T.; de Gramont, A.; Vernerey, D.; Chibaudel, B.; Bonnetain, F.; Tijeras-Raballand, A.; Scriva, A.; Hickish, T.; Tabernero, J.; Van Laethem, J.L.; et al. Adjuvant Fluorouracil, Leucovorin, and Oxaliplatin in Stage II to III Colon Cancer: Updated 10-Year Survival and Outcomes According to BRAF Mutation and Mismatch Repair Status of the MOSAIC Study. J. Clin. Oncol. 2015, 33, 4176–4187. [Google Scholar] [CrossRef] [PubMed]
- Tougeron, D.; Mouillet, G.; Trouilloud, I.; Lecomte, T.; Coriat, R.; Aparicio, T.; Des Guetz, G.; Lecaille, C.; Artru, P.; Sickersen, G.; et al. Efficacy of Adjuvant Chemotherapy in Colon Cancer With Microsatellite Instability: A Large Multicenter AGEO Study. J. Natl. Cancer Inst. 2016, 108, djv438. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Spira, A.; Disis, M.L.; Schiller, J.T.; Vilar, E.; Rebbeck, T.R.; Bejar, R.; Ideker, T.; Arts, J.; Yurgelun, M.B.; Mesirov, J.P.; et al. Leveraging premalignant biology for immune-based cancer prevention. Proc. Natl. Acad. Sci. USA 2016, 113, 10750–10758. [Google Scholar] [CrossRef] [PubMed]
- Kloor, M.; von Knebel Doeberitz, M. The Immune Biology of Microsatellite-Unstable Cancer. Trends Cancer 2016, 2, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Stigliano, V.; Assisi, D.; Cosimelli, M.; Palmirotta, R.; Giannarelli, D.; Mottolese, M.; Mete, L.S.; Mancini, R.; Casale, V. Survival of hereditary non-polyposis colorectal cancer patients compared with sporadic colorectal cancer patients. J. Exp. Clin. Cancer Res. 2008, 27, 39. [Google Scholar] [CrossRef] [Green Version]
- Buckowitz, A.; Knaebel, H.P.; Benner, A.; Blaker, H.; Gebert, J.; Kienle, P.; von Knebel Doeberitz, M.; Kloor, M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br. J. Cancer 2005, 92, 1746–1753. [Google Scholar] [CrossRef] [Green Version]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Maby, P.; Tougeron, D.; Hamieh, M.; Mlecnik, B.; Kora, H.; Bindea, G.; Angell, H.K.; Fredriksen, T.; Elie, N.; Fauquembergue, E.; et al. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 2015, 75, 3446–3455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Boussiotis, V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N. Engl. J. Med. 2016, 375, 1767–1778. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Li, L.; Jiang, X.; Li, Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J. Hematol. Oncol. 2019, 12, 54. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): Final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022, 23, 659–670. [Google Scholar] [CrossRef]
- Andre, T.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Nivolumab plus low-dose ipilimumab in previously treated patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: 4-year follow-up from CheckMate 142. Ann. Oncol. 2022, 33, 1052–1060. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, T.; Cai, X.; Dong, J.; Xia, C.; Zhou, Y.; Ding, R.; Yang, R.; Tan, J.; Zhang, L.; et al. Neoadjuvant Immunotherapy for MSI-H/dMMR Locally Advanced Colorectal Cancer: New Strategies and Unveiled Opportunities. Front. Immunol. 2022, 13, 795972. [Google Scholar] [CrossRef]
- Rousseau, B.; Foote, M.B.; Maron, S.B.; Diplas, B.H.; Lu, S.; Argiles, G.; Cercek, A.; Diaz, L.A., Jr. The Spectrum of Benefit from Checkpoint Blockade in Hypermutated Tumors. N. Engl. J. Med. 2021, 384, 1168–1170. [Google Scholar] [CrossRef]
- Lanzi, A.; Pages, F.; Lagorce-Pages, C.; Galon, J. The consensus immunoscore: Toward a new classification of colorectal cancer. Oncoimmunology 2020, 9, 1789032. [Google Scholar] [CrossRef]
- von Knebel Doeberitz, M.; Kloor, M. Towards a vaccine to prevent cancer in Lynch syndrome patients. Fam. Cancer 2013, 12, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Kloor, M.; Reuschenbach, M.; Pauligk, C.; Karbach, J.; Rafiyan, M.R.; Al-Batran, S.E.; Tariverdian, M.; Jager, E.; von Knebel Doeberitz, M. A Frameshift Peptide Neoantigen-Based Vaccine for Mismatch Repair-Deficient Cancers: A Phase I/IIa Clinical Trial. Clin. Cancer Res. 2020, 26, 4503–4510. [Google Scholar] [CrossRef]
- Houot, R.; Schultz, L.M.; Marabelle, A.; Kohrt, H. T-cell-based Immunotherapy: Adoptive Cell Transfer and Checkpoint Inhibition. Cancer Immunol. Res. 2015, 3, 1115–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Martino, M.T.; Riillo, C.; Scionti, F.; Grillone, K.; Polera, N.; Caracciolo, D.; Arbitrio, M.; Tagliaferri, P.; Tassone, P. miRNAs and lncRNAs as Novel Therapeutic Targets to Improve Cancer Immunotherapy. Cancers 2021, 13, 1587. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Wang, Z.; Lu, Z.; Xia, J.; Xie, Z.; Jiao, M.; Liu, R.; Chu, Y. MicroRNAs: Immune modulators in cancer immunotherapy. Immunother. Adv. 2021, 1, ltab006. [Google Scholar]
- Szczepanek, J.; Skorupa, M.; Tretyn, A. MicroRNA as a Potential Therapeutic Molecule in Cancer. Cells. 2022, 11, 1008. [Google Scholar] [CrossRef]
- Shin, V.Y.; Chu, K.M. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J. Gastroenterol. 2014, 20, 10432–10439. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; Kerin, M.J. MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 2010, 10, 543–550. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Swarup, V.; Rajeswari, M.R. Circulating (cell-free) nucleic acids--a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef] [Green Version]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievanen, T.; Korhonen, T.M.; Jokela, T.; Ahtiainen, M.; Lahtinen, L.; Kuopio, T.; Lepisto, A.; Sillanpaa, E.; Mecklin, J.P.; Seppala, T.T.; et al. Systemic circulating microRNA landscape in Lynch syndrome. Int. J. Cancer 2023, 152, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Mori, M. MicroRNAs as Therapeutic Targets and Colorectal Cancer Therapeutics. Adv. Exp. Med. Biol. 2016, 937, 239–247. [Google Scholar] [PubMed]
- Lin, P.L.; Wu, D.W.; Huang, C.C.; He, T.Y.; Chou, M.C.; Sheu, G.T.; Lee, H. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of beta-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis 2014, 35, 2175–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Liu, D.; Guan, S.; Dong, M. Diagnostic role of circulating MiR-21 in colorectal cancer: A update meta-analysis. Ann. Med. 2021, 53, 87–102. [Google Scholar] [CrossRef]
- Hao, Y.J.; Yang, C.Y.; Chen, M.H.; Chang, L.W.; Lin, C.P.; Lo, L.C.; Huang, S.C.; Lyu, Y.Y.; Jiang, J.K.; Tseng, F.G. Potential Values of Circulating microRNA-21 to Predict Early Recurrence in Patients with Colorectal Cancer after Treatments. J. Clin. Med. 2022, 11, 2400. [Google Scholar] [CrossRef]
- Cheong, J.K.; Tang, Y.C.; Zhou, L.; Cheng, H.; Too, H.P. Advances in quantifying circulatory microRNA for early disease detection. Curr. Opin. Biotechnol. 2022, 74, 256–262. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.; Winkler, M. The promise of microRNA replacement therapy. Cancer Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef] [Green Version]
- Seto, A.G. The road toward microRNA therapeutics. Int. J. Biochem. Cell Biol. 2010, 42, 1298–1305. [Google Scholar] [CrossRef]
- Tassone, P.; Di Martino, M.T.; Arbitrio, M.; Fiorillo, L.; Staropoli, N.; Ciliberto, D.; Cordua, A.; Scionti, F.; Bertucci, B.; Salvino, A.; et al. Safety and activity of the first-in-class locked nucleic acid (LNA) miR-221 selective inhibitor in refractory advanced cancer patients: A first-in-human, phase 1, open-label, dose-escalation study. J. Hematol. Oncol. 2023, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xu, W.; Gong, F.; Chi, B.; Chen, J.; Zhou, L. MicroRNA-155 regulates the proliferation, cell cycle, apoptosis and migration of colon cancer cells and targets CBL. Exp. Ther. Med. 2017, 14, 4053–4060. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Study Title | ID | Status | Last Update Posted |
|---|---|---|---|
| Project CADENCE (CAncer Detected Early caN be CurEd) (CADENCE) | NCT05633342 | Recruiting | 20-03-2023 |
| Predictive and Prognostic Value of Inflammatory Markers and microRNA in Stage IV Colorectal Cancer | NCT04149613 | Enrolling by invitation | 20-07-2022 |
| Establishment of Molecular Classification Models for Early Diagnosis of Digestive System Cancers | NCT05431621 | Recruiting | 24-06-2022 |
| Validation of a microRNA-based Fecal (miRFec) Test for Colorectal Cancer Screening | NCT05346757 | Recruiting | 26-04-2022 |
| Timisnar-Biomarkers Substudy (Timisnar-mirna) | NCT03962088 | Recruiting | 28-07-2021 |
| microRNAs Tool for Stratifying Stage II Colon Cancer | NCT0263508 | Recruiting | 30-12-2015 |
| A 6 microRNA Tool for Stratifying Stage II Colon Cancer of Receiving Adjuvant Chemotherapy | NCT02466113 | Not yet recruiting | 09-06-2015 |
| Quantifying Micro RNA Levels of Colon (CRC) | NCT01712958 | Unknown status | 24-10-2012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ascrizzi, S.; Arillotta, G.M.; Grillone, K.; Caridà, G.; Signorelli, S.; Ali, A.; Romeo, C.; Tassone, P.; Tagliaferri, P. Lynch Syndrome Biopathology and Treatment: The Potential Role of microRNAs in Clinical Practice. Cancers 2023, 15, 3930. https://doi.org/10.3390/cancers15153930
Ascrizzi S, Arillotta GM, Grillone K, Caridà G, Signorelli S, Ali A, Romeo C, Tassone P, Tagliaferri P. Lynch Syndrome Biopathology and Treatment: The Potential Role of microRNAs in Clinical Practice. Cancers. 2023; 15(15):3930. https://doi.org/10.3390/cancers15153930
Chicago/Turabian StyleAscrizzi, Serena, Grazia Maria Arillotta, Katia Grillone, Giulio Caridà, Stefania Signorelli, Asad Ali, Caterina Romeo, Pierfrancesco Tassone, and Pierosandro Tagliaferri. 2023. "Lynch Syndrome Biopathology and Treatment: The Potential Role of microRNAs in Clinical Practice" Cancers 15, no. 15: 3930. https://doi.org/10.3390/cancers15153930
APA StyleAscrizzi, S., Arillotta, G. M., Grillone, K., Caridà, G., Signorelli, S., Ali, A., Romeo, C., Tassone, P., & Tagliaferri, P. (2023). Lynch Syndrome Biopathology and Treatment: The Potential Role of microRNAs in Clinical Practice. Cancers, 15(15), 3930. https://doi.org/10.3390/cancers15153930








