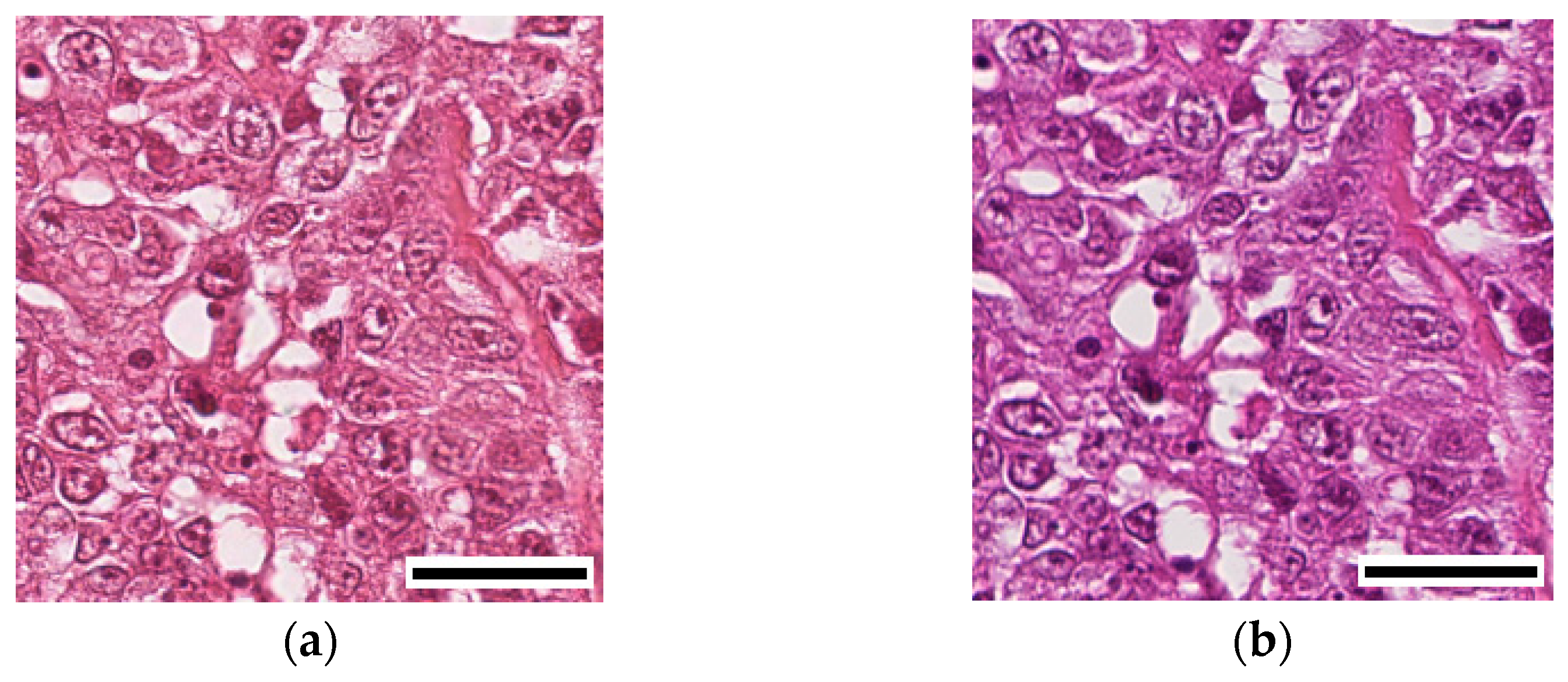
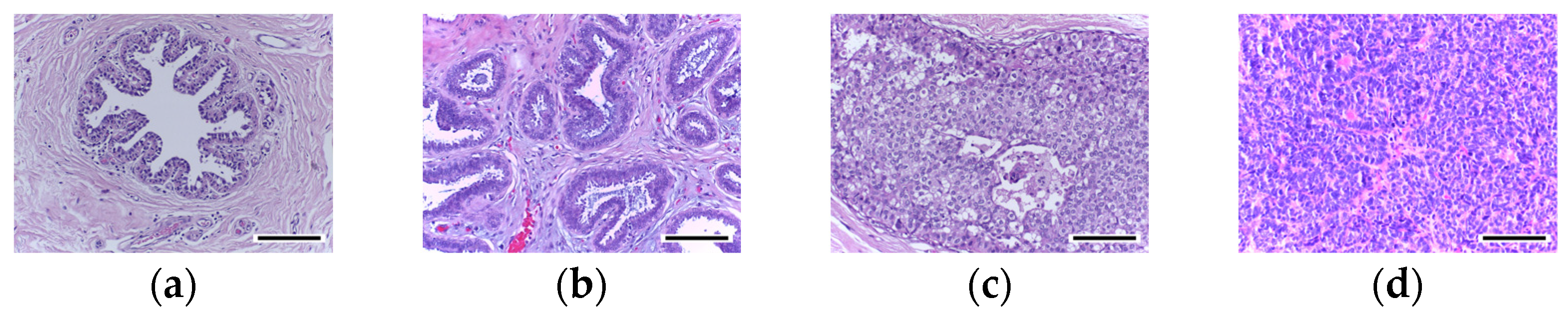
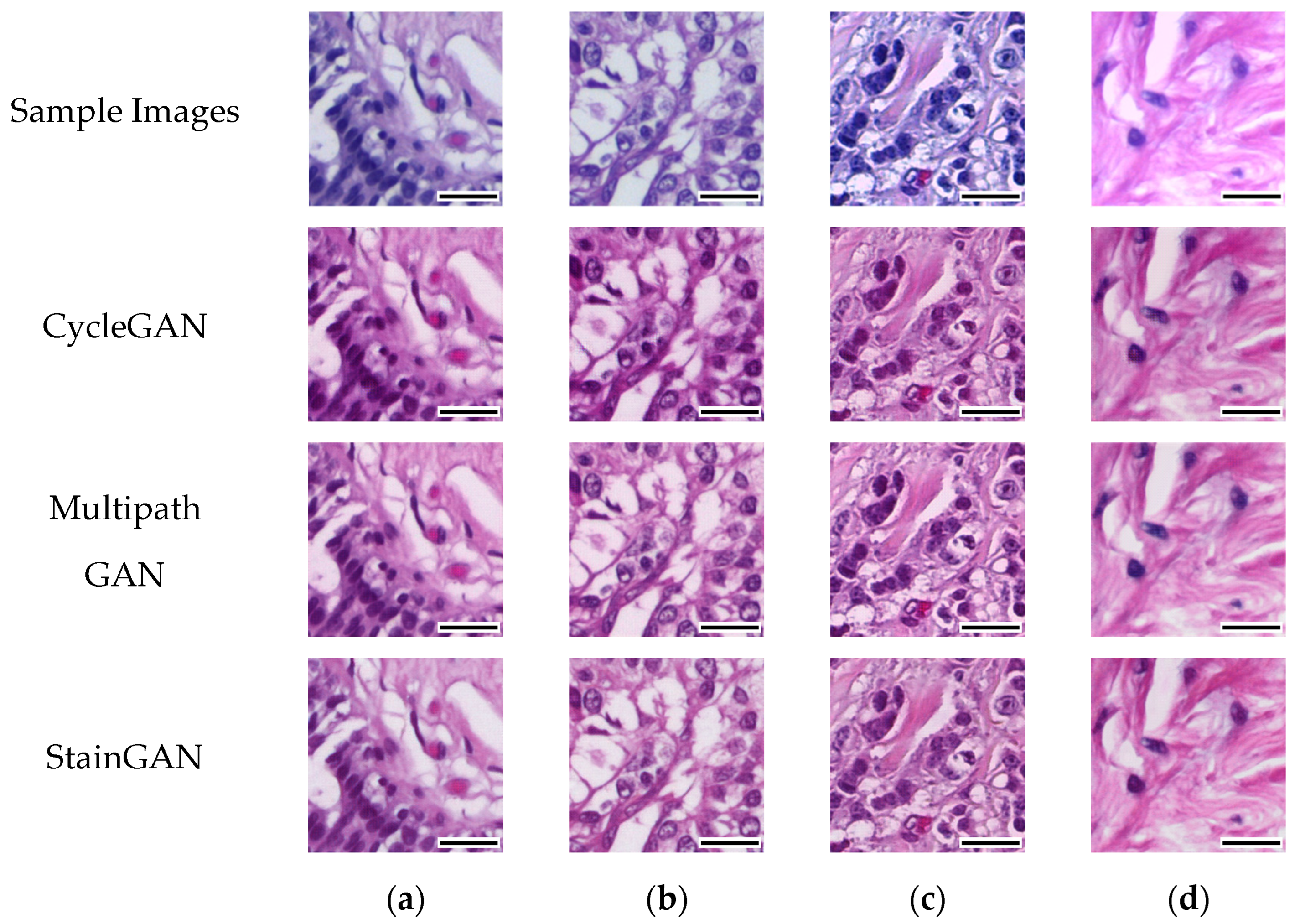
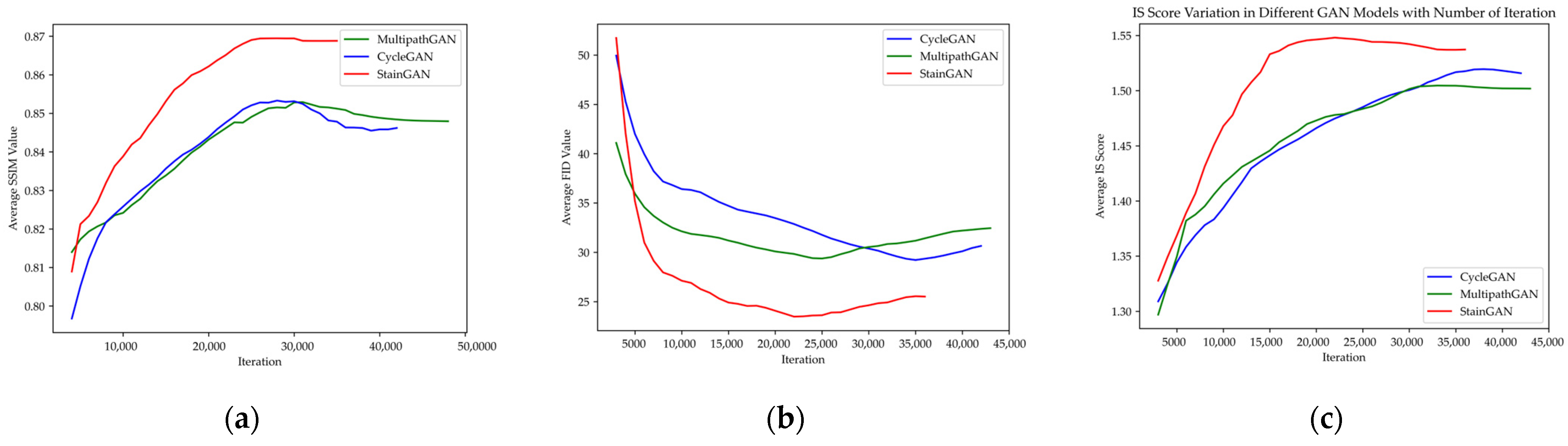
3.2. Deep Learning Model Performance in Cancer Classification
In the image classification phase, various DL models, including MobileNet, XceptionNet, InceptionV3, ResNet50, VGG16, and VGG19, were employed to classify the stain-normalized histopathological images into different cancer categories. The evaluation of the models’ performance with different image sizes used and involved metrics such as accuracy, precision, recall, and F1-score [
8,
23,
33,
34].
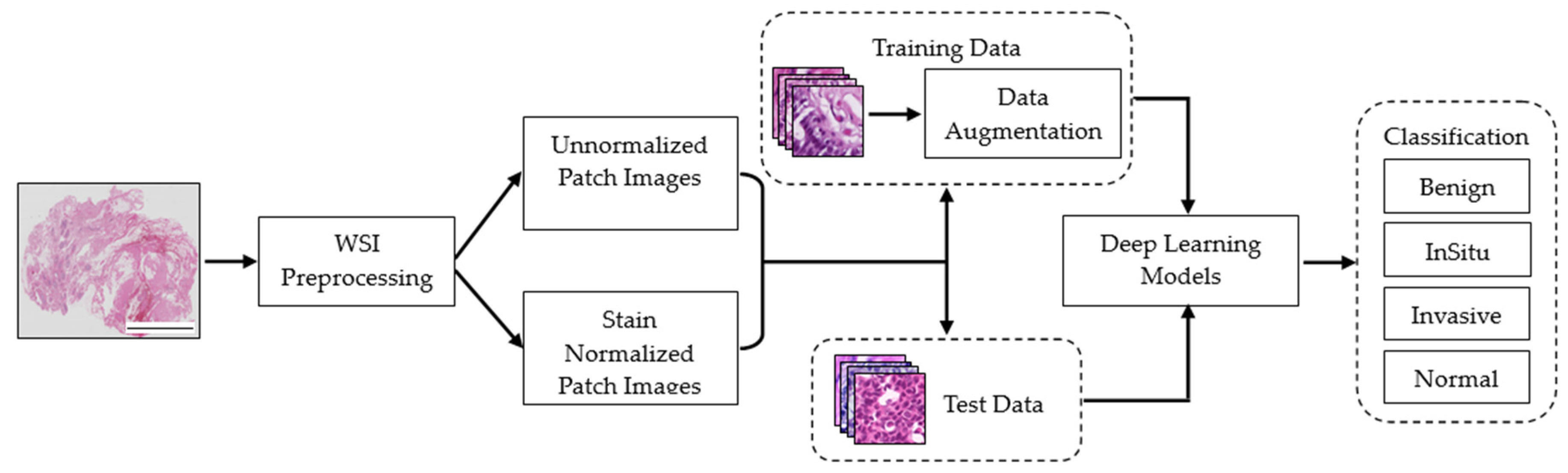
Table 3 summarizes the classification performance of different DL models on the dataset with varying image sizes (128 × 128, 256 × 256, and 512 × 512) without stain normalization. The primary objective of this table is to demonstrate the models’ effectiveness in cancer classification when stain normalization is not employed.
When analyzing the performance metrices, it is evident that VGG16 consistently achieved the highest accuracy scores across all image sizes. Specifically, it attained an accuracy of 68.22% on the 128 × 128 image size, 71.59% on the 256 × 256 image size, and 76.80% on the 512 × 512 image size. Similarly, among the models examined, Xception demonstrated strong performance, particularly excelling on the 256 × 256 and 512 × 512 image sizes with an accuracy of 68.92% and 75.25%, respectively.
In contrast, ResNet50 exhibited comparatively lower accuracy scores across all image sizes, suggesting a relatively weaker performance on this unstain-normalized dataset. These findings emphasize the influence of image size on the models’ classification performance. Notably, larger image resolutions, such as the 512 × 512 size, tend to yield higher accuracy scores, potentially augmenting the models’ ability to discern subtle patterns and features within the histopathological images.
Table 4 summarizes the performance of different DL models on the dataset with varying image sizes (128 × 128, 256 × 256, and 512 × 512) with stain normalization. The primary objective of this table is to demonstrate the models’ effectiveness in cancer classification when stain normalization is employed.
The analysis of the results from the provided
Table 4 reveals significant insights into the performance of DL models trained on stain-normalized datasets at different image sizes. Across all image sizes, VGG16 consistently achieved the highest accuracy scores on the stain-normalized dataset. It obtained accuracy values of 74.24% for the 128 × 128 image size, 85.29% for the 256 × 256 image size, and 88.64% for the 512 × 512 image size. These results demonstrate the robustness of VGG16 in accurately classifying cancer samples when stain normalization is applied. The XceptionNet exhibited strong performance on the stain-normalized dataset, particularly excelling on the 256 × 256 and 512 × 512 image sizes with accuracies of 82.92% and 85.13%, respectively. This suggests that XceptionNet is effective in capturing relevant features and patterns in histopathological images, even after stain normalization.
In contrast, ResNet50 showed relatively lower accuracy scores across all image sizes on the stain-normalized dataset, indicating its comparatively weaker performance in this context. This suggests that ResNet50 might struggle to fully exploit the benefits of stain normalization for improving classification accuracy. Examining precision, recall, and F1-score, VGG16 consistently achieved high scores across all image sizes on the stain-normalized dataset. This demonstrates that VGG16 not only achieved high accuracy but also exhibited a good balance between true positives and false positives, resulting in high precision and recall values.
The results emphasize the influence of image size on the models’ performance, when stain normalization is applied. Larger image resolutions, such as the 512 × 512 size, tend to yield higher accuracy scores, indicating the potential enhancement in capturing subtle patterns and features within stain-normalized histopathological images.
The analysis of the results presented in
Table 3 and
Table 4 unveiled a notable enhancement in the classification accuracy of the DL models upon the application of stain normalization techniques. The incorporation of stain-normalized images, which were generated using stain normalization GANs, ensured a consistent visual depiction of tissue structures across diverse samples. This consistency, in terms of color and intensity, played a crucial role in enabling the DL models to extract more meaningful features. Consequently, the models exhibited improved accuracy in cancer classification tasks. These findings emphasize the effectiveness of stain normalization in standardizing the image data and enhancing the models’ capacity to discern relevant patterns and structures, thus leading to more accurate classification outcomes.
3.3. Computational Complexity Analysis
The computational complexity analysis aimed to investigate the impact of input image sizes and batch sizes on the resource utilization of DL models used in cancer image classification. The analysis involved a comprehensive comparison of various performance metrics, including the number of parameters and image size in the DL models, processing speed in relation to both image size and batch size, FLOPs (floating-point operations per second) relative to image size, and the correlation between image size and batch size with GPU usage. This evaluation encompassed diverse DL models, each employing different input image sizes and batch sizes.
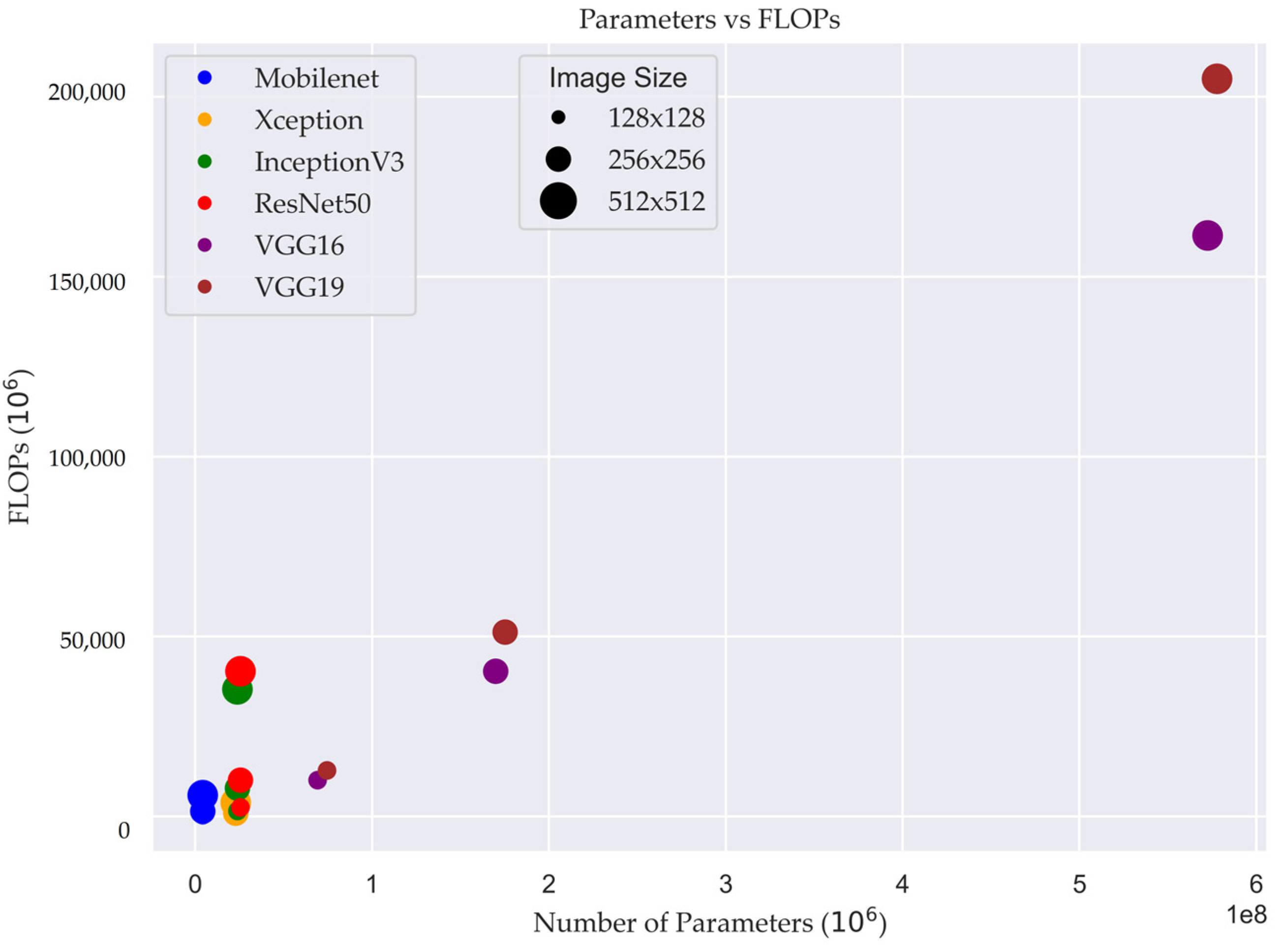
Table 5 provides information on different models, including their respective image sizes, number of parameters, and FLOPs (floating-point operations) measured in millions. The FLOPs served as a measure of computational complexity, providing insights into the computational demands of the models.
Through meticulous analysis of these performance metrics, this investigation yielded invaluable insights into the trade-offs, complexities, and resource requirements associated with DL models deployed in breast cancer image classification tasks that encompass diverse input image sizes and batch sizes.
The experiments were conducted using a computer system with specific specifications to ensure efficient execution of the DL experiments. The computer system used for these experiments was equipped with an Intel Core i5 processor running at 3.5 GHz, 64 GB of RAM, and a high-performance NVIDIA GeForce RTX 4090 graphics card.
The choice of this computer system was driven by the need for substantial computational power to handle the large-scale DL tasks involved in training and evaluating the models. The inclusion of the NVIDIA GeForce RTX 4090 graphics card ensured accelerated training and inference processes, leveraging the card’s parallel computing capabilities.
Figure 6 illustrates the relationship between the number of FLOPs and the number of parameters in different DL models. The FLOPs metric provides insights into the computational complexity of the models, reflecting the number of arithmetic operations required for processing the input data.
In
Figure 6, the size of the plot demonstrates size of input image and how the number of FLOPs changes as the number of parameters varies across different models. Each point on the graph represents a specific model configuration, with the x-axis denoting the number of parameters and the y-axis representing the corresponding number of FLOPs.
In the case of the models MobileNet, Xception, InceptionV3, and ResNet50, the number of training parameters remained constant regardless of the input image size. However, the number of floating-point operations (FLOPs) performed during model inference varied based on the input image size. This means that as the size of the input image increased, the computational workload in terms of FLOPs also increased. This insight is valuable for optimizing computational efficiency and resource allocation when utilizing these models, as it allows for better understanding of the computational requirements associated with different input sizes.
To evaluate the effect of increased image sizes on classification performance, the accuracy of each model was measured using the test dataset. Additionally, the processing speed, quantified as the number of images processed per second (IPS), was examined to identify disparities in computational efficiency. The findings of this investigation are summarized in
Table 6, offering a comprehensive overview of diverse DL models. The table presents relevant information such as image sizes, the number of images processed per second, and corresponding batch and images sizes. By conducting a comparative assessment of processing speeds across different image sizes and batch sizes, valuable insights were obtained concerning the computational efficiency and capability of each model to handle varying workloads.
The results presented in
Table 3 and
Table 4 provide evidence that increasing the image size has a positive impact on cancer classification performance of the DL models. The larger image sizes allow for capturing more detailed information, leading to improved accuracy in classification. However, it is important to consider the potential challenges associated with increasing image size, such as computational complexity and increased training and inference time.
To further investigate the impact on processing speed, we examined the relationship between batch size and image size on DL model performance. We explored how varying these parameters influenced the computational demands of the models during training and inference. By analyzing the processing speed, we aimed to identify an optimal balance between image size and batch size that maximizes both classification accuracy and computational efficiency.
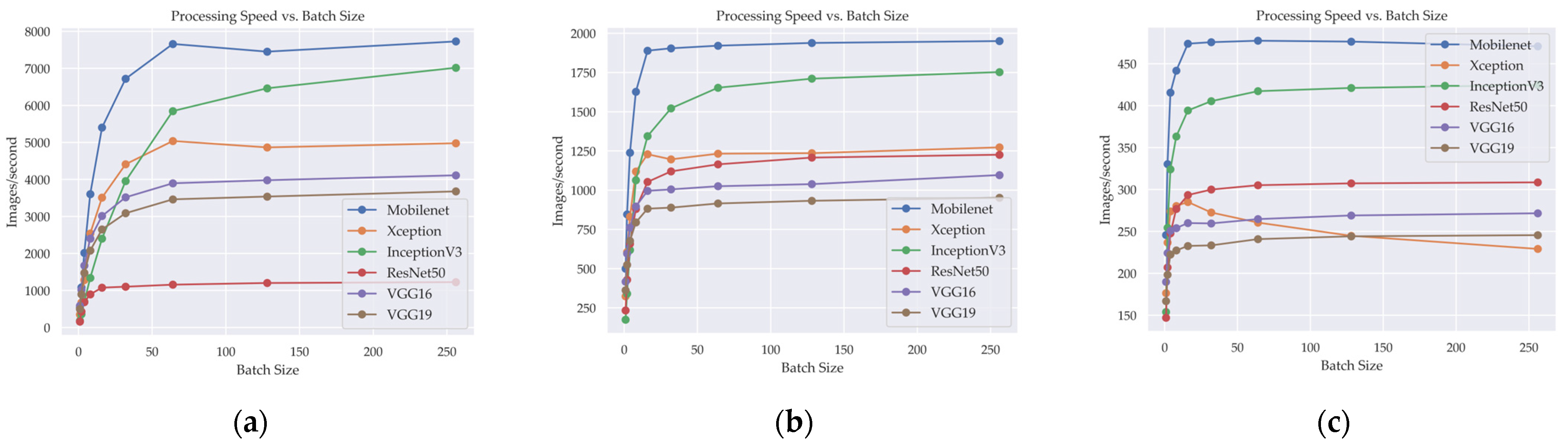
Table 6 provides information about the processing speed of different DL models measured in IPS for various image sizes (128 × 128, 256 × 256, 512 × 512) and batch sizes (1, 2, 4, 8, 16, 32, 64, 128, 256).
Figure 7 illustrates the relationship between the number of IPS and the batch size in various DL models when image size is changed. The IPS metric serves as a valuable indicator of the computational efficiency of the models, quantifying the number of images processed within a second across different image sizes and batch sizes.
Analyzing the results, it is evident that increasing the image size generally leads to a decrease in processing speed across all models. This is expected since larger images contain more pixels and require more computational resources, resulting in a reduced number of images processed per second.
Furthermore,
Table 6 shows that the batch size also influences the processing speed. Generally, as the batch size increases, the processing speed improves, indicating better utilization of parallel processing capabilities. However, there is a diminishing return in speed improvements beyond a certain batch size, as the models may experience limitations in memory or computational capacity.
Considering specific models, it can be observed that MobileNet generally achieves the highest processing speeds compared to other models, particularly at larger image sizes and batch size. On the other hand, Inception3 also consistently demonstrates faster processing speeds across different image sizes and batch sizes.
This relationship provides meaningful insights into the models’ ability to handle larger workloads and deliver faster processing speeds, which are crucial considerations for optimizing DL model performance in real-world applications.
Furthermore, our analysis included an examination of memory utilization to investigate the impact of larger input image sizes and varying batch sizes on the utilization of Graphics Processing Unit (GPU) memory.
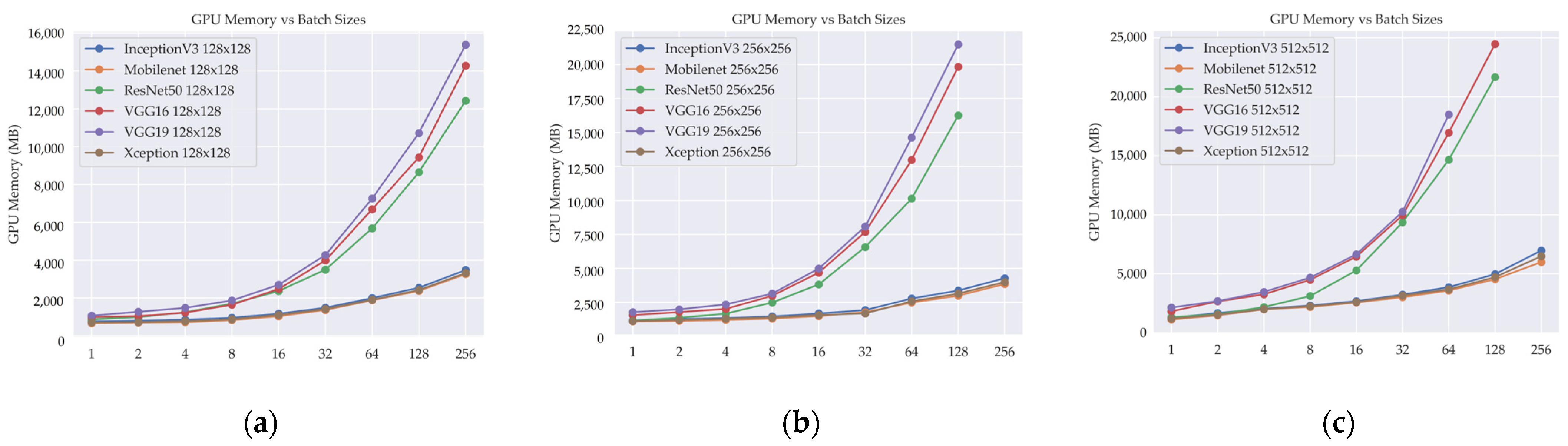
Table 7 provides insights into GPU memory utilization in DL models, highlighting the impact of image and batch sizes on resource demands.
Among the DL models analyzed, MobileNet consistently demonstrates lower GPU memory requirements compared to others, irrespective of image size or batch size. Xception and InceptionV3 also exhibit relatively low GPU memory usage, with InceptionV3 occasionally showing slightly higher requirements than Xception. ResNet50 generally demands more GPU memory than MobileNet, Xception, and InceptionV3. Notably, VGG16 and VGG19 exhibit the highest GPU memory usage across the analyzed models, even with smaller image sizes and batch sizes. These findings emphasize the importance of considering GPU memory limitations when selecting a DL model, as higher memory requirements may restrict the feasible batch size or image size. Employing optimization techniques such as model pruning can be beneficial in reducing the GPU memory footprint.
A subset of data corresponding to specific batch sizes is absent in the 256 × 256 and 512 × 512 image resolutions for certain DL models. This discrepancy arises from the inherent limitations of computer GPU memory. The constrained memory capacity of the GPU hindered the feasibility of processing and storing the entire dataset for these batch sizes at the aforementioned higher image resolutions. Consequently, the absence of data points in the experimental results directly stems from these hardware limitations.
Figure 8 illustrates the relationship between the number of GPU memory utilization and the batch size in different DL models when image size is changed. The GPU memory utilization metric serves as a valuable indicator of the computational resource demand of the models, quantifying the amount of GPU memory used during the training of DL models for different image sizes and batch sizes.
The absence of data for certain batch sizes of DL models in the 256 × 256 and 512 × 512 image sizes is attributed to the inability to complete the processing task due to insufficient GPU memory. These larger image sizes necessitate a substantial amount of memory for processing, and when the allocated GPU memory falls short, the task cannot be executed successfully. This limitation arises from the inherent physical limitations of the GPU memory capacity, which imposes restrictions on the sizes of images that can be processed. As a result, data collection and analysis for batch sizes in these image sizes were not possible due to the impracticality of storing and manipulating the necessary data within the available memory resources. This underscores the critical importance of effective memory resource management and considering the hardware limitations when working with DL models that involve large image sizes and batch sizes. Employing memory optimization techniques and adopting memory-efficient architectures can help alleviate these constraints and enable the processing of larger image sizes within the constraints of the available GPU memory.
These insights played a crucial role in identifying potential challenges and limitations related to GPU memory capacity, providing invaluable guidance for optimizing the selection of image sizes and batch sizes. The ultimate goal of these optimization efforts was to maximize the effective utilization of available computational resources and ensure the smooth execution of the DL models. This approach aimed to strike a balance between resource efficiency and computational performance, allowing for efficient utilization of the GPU and facilitating optimal model performance.