Assessing the Performance of a Novel Stool-Based Microbiome Test That Predicts Response to First Line Immune Checkpoint Inhibitors in Multiple Cancer Types
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Inclusion/Exclusion Criteria
2.2. Study Design and Treatments
2.3. Questionnaire Data
2.4. Sample Collection
2.5. DNA Extraction and Sequencing
2.6. Bioinformatic Analysis
2.7. BiomeOne® Response Prediction
2.8. Statistical Analyses
3. Results
3.1. Patients’ Demographics
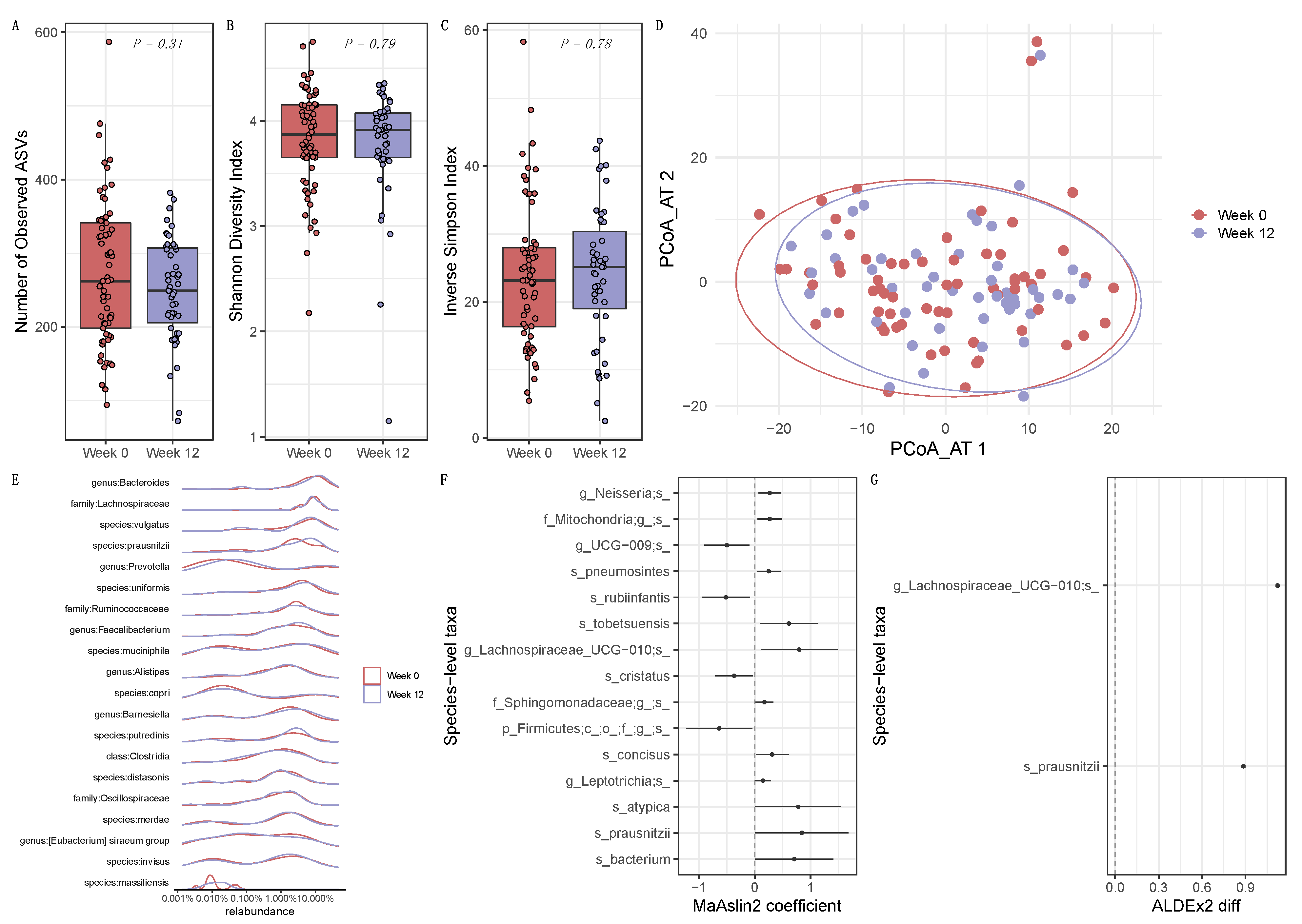
3.2. Longitudinal Influence of ICI Treatment on the Intestinal Microbiome
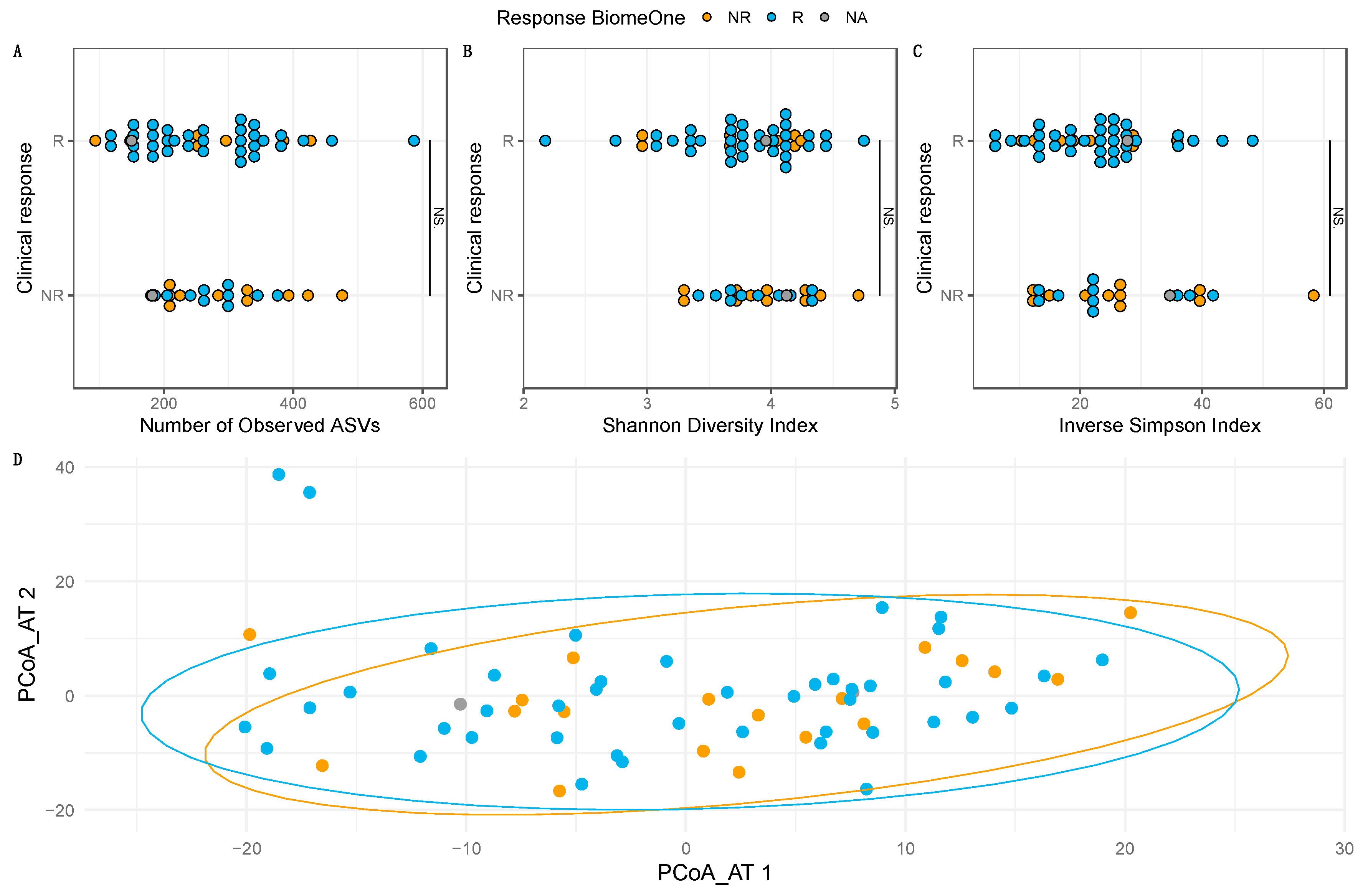
3.3. Prediction of Clinical Outcome of ICI Therapy Using the Stool-Based Microbiome Test BiomeOne® at Baseline
3.4. Baseline Compositional Differences May Drive Clinical Response
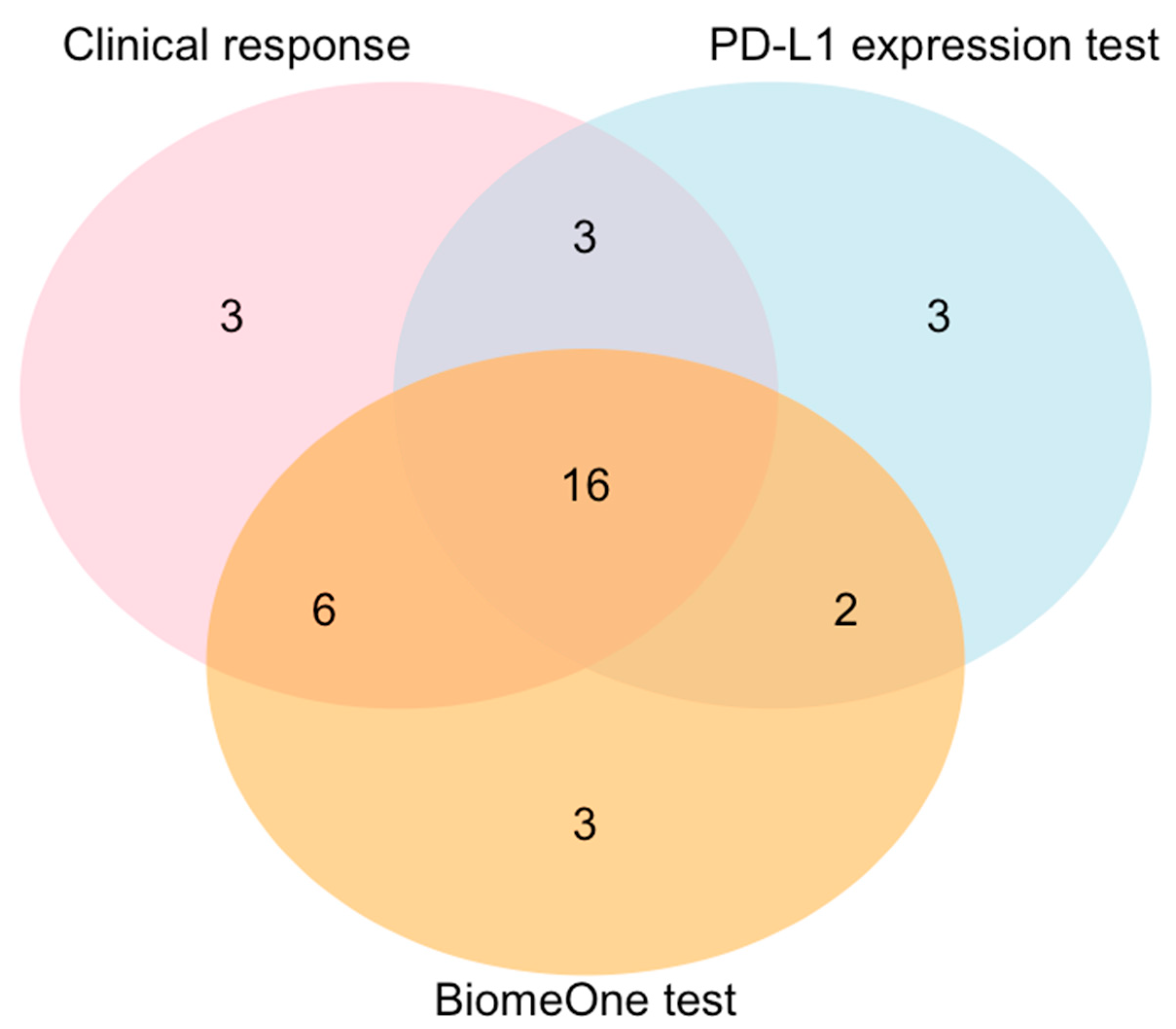
3.5. BiomeOne® Exhibited Higher Sensitivity in Predicting Response Than the PD-L1 Expression Test
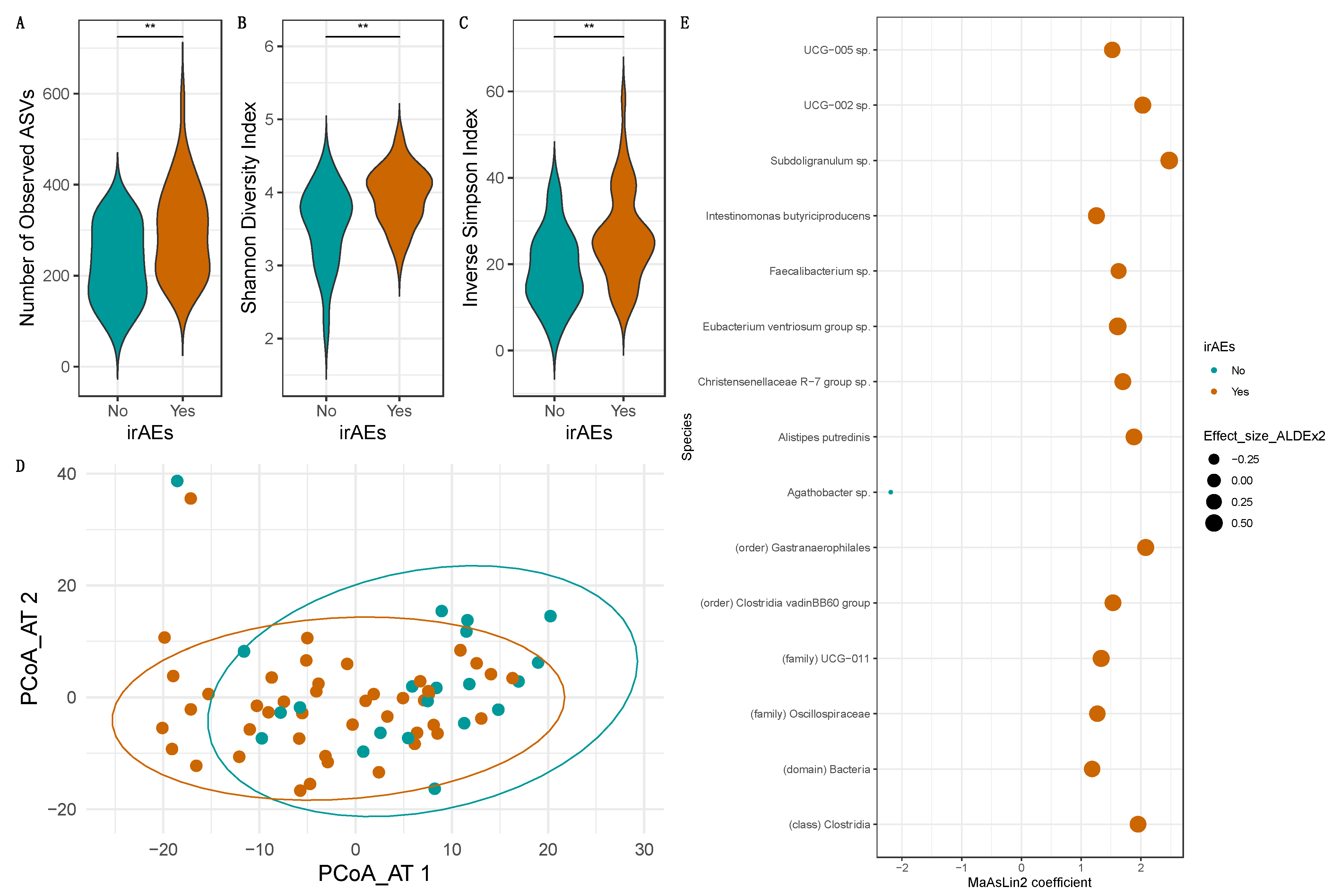
3.6. Occurrence/Severity of irAEs Depends Heavily on the Baseline Intestinal Microbiome Composition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Andrews, M.C.; Duong, C.P.M.; Gopalakrishnan, V.; Iebba, V.; Chen, W.-S.; Derosa, L.; Khan, A.W.; Cogdill, A.P.; White, M.G.; Wong, M.C.; et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 2021, 27, 1432–1441. [Google Scholar] [CrossRef]
- Chau, J.; Yadav, M.; Ben Liu, B.; Furqan, M.; Dai, Q.; Shahi, S.; Gupta, A.; Mercer, K.N.; Eastman, E.; Abu Hejleh, T.; et al. Prospective correlation between the patient microbiome with response to and development of immune-mediated adverse effects to immunotherapy in lung cancer. BMC Cancer 2021, 21, 808. [Google Scholar] [CrossRef]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Khan, A.W.; Zhang, X.; White, M.G.; Peterson, C.B.; et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; le Chatelier, E.; DeRosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Hakozaki, T.; Richard, C.; Elkrief, A.; Hosomi, Y.; Benlaïfaoui, M.; Mimpen, I.; Terrisse, S.; Derosa, L.; Zitvogel, L.; Routy, B.; et al. The Gut Microbiome Associates with Immune Checkpoint Inhibition Outcomes in Patients with Advanced Non–Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Wilson, M.; Moran, U.; Pavlick, A.; Izsak, A.; Wechter, T.; Weber, J.S.; Osman, I.; Ahn, J. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 2019, 11, 61. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Fang, Z.; Xue, Y.; Zhang, J.; Zhu, J.; Gao, R.; Yao, S.; Ye, Y.; Wang, S.; Lin, C.; et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 2020, 11, 1030–1042. [Google Scholar] [CrossRef]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2020, 371, 602–609. [Google Scholar] [CrossRef]
- Cascone, T.; William, W.N., Jr.; Weissferdt, A.; Leung, C.H.; Lin, H.Y.; Pataer, A.; Godoy, M.C.B.; Carter, B.W.; Federico, L.; Reuben, A.; et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: The phase 2 randomized NEOSTAR trial. Nat. Med. 2021, 27, 504–514. [Google Scholar] [CrossRef]
- Gopalakrishnan, V. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, J.A.; Davar, D.; Rodrigues, R.R.; Badger, J.H.; Fang, J.R.; Cole, A.M.; Balaji, A.K.; Vetizou, M.; Prescott, S.M.; Fernandes, M.R.; et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 2022, 28, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2019, 362, eaar3593. [Google Scholar] [CrossRef]
- Komiya, K.; Nakamura, T.; Abe, T.; Ogusu, S.; Nakashima, C.; Takahashi, K.; Kimura, S.; Sueoka-Aragane, N. Discontinuation due to immune-related adverse events is a possible predictive factor for immune checkpoint inhibitors in patients with non-small cell lung cancer. Thorac. Cancer 2019, 10, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Bilger, G.; Girard, N.; Doubre, H.; Levra, M.G.; Giroux-Leprieur, E.; Giraud, F.; Decroisette, C.; Carton, M.; Massiani, M.A. Discontinuation of immune checkpoint inhibitor (ICI) above 18 months of treatment in real-life patients with advanced non-small cell lung cancer (NSCLC): INTEPI, a multicentric retrospective study. Cancer Immunol. Immunother. 2021, 71, 1719–1731. [Google Scholar] [CrossRef]
- Iivanainen, S.; Koivunen, J.P. Possibilities of Improving the Clinical Value of Immune Checkpoint Inhibitor Therapies in Cancer Care by Optimizing Patient Selection. Int. J. Mol. Sci. 2020, 21, 556. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Powles, T. Re: Nivolumab plus Ipilimumab Versus Sunitinib in Advanced Renal-cell Carcinoma. Eur. Urol. 2018, 74, 679–680. [Google Scholar] [CrossRef]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, Z.-D.; Abu-Sbeih, H.; Sanchez, C.A.; Chang, C.-C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- Robinson, I.; Schmidinger, M.; Hochmair, M.; Ay, L.; Absenger, G.; Pichler, M.; Nguyen, V.; Richtig, E.; Rainer, B.; Jansen, C.; et al. 117P BiomeOne: Multi-centric validation of a novel microbiome-based biomarker to predict response to cancer immunotherapy. Ann. Oncol. 2022, 33, S592. [Google Scholar] [CrossRef]
- Robinson, I.; Hochmair, M.; Ay, L.; Absenger, G.; Jansen, C.; Pacifico, C.; Sladek, B.; Knabl, A.; Gasche, N.; Valipour, A. 55P Comparison of BiomeOne and PD-L1 expression tests as a predictor for response to immune checkpoint inhibitors (ICI) in patients with non-small cell lung cancer (NSCLC). Ann. Oncol. 2022, 33, S1400. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Cebula, A.; Seweryn, M.; Rempala, G.A.; Pabla, S.S.; McIndoe, R.A.; Denning, T.L.; Bry, L.; Kraj, P.; Kisielow, P.; Ignatowicz, L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013, 497, 258–262. [Google Scholar] [CrossRef]
- Botticelli, A.; Putignani, L.; Zizzari, I.; Del Chierico, F.; Reddel, S.; Di Pietro, F.; Quagliarello, A.; Onesti, C.E.; Raffaele, G.; Mazzuca, F.; et al. Changes of microbiome profile during nivolumab treatment in NSCLC patients. J. Clin. Oncol. 2018, 36, e15020. [Google Scholar] [CrossRef]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated with Favorable Responses to Anti–Programmed Death 1 Immunotherapy in Chinese Patients with NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and Anticancer Immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef]
- Liberini, V.; Mariniello, A.; Righi, L.; Capozza, M.; Delcuratolo, M.D.; Terreno, E.; Farsad, M.; Volante, M.; Novello, S.; Deandreis, D. NSCLC Biomarkers to Predict Response to Immunotherapy with Checkpoint Inhibitors (ICI): From the Cells to In Vivo Images. Cancers 2021, 13, 4543. [Google Scholar] [CrossRef]
- Haratani, K.; Hayashi, H.; Chiba, Y.; Kudo, K.; Yonesaka, K.; Kato, R.; Kaneda, H.; Hasegawa, Y.; Tanaka, K.; Takeda, M.; et al. Association of Immune-Related Adverse Events with Nivolumab Efficacy in Non–Small-Cell Lung Cancer. JAMA Oncol. 2018, 4, 374–378. [Google Scholar] [CrossRef]
- Lee, K.A.; Thomas, A.M.; Bolte, L.A.; Björk, J.R.; de Ruijter, L.K.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; Board, R.; Calbet-Llopart, N.; et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 2022, 28, 535–544. [Google Scholar] [CrossRef] [PubMed]
| Total (n = 65) | R (n = 43) | NR (n = 22) | p-Value | ||
|---|---|---|---|---|---|
| Sex | |||||
| Male | 38 (58.46) | 24 (55.81) | 14 (63.64) | 0.60 | |
| Female | 27 (41.54) | 19 (44.19) | 8 (36.36) | ||
| Age (mean ± SD, years) | 66.57 ± 8.78 | 65.38 ± 8.69 | 68.91 ± 8.67 | 0.12 | |
| Cancer type | |||||
| NSCLC | Stage III | 11 (16.92) | 10 (23.26) | 1 (4.54) | 0.07 |
| Stage IV | 31 (47.69) | 21 (48.84) | 10 (45.45) | ||
| RCC | Stage III | 4 (6.15) | 3 (6.98) | 1 (4.54) | |
| Stage IV | 12 (18.46) | 7 (16.28) | 5 (22.73) | ||
| Melanoma | Stage III | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Stage IV | 7 (10.77) | 2 (4.65) | 5 (22.73) | ||
| Treatment | |||||
| Anti-PD-1/PD-L1 | 62 (95.38) | 43 (100.00) | 19 (86.36) | 0.04 | |
| Anti-PD-1/CTLA-4 | 3 (4.62) | 0 (0.00) | 3 (13.64) | ||
| irAEs | |||||
| No | 21 (32.31) | 20 (46.51) | 1 (4.55) | <0.01 | |
| Yes (≥grade 1) | 44 (67.69) | 23 (53.49) | 21 (95.45) |
| Rs (CR + PR) | NRs (SD + PD) | Total | |
|---|---|---|---|
| BiomeOne result > 50% | 34 | 10 | 44 |
| BiomeOne result ≤ 50% | 8 | 11 | 19 |
| Total | 42 | 21 | 63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, I.; Hochmair, M.J.; Schmidinger, M.; Absenger, G.; Pichler, M.; Nguyen, V.A.; Richtig, E.; Rainer, B.M.; Ay, L.; Jansen, C.; et al. Assessing the Performance of a Novel Stool-Based Microbiome Test That Predicts Response to First Line Immune Checkpoint Inhibitors in Multiple Cancer Types. Cancers 2023, 15, 3268. https://doi.org/10.3390/cancers15133268
Robinson I, Hochmair MJ, Schmidinger M, Absenger G, Pichler M, Nguyen VA, Richtig E, Rainer BM, Ay L, Jansen C, et al. Assessing the Performance of a Novel Stool-Based Microbiome Test That Predicts Response to First Line Immune Checkpoint Inhibitors in Multiple Cancer Types. Cancers. 2023; 15(13):3268. https://doi.org/10.3390/cancers15133268
Chicago/Turabian StyleRobinson, Irina, Maximilian Johannes Hochmair, Manuela Schmidinger, Gudrun Absenger, Martin Pichler, Van Anh Nguyen, Erika Richtig, Barbara Margaretha Rainer, Leyla Ay, Christian Jansen, and et al. 2023. "Assessing the Performance of a Novel Stool-Based Microbiome Test That Predicts Response to First Line Immune Checkpoint Inhibitors in Multiple Cancer Types" Cancers 15, no. 13: 3268. https://doi.org/10.3390/cancers15133268
APA StyleRobinson, I., Hochmair, M. J., Schmidinger, M., Absenger, G., Pichler, M., Nguyen, V. A., Richtig, E., Rainer, B. M., Ay, L., Jansen, C., Pacífico, C., Knabl, A., Sladek, B., Gasche, N., & Valipour, A. (2023). Assessing the Performance of a Novel Stool-Based Microbiome Test That Predicts Response to First Line Immune Checkpoint Inhibitors in Multiple Cancer Types. Cancers, 15(13), 3268. https://doi.org/10.3390/cancers15133268








