Luciferase Expressing Preclinical Model Systems Representing the Different Molecular Subtypes of Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Generation of Luciferase Expressing Clones
2.2. Spheroid Preparation, Growth Kinetics, Drug Treatment
2.3. RNA Preparation, Microarray Analysis, Molecular Subtyping and Classification
2.4. Animal Studies, Treatment, Imaging
3. Results
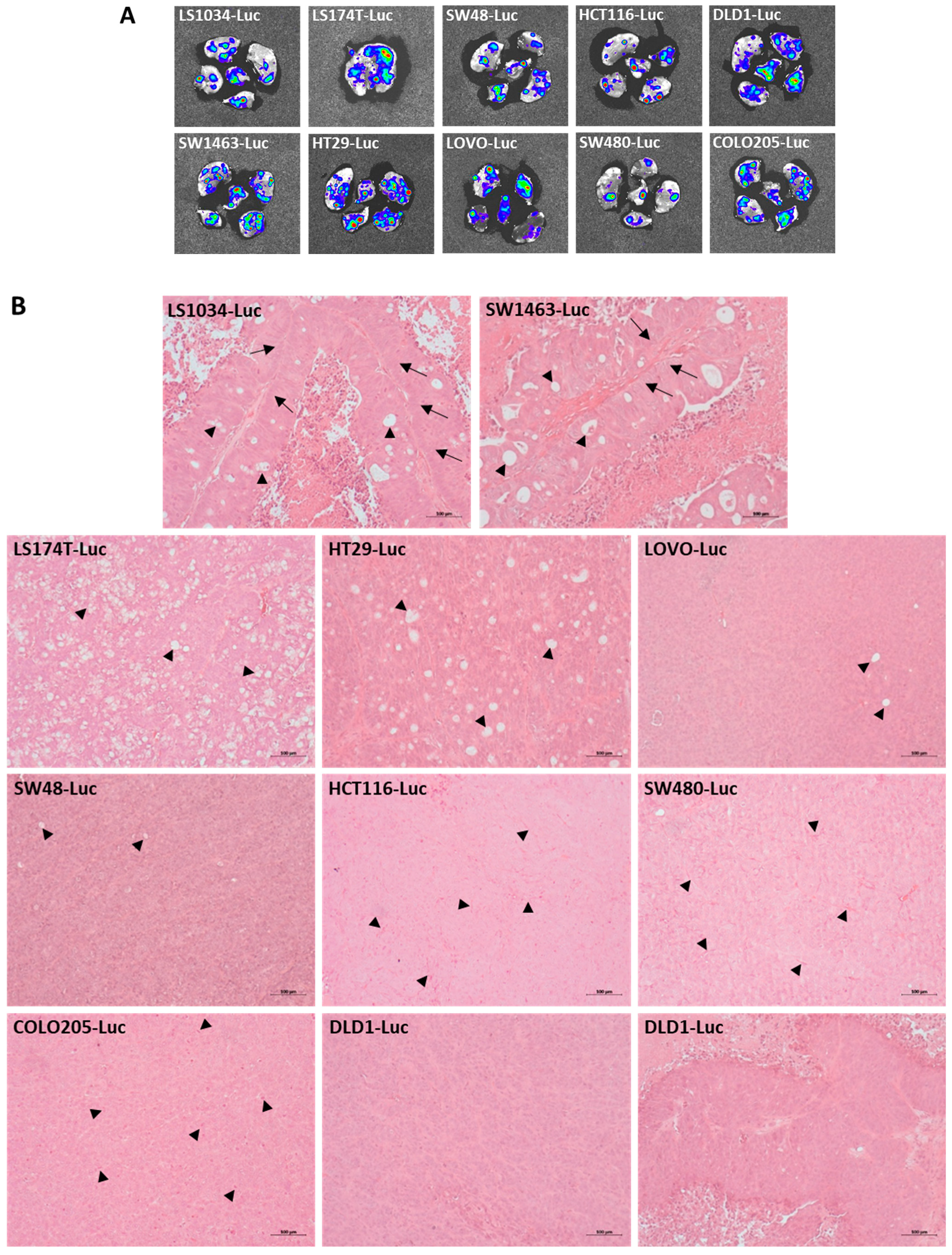
3.1. Luciferase-Labelled CRC Cell Line Clones form Subcutaneous Xenograft Tumors with Various Differentiation Characteristics and Metastasize to the Lung
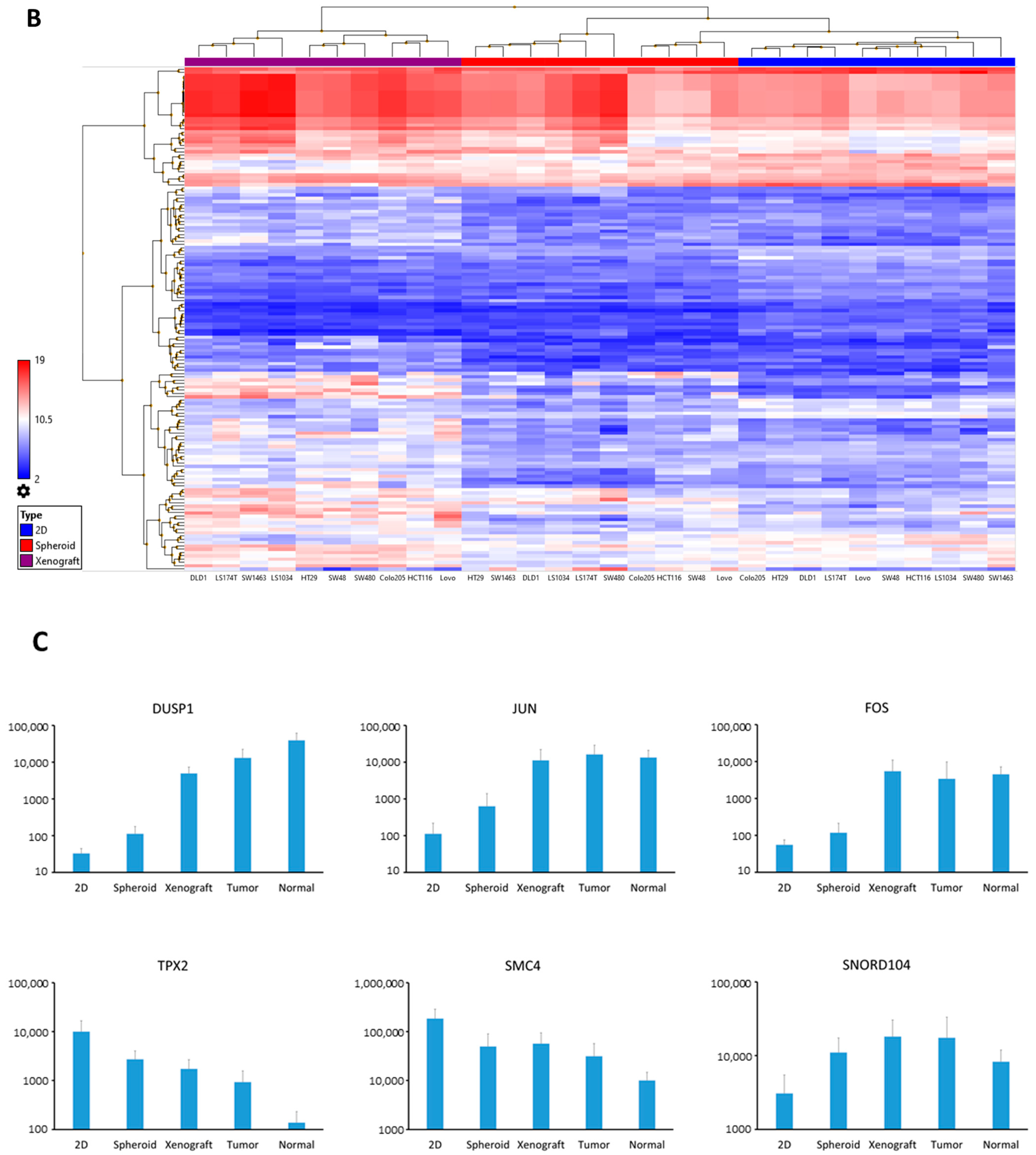
3.2. Luciferase Expressing Cell Lines as Well as Their Derived Spheroids and Xenograft Tumors Represent Molecular Characteristics of CRC
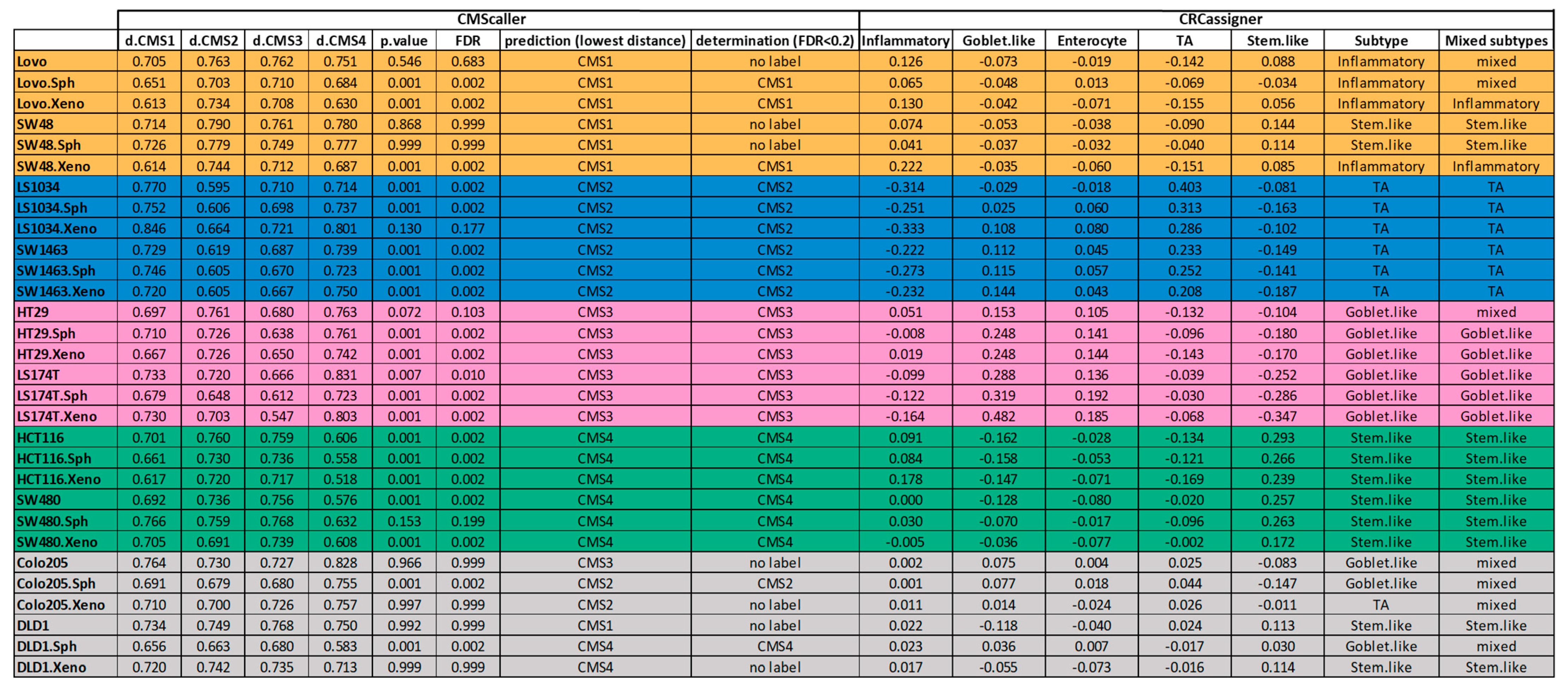
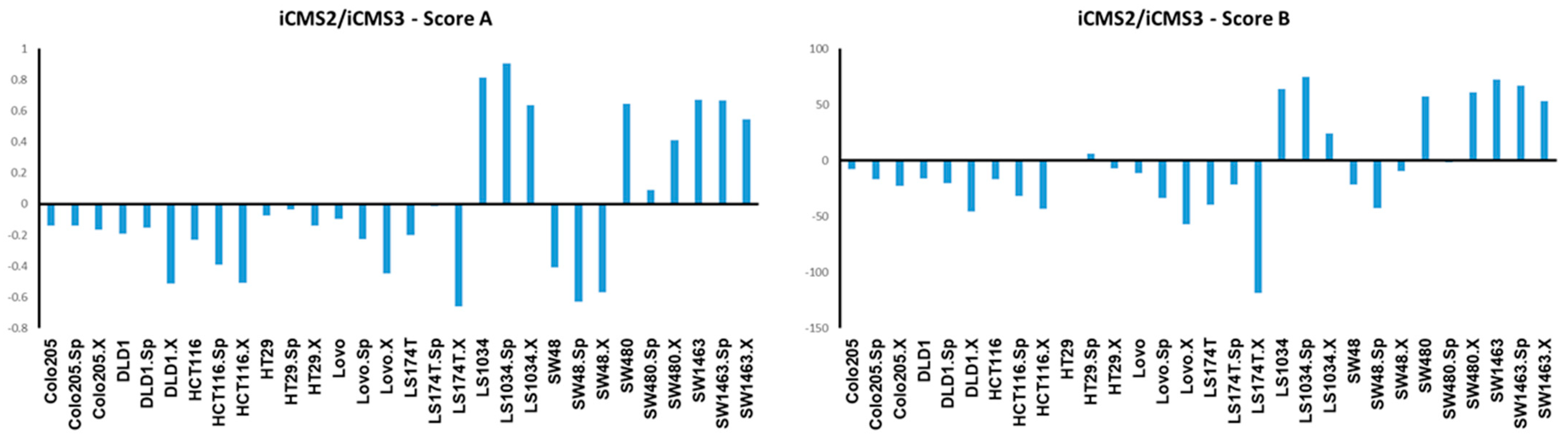
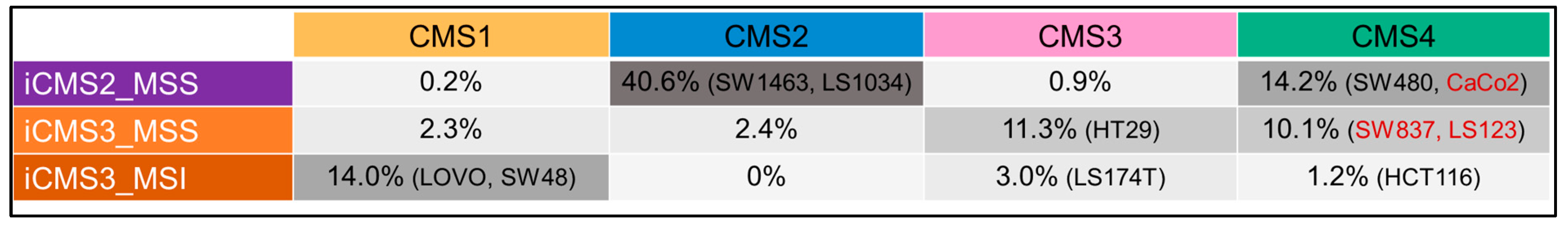
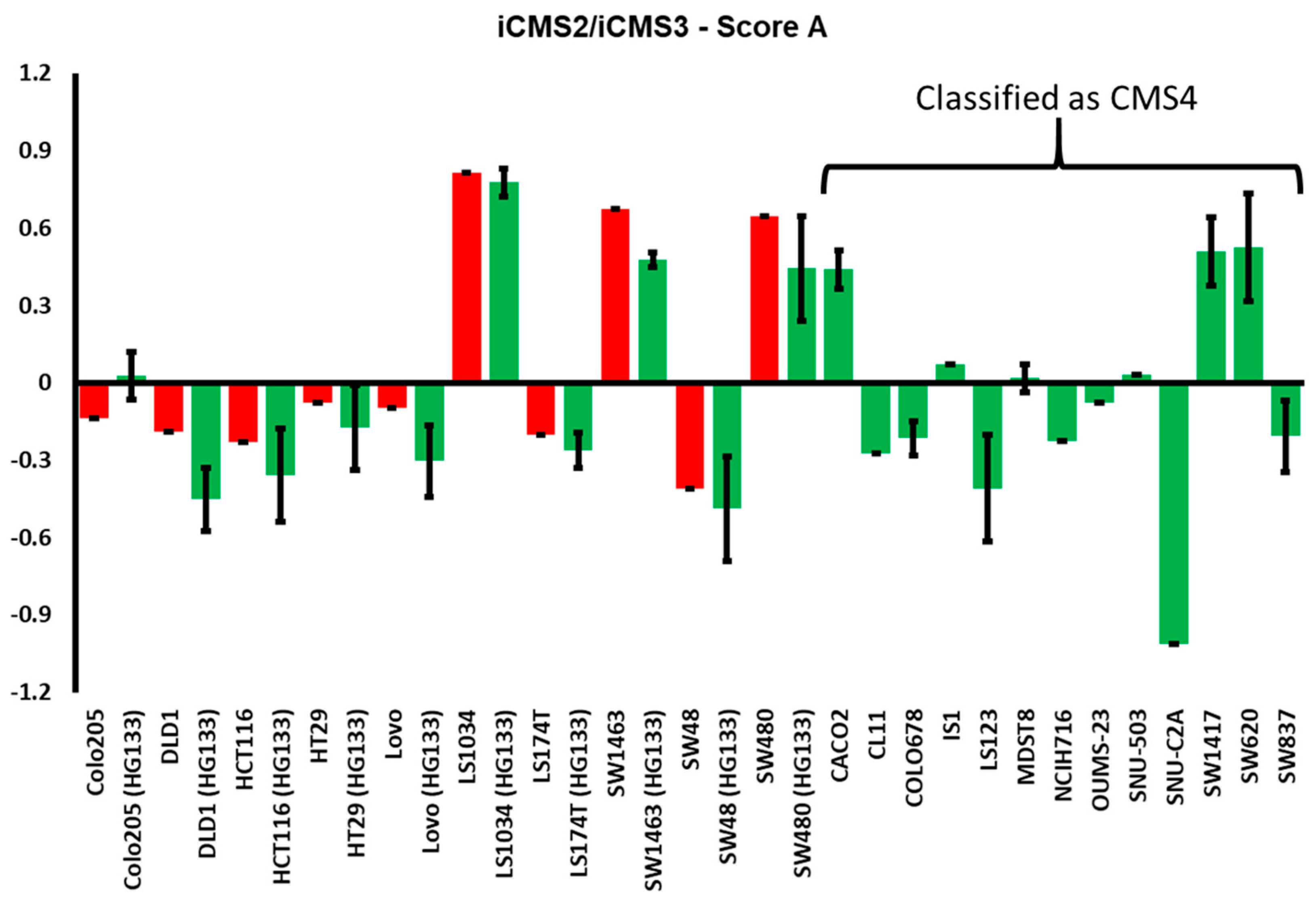
3.3. Luciferase Expressing Cell Lines and Their Derived Spheroids and Xenograft Tumors Represent the Main Subtypes of CRC According to the CMS Classification
3.4. Establishing a Bioluminescence Based Cytotoxicity Assay for Spheroid Models
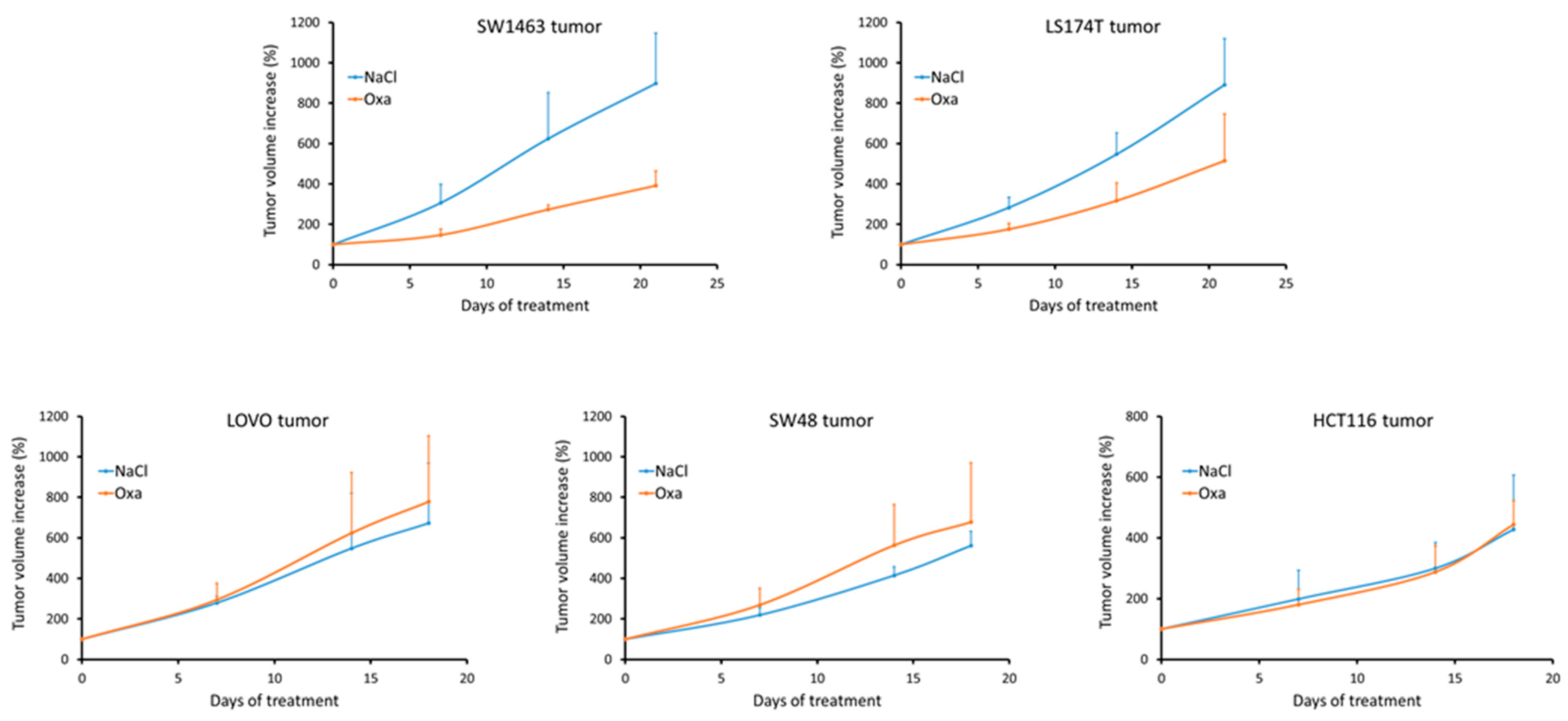
3.5. Analyzing Response to Oxaliplatin in Nude Mice Xenograft Tumors
3.6. Identification of CAV1 as a Putative Marker of Oxaliplatin Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Li, H.; Ning, S.; Ghandi, M.; Kryukov, G.V.; Gopal, S.; Deik, A.; Souza, A.; Pierce, K.; Keskula, P.; Hernandez, D.; et al. The landscape of cancer cell line metabolism. Nat. Med. 2019, 25, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Demere, Z.; Nair, K.; Ali, A.; Ferraro, G.B.; Natoli, T.; Deik, A.; Petronio, L.; Tang, A.A.; Zhu, C.; et al. A metastasis map of human cancer cell lines. Nature 2020, 588, 331–336. [Google Scholar] [CrossRef]
- Kinker, G.S.; Greenwald, A.C.; Tal, R.; Orlova, Z.; Cuoco, M.S.; McFarland, J.M.; Warren, A.; Rodman, C.; Roth, J.A.; Bender, S.A.; et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 2020, 52, 1208–1218. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef]
- Mouradov, D.; Sloggett, C.; Jorissen, R.N.; Love, C.G.; Li, S.; Burgess, A.W.; Arango, D.; Strausberg, R.L.; Buchanan, D.; Wormald, S.; et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014, 74, 3238–3247. [Google Scholar] [CrossRef] [PubMed]
- Budinska, E.; Popovici, V.; Tejpar, S.; D’Ario, G.; Lapique, N.; Sikora, K.O.; Di Narzo, A.F.; Yan, P.; Hodgson, J.G.; Weinrich, S.; et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J. Pathol. 2013, 231, 63–76. [Google Scholar] [CrossRef]
- De Sousa, E.M.F.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; de Rooij, L.P.; de Jong, J.H.; de Boer, O.J.; van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef]
- Marisa, L.; de Reynies, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef]
- Roepman, P.; Schlicker, A.; Tabernero, J.; Majewski, I.; Tian, S.; Moreno, V.; Snel, M.H.; Chresta, C.M.; Rosenberg, R.; Nitsche, U.; et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int. J. Cancer 2014, 134, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.; Lannon, W.A.; Grotzinger, C.; Del Rio, M.; et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Schlicker, A.; Beran, G.; Chresta, C.M.; McWalter, G.; Pritchard, A.; Weston, S.; Runswick, S.; Davenport, S.; Heathcote, K.; Castro, D.A.; et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med. Genom. 2012, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Medico, E.; Russo, M.; Picco, G.; Cancelliere, C.; Valtorta, E.; Corti, G.; Buscarino, M.; Isella, C.; Lamba, S.; Martinoglio, B.; et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat. Commun. 2015, 6, 7002. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Isella, C.; Terrasi, A.; Bellomo, S.E.; Petti, C.; Galatola, G.; Muratore, A.; Mellano, A.; Senetta, R.; Cassenti, A.; Sonetto, C.; et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015, 47, 312–319. [Google Scholar] [CrossRef]
- Linnekamp, J.F.; Hooff, S.R.V.; Prasetyanti, P.R.; Kandimalla, R.; Buikhuisen, J.Y.; Fessler, E.; Ramesh, P.; Lee, K.; Bochove, G.G.W.; de Jong, J.H.; et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ. 2018, 25, 616–633. [Google Scholar] [CrossRef]
- Eide, P.W.; Bruun, J.; Lothe, R.A.; Sveen, A. CMScaller: An R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 2017, 7, 16618. [Google Scholar] [CrossRef] [PubMed]
- Eide, P.W.; Moosavi, S.H.; Eilertsen, I.A.; Brunsell, T.H.; Langerud, J.; Berg, K.C.G.; Rosok, B.I.; Bjornbeth, B.A.; Nesbakken, A.; Lothe, R.A.; et al. Metastatic heterogeneity of the consensus molecular subtypes of colorectal cancer. NPJ Genom. Med. 2021, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.C.G.; Eide, P.W.; Eilertsen, I.A.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjornslett, M.; Meza-Zepeda, L.A.; Eknaes, M.; Lind, G.E.; et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 2017, 16, 116. [Google Scholar] [CrossRef]
- Sveen, A.; Bruun, J.; Eide, P.W.; Eilertsen, I.A.; Ramirez, L.; Murumagi, A.; Arjama, M.; Danielsen, S.A.; Kryeziu, K.; Elez, E.; et al. Colorectal Cancer Consensus Molecular Subtypes Translated to Preclinical Models Uncover Potentially Targetable Cancer Cell Dependencies. Clin. Cancer Res. 2018, 24, 794–806. [Google Scholar] [CrossRef]
- Joanito, I.; Wirapati, P.; Zhao, N.; Nawaz, Z.; Yeo, G.; Lee, F.; Eng, C.L.P.; Macalinao, D.C.; Kahraman, M.; Srinivasan, H.; et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 2022, 54, 963–975. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.S.; Barros, A.S.; Costa, E.C.; Moreira, A.F.; Correia, I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019, 116, 206–226. [Google Scholar] [CrossRef]
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M.; Kohrt, D.; Florentine, C.M.; Grossel, M.J. A Firefly Luciferase Dual Color Bioluminescence Reporter Assay Using Two Substrates To Simultaneously Monitor Two Gene Expression Events. Sci. Rep. 2018, 8, 5990. [Google Scholar] [CrossRef]
- Luetzkendorf, J.; Mueller, L.P.; Mueller, T.; Caysa, H.; Nerger, K.; Schmoll, H.J. Growth inhibition of colorectal carcinoma by lentiviral TRAIL-transgenic human mesenchymal stem cells requires their substantial intratumoral presence. J. Cell Mol. Med. 2010, 14, 2292–2304. [Google Scholar] [CrossRef]
- Mueller, T.; Pfankuchen, D.B.; Wantoch von Rekowski, K.; Schlesinger, M.; Reipsch, F.; Bendas, G. The Impact of the Low Molecular Weight Heparin Tinzaparin on the Sensitization of Cisplatin-Resistant Ovarian Cancers-Preclinical In Vivo Evaluation in Xenograft Tumor Models. Molecules 2017, 22, 728. [Google Scholar] [CrossRef]
- Zhang, B.; Babu, K.R.; Lim, C.Y.; Kwok, Z.H.; Li, J.; Zhou, S.; Yang, H.; Tay, Y. A comprehensive expression landscape of RNA-binding proteins (RBPs) across 16 human cancer types. RNA Biol. 2020, 17, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y. Nearest template prediction: A single-sample-based flexible class prediction with confidence assessment. PLoS ONE 2010, 5, e15543. [Google Scholar] [CrossRef]
- Wagner, K.W.; Punnoose, E.A.; Januario, T.; Lawrence, D.A.; Pitti, R.M.; Lancaster, K.; Lee, D.; von Goetz, M.; Yee, S.F.; Totpal, K.; et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 2007, 13, 1070–1077. [Google Scholar] [CrossRef]
- Hook, K.E.; Garza, S.J.; Lira, M.E.; Ching, K.A.; Lee, N.V.; Cao, J.; Yuan, J.; Ye, J.; Ozeck, M.; Shi, S.T.; et al. An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF-03814735. Mol. Cancer Ther. 2012, 11, 710–719. [Google Scholar] [CrossRef]
- Bull, J.A.; Mech, F.; Quaiser, T.; Waters, S.L.; Byrne, H.M. Mathematical modelling reveals cellular dynamics within tumour spheroids. PLoS Comput. Biol. 2020, 16, e1007961. [Google Scholar] [CrossRef]
- Mukomoto, R.; Nashimoto, Y.; Terai, T.; Imaizumi, T.; Hiramoto, K.; Ino, K.; Yokokawa, R.; Miura, T.; Shiku, H. Oxygen consumption rate of tumour spheroids during necrotic-like core formation. Analyst 2020, 145, 6342–6348. [Google Scholar] [CrossRef]
- Tindall, M.J.; Dyson, L.; Smallbone, K.; Maini, P.K. Modelling acidosis and the cell cycle in multicellular tumour spheroids. J. Theor. Biol. 2012, 298, 107–115. [Google Scholar] [CrossRef]
- Reipsch, F.; Biersack, B.; Lucas, H.; Schobert, R.; Mueller, T. Imidazole Analogs of Vascular-Disrupting Combretastatin A-4 with Pleiotropic Efficacy against Resistant Colorectal Cancer Models. Int. J. Mol. Sci. 2021, 22, 13082. [Google Scholar] [CrossRef]
- Ketteler, J.; Klein, D. Caveolin-1, cancer and therapy resistance. Int. J. Cancer 2018, 143, 2092–2104. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, N.; Huang, C.; Bao, Y.; Jiang, Y.; Zhu, G. Downregulation of caveolin-1 increases the sensitivity of drug-resistant colorectal cancer HCT116 cells to 5-fluorouracil. Oncol. Lett. 2017, 13, 483–487. [Google Scholar] [CrossRef]
- Luo, F.; Li, J.; Liu, J.; Liu, K. Stabilizing and upregulating Axin with tankyrase inhibitor reverses 5-fluorouracil chemoresistance and proliferation by targeting the WNT/caveolin-1 axis in colorectal cancer cells. Cancer Gen. Ther. 2022, 29, 1707–1719. [Google Scholar] [CrossRef]
- Rodel, F.; Capalbo, G.; Rodel, C.; Weiss, C. Caveolin-1 as a prognostic marker for local control after preoperative chemoradiation therapy in rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2009, 73, 846–852. [Google Scholar] [CrossRef]
- Okita, A.; Takahashi, S.; Ouchi, K.; Inoue, M.; Watanabe, M.; Endo, M.; Honda, H.; Yamada, Y.; Ishioka, C. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget 2018, 9, 18698–18711. [Google Scholar] [CrossRef] [PubMed]
- Stintzing, S.; Wirapati, P.; Lenz, H.J.; Neureiter, D.; Fischer von Weikersthal, L.; Decker, T.; Kiani, A.; Kaiser, F.; Al-Batran, S.; Heintges, T.; et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019, 30, 1796–1803. [Google Scholar] [CrossRef]
- Lenz, H.J.; Ou, F.S.; Venook, A.P.; Hochster, H.S.; Niedzwiecki, D.; Goldberg, R.M.; Mayer, R.J.; Bertagnolli, M.M.; Blanke, C.D.; Zemla, T.; et al. Impact of Consensus Molecular Subtype on Survival in Patients With Metastatic Colorectal Cancer: Results From CALGB/SWOG 80405 (Alliance). J. Clin. Oncol. 2019, 37, 1876–1885. [Google Scholar] [CrossRef] [PubMed]
- Ten Hoorn, S.; de Back, T.R.; Sommeijer, D.W.; Vermeulen, L. Clinical Value of Consensus Molecular Subtypes in Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2021, 114, 503–516. [Google Scholar] [CrossRef]
- Del Rio, M.; Mollevi, C.; Bibeau, F.; Vie, N.; Selves, J.; Emile, J.F.; Roger, P.; Gongora, C.; Robert, J.; Tubiana-Mathieu, N.; et al. Molecular subtypes of metastatic colorectal cancer are associated with patient response to irinotecan-based therapies. Eur. J. Cancer 2017, 76, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Pogue-Geile, K.L.; Gavin, P.G.; Yothers, G.; Kim, S.R.; Johnson, N.L.; Lipchik, C.; Allegra, C.J.; Petrelli, N.J.; O’Connell, M.J.; et al. Clinical Outcome From Oxaliplatin Treatment in Stage II/III Colon Cancer According to Intrinsic Subtypes: Secondary Analysis of NSABP C-07/NRG Oncology Randomized Clinical Trial. JAMA Oncol. 2016, 2, 1162–1169. [Google Scholar] [CrossRef]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef]
- Losada, A.; Hirano, T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes Dev. 2005, 19, 1269–1287. [Google Scholar] [CrossRef]
- Neumayer, G.; Belzil, C.; Gruss, O.J.; Nguyen, M.D. TPX2: Of spindle assembly, DNA damage response, and cancer. Cell Mol. Life Sci. 2014, 71, 3027–3047. [Google Scholar] [CrossRef]
- Manios, K.; Tsiambas, E.; Stavrakis, I.; Stamatelopoulos, A.; Kavantzas, N.; Agrogiannis, G.; Lazaris, A.C. c-Fos/c-Jun transcription factors in non-small cell lung carcinoma. J. BUON 2020, 25, 2141–2143. [Google Scholar]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef] [PubMed]
- Tsiambas, E.; Mastronikolis, N.; Fotiades, P.P.; Kyrodimos, E.; Chrysovergis, A.; Papanikolaou, V.; Mastronikolis, S.; Peschos, D.; Roukas, D.; Ragos, V. c-Jun/c-Fos complex in laryngeal squamous cell carcinoma. J. BUON 2020, 25, 618–620. [Google Scholar] [PubMed]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, J.; Zhou, J.Y.; Liu, Y.; Wu, G.S. Mitogen-activated protein kinase phosphatase-1 is required for cisplatin resistance. Cancer Res. 2006, 66, 8870–8877. [Google Scholar] [CrossRef]
- Lu, B.; Chen, X.; Liu, X.; Chen, J.; Qin, H.; Chen, S.; Zhao, Y. C/D box small nucleolar RNA SNORD104 promotes endometrial cancer by regulating the 2’-O-methylation of PARP1. J. Transl. Med. 2022, 20, 618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xin, M.; Wang, P. Identification of a novel snoRNA expression signature associated with overall survival in patients with lung adenocarcinoma: A comprehensive analysis based on RNA sequencing dataset. Math. Biosci. Eng. 2021, 18, 7837–7860. [Google Scholar] [CrossRef]
- LaBonia, G.J.; Lockwood, S.Y.; Heller, A.A.; Spence, D.M.; Hummon, A.B. Drug penetration and metabolism in 3D cell cultures treated in a 3D printed fluidic device: Assessment of irinotecan via MALDI imaging mass spectrometry. Proteomics 2016, 16, 1814–1821. [Google Scholar] [CrossRef]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef]
- Ramgolam, K.; Lauriol, J.; Lalou, C.; Lauden, L.; Michel, L.; de la Grange, P.; Khatib, A.M.; Aoudjit, F.; Charron, D.; Alcaide-Loridan, C.; et al. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PLoS ONE 2011, 6, e18784. [Google Scholar] [CrossRef] [PubMed]
- Sant, S.; Johnston, P.A. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov. Today Technol. 2017, 23, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Monch, D.; Maass, A.; Gromoll, C.; Hehr, T.; Leibold, T.; Schlitt, H.J.; Dahlke, M.H.; Renner, P. Three dimensional cultivation increases chemo- and radioresistance of colorectal cancer cell lines. PLoS ONE 2021, 16, e0244513. [Google Scholar] [CrossRef] [PubMed]
- Hammond, W.A.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 57–84. [Google Scholar] [CrossRef] [PubMed]

| Cell Line | Mean Monolayer | SD Monolayer | Mean Spheroid | SD Spheroid | p-Value | Significance |
|---|---|---|---|---|---|---|
| LOVO | 0.13 | 0.02 | 0.37 | 0.09 | 0.0009 | *** |
| SW48 | 0.08 | 0.00 | 0.49 | 0.12 | 0.0005 | *** |
| LS1034 | 0.30 | 0.05 | 0.37 | 0.14 | 0.3537 | ns |
| SW1463 | 0.09 | 0.04 | 0.10 | 0.03 | 0.7680 | ns |
| HT29 | 0.26 | 0.04 | 2.12 | 0.97 | 0.0053 | ** |
| LS174T | 0.11 | 0.04 | 0.18 | 0.04 | 0.0778 | ns |
| HCT116 | 0.24 | 0.01 | 1.39 | 0.33 | 0.0003 | *** |
| SW480 | 0.43 | 0.12 | 2.40 | 0.95 | 0.0035 | ** |
| COLO205 | 0.27 | 0.12 | 12.44 | 6.77 | 0.0159 | * |
| DLD1 | 2.02 | 0.36 | 4.10 | 0.84 | 0.0012 | ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotermund, A.; Staege, M.S.; Brandt, S.; Luetzkendorf, J.; Lucas, H.; Mueller, L.P.; Mueller, T. Luciferase Expressing Preclinical Model Systems Representing the Different Molecular Subtypes of Colorectal Cancer. Cancers 2023, 15, 4122. https://doi.org/10.3390/cancers15164122
Rotermund A, Staege MS, Brandt S, Luetzkendorf J, Lucas H, Mueller LP, Mueller T. Luciferase Expressing Preclinical Model Systems Representing the Different Molecular Subtypes of Colorectal Cancer. Cancers. 2023; 15(16):4122. https://doi.org/10.3390/cancers15164122
Chicago/Turabian StyleRotermund, Arne, Martin S. Staege, Sarah Brandt, Jana Luetzkendorf, Henrike Lucas, Lutz P. Mueller, and Thomas Mueller. 2023. "Luciferase Expressing Preclinical Model Systems Representing the Different Molecular Subtypes of Colorectal Cancer" Cancers 15, no. 16: 4122. https://doi.org/10.3390/cancers15164122
APA StyleRotermund, A., Staege, M. S., Brandt, S., Luetzkendorf, J., Lucas, H., Mueller, L. P., & Mueller, T. (2023). Luciferase Expressing Preclinical Model Systems Representing the Different Molecular Subtypes of Colorectal Cancer. Cancers, 15(16), 4122. https://doi.org/10.3390/cancers15164122







