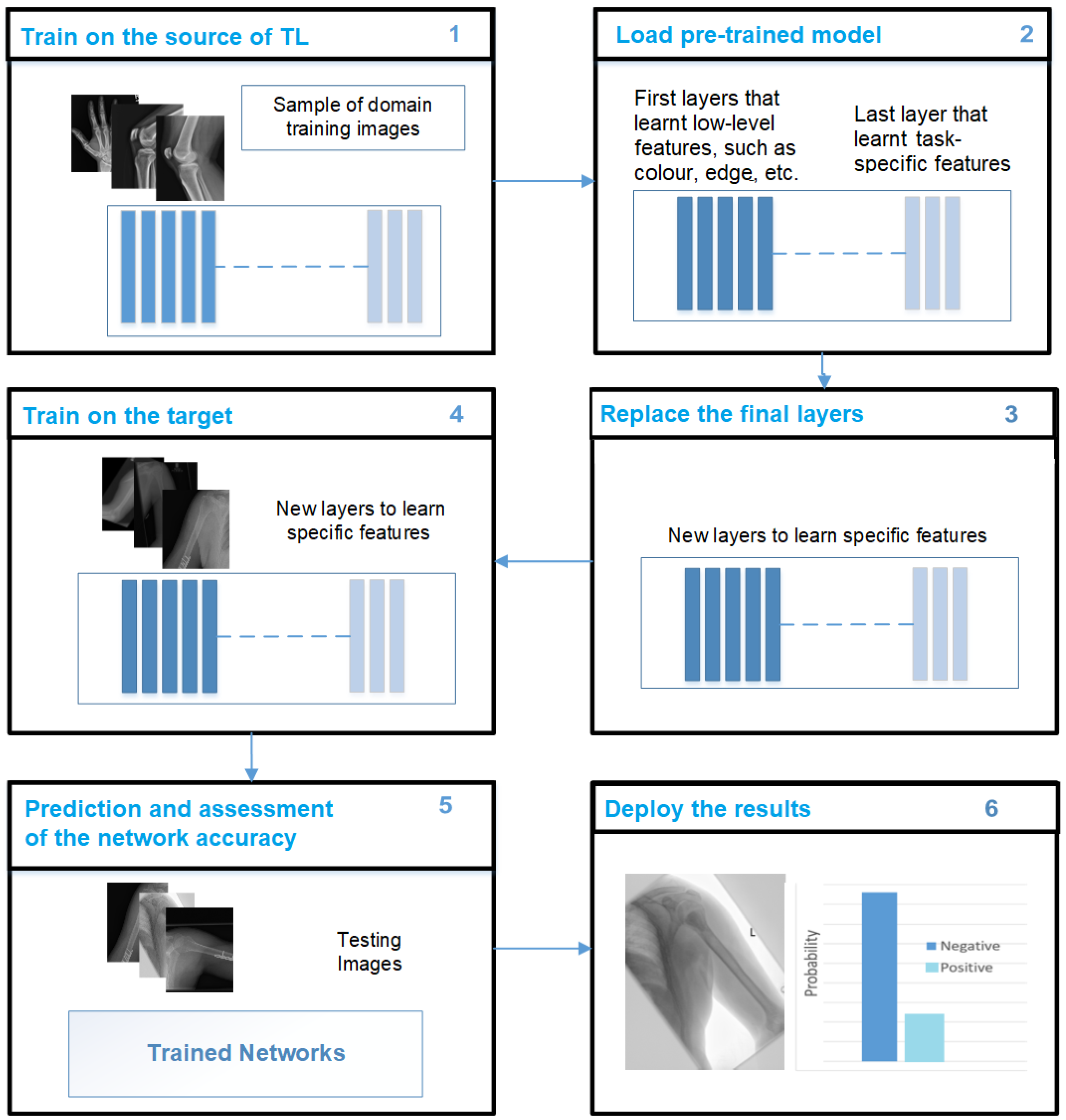
1. Introduction
X-ray medical images are widely recognised as powerful tools for identifying abnormalities in bone classification. They provide valuable information on the structure and condition of bones, helping to diagnose and treat various skeletal disorders. However, one of the most-significant challenges faced in the use of artificial intelligence (AI) algorithms for medical image analysis is the scarcity of data available for training and validation [
1].
The limited availability of labelled medical images poses a substantial obstacle to the development of accurate and reliable AI models for bone classification tasks. Obtaining a large and diverse medical image dataset is crucial to the effective training of AI algorithms because the performance and generalisation capabilities of AI models can be compromised without sufficient data.
Addressing the issue of data scarcity in medical imaging is essential to unlocking the full potential of AI to improve bone classification accuracy and aid clinical decision-making. Researchers and practitioners continue to explore methods such as active learning (AL), synthetic data generation [
2], generative data augmentation [
3], transfer learning (TL) [
4], and collaboration between institutions to overcome the challenges posed by limited medical image datasets. By expanding the availability and quality of annotated medical images, the performance and robustness of AI algorithms for bone classification will be enhanced and ultimately improve patient care in the medical imaging domain [
5].
Despite advancements in medical applications and the progress made in computer vision, detecting abnormalities in the humerus and wrist using X-ray images remains a challenge [
6]. The complexity of bone structures, subtle variations in abnormalities, and the inherent limitations of X-ray imaging techniques contribute to this challenge. Ongoing research and development efforts focus on using AI and machine learning (ML) techniques to improve the accuracy and efficiency of humerus and wrist abnormality detection in X-ray images. ML models can be trained on large datasets of annotated X-ray images, which enables them to recognise patterns and detect subtle pathological indicators.
By integrating AI and ML technologies into the field of orthopaedics, medical professionals can benefit from improved accuracy and efficiency when diagnosing and treating patients with bone conditions. Deep learning (DL) is a branch of AI techniques that has demonstrated exceptional abilities in accurately, reliably, and rapidly classifying medical images into binary and multiclass categories [
7,
8]. DL has become the gold standard in medical image analysis and has demonstrated remarkable performance in various areas, such as radiology [
9,
10], dermatology [
11], pathology [
12], and ophthalmology [
13,
14]. These applications, which span different medical fields, are based on human experience, thus making DL a valuable tool in a competitive domain.
The requirement for large amounts of labelled data is a significant challenge to the development of high-performing DL models. However, the scarcity and imbalance of medical image datasets pose significant challenges due to the cost effectiveness and time consumption associated with DL approaches. Despite these challenges, DL models have consistently demonstrated impressive performance in classifying medical images. Krizhevsky et al. [
15] introduced a model based on a convolutional neural network (CNN) architecture that represented a significant milestone in the history of DL and computer vision. Their developing work demonstrated the potential of deep CNNs in image-classification tasks, thus setting new standards for accuracy and inspiring further research and innovation in the ImageNet Large Scale Visual Recognition Challenge (ILSVRC), a critical image classification competition. Several studies have successfully used TL techniques in DL models to address data scarcity. TL involves using models that were pre-trained on large-scale datasets and fine-tuning them on target medical image datasets [
16]. This approach has been shown to be effective in improving the performance of DL models in various studies [
1]. For example, Fang et al. [
17] used fine-tuning and feature augmentation methods and an area under the curve (AUC) of 0.73. However, unbalanced data remain a limitation in certain studies [
18]. Musculoskeletal radiographs (MURA) is a dataset designed specifically for musculoskeletal medical imaging, as it comprises a large collection of radiographs (X-rays). The dataset covers a diverse set of musculoskeletal abnormalities, as well as normal cases, which makes it a valuable resource for training and evaluating medical image classifiers [
19]. The use of TL techniques for medical image classification has increased significantly, and this trend highlights the growing recognition of TL as a valuable strategy for handling data scarcity and improving the performance of DL models in the medical field.
CNNs have been widely used to classify input data as various states of disease [
20]. CNNs’ deep architecture feedforward neural networks serve as the basis for many deep neural network models (DNN) in the medical field [
20]. In addition to CNNs, other types of neural networks have also been used, such as recurrent neural networks (RNNs) with variations such as long short-term memory (LSTM), transformers, and generative adversarial networks (GANs) [
21]. CNNs have proven to be particularly effective for image-processing and -classification tasks. One of their key strengths lies in their ability to extract meaningful patterns and characteristics from images regardless of scaling, mirroring, rotation, or translation [
22]. This property makes CNNs highly suitable for medical image analysis, where the accurate identification and classification of image characteristics are crucial for diagnosis and treatment.
Furthermore, it should be noted that most studies that focus on humerus and wrist abnormalities do not thoroughly evaluate the “black box” explanation [
23]. However, the lack of model explainability associated with black box methods is considered a significant obstacle to clinical adoption and user confidence [
24]. To identify biases and ensure the reliability of DL applications, it is essential to explain the decision-making processes of the models. The use of TL is specifically recommended to address the issue of data scarcity and inconsistency in the medical field. TL leverages pre-trained DL models by using source datasets and fine-tuning them for target tasks. TL has had a positive impact on the medical field, especially in scenarios where limited data are available. Given the challenges of gathering medical imaging data, TL has become a crucial tool in medical image analysis. The ImageNet Large Scale Visual Recognition Challenge (ILSVRC-2012) competition dataset is widely recognised and widely used to improve the performance of various image-processing tasks, including classification, segmentation, and detection [
25,
26,
27]. Although ImageNet has improved model performance, it is essential to note that medical images differ significantly from the natural images represented in ImageNet. These differences encompass various aspects, such as shape, colour, resolution, and dimensionality.
Models pre-trained on ImageNet are limited in terms of performance enhancement when dealing with medical images due to domain mismatch. Several authors have explained how TL using the same domain improved the performance of DL models in medical imaging applications [
16,
28,
29].
Alternatively, the fusion technique could be used as an effective way to merge the features extracted by various CNN models for further enhancement. However, supporting the models’ results using the appropriate tools is necessary in order to trust the DL outcome [
30].
The trade-off lies in the ability of DL models to leverage large amounts of data and learn complex patterns, whereas traditional techniques may have been more suitable for cases with limited data availability [
31].
This paper aims to address the problem of data scarcity and the mismatch features of TL. Furthermore, it also addresses feature generalisation in training ML classifiers. Therefore, in this paper:
We propose a new TL approach to address the issue of data scarcity and the drawbacks of previous TL methods in medical imaging applications.
An improved feature fusion is proposed to increase trust in the final decision.
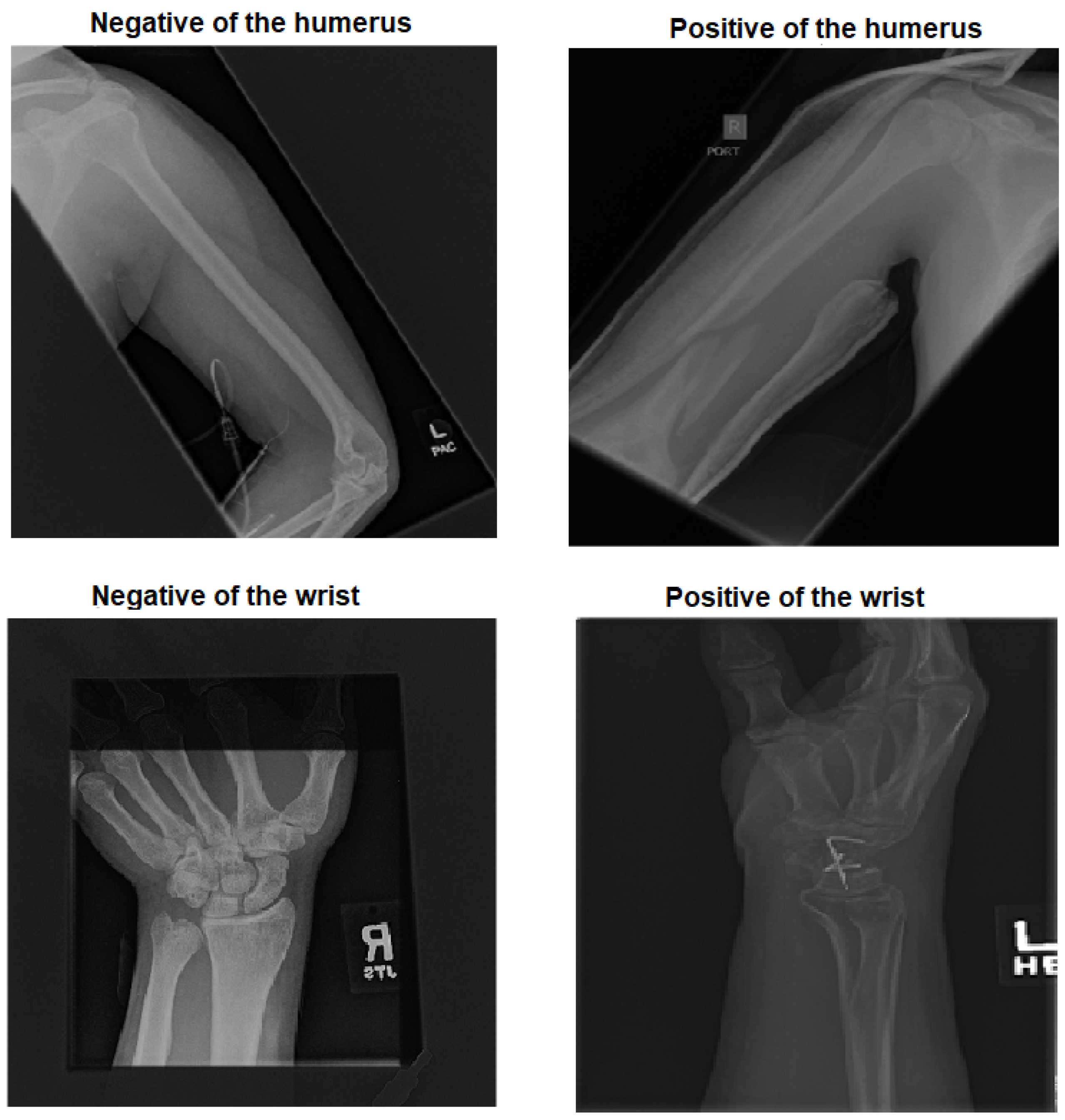
We employed two pre-trained ImageNet models for experimenting with two X-ray tasks to detect abnormalities in the humerus and wrist.
We applied a feature fusion strategy to train multiple ML classifiers in two different training scenarios.
We achieved an accuracy of 87.85%, an F1-score of 87.63%, and a Cohen’s Kappa coefficient of 75.69% in humerus classification. For wrist classification, our approach achieved an accuracy of 85.58%, an F1-score of 82.70%, and a Cohen’s Kappa coefficient of 70.46%.
We briefly review the most-recent DL techniques from the MURA dataset.
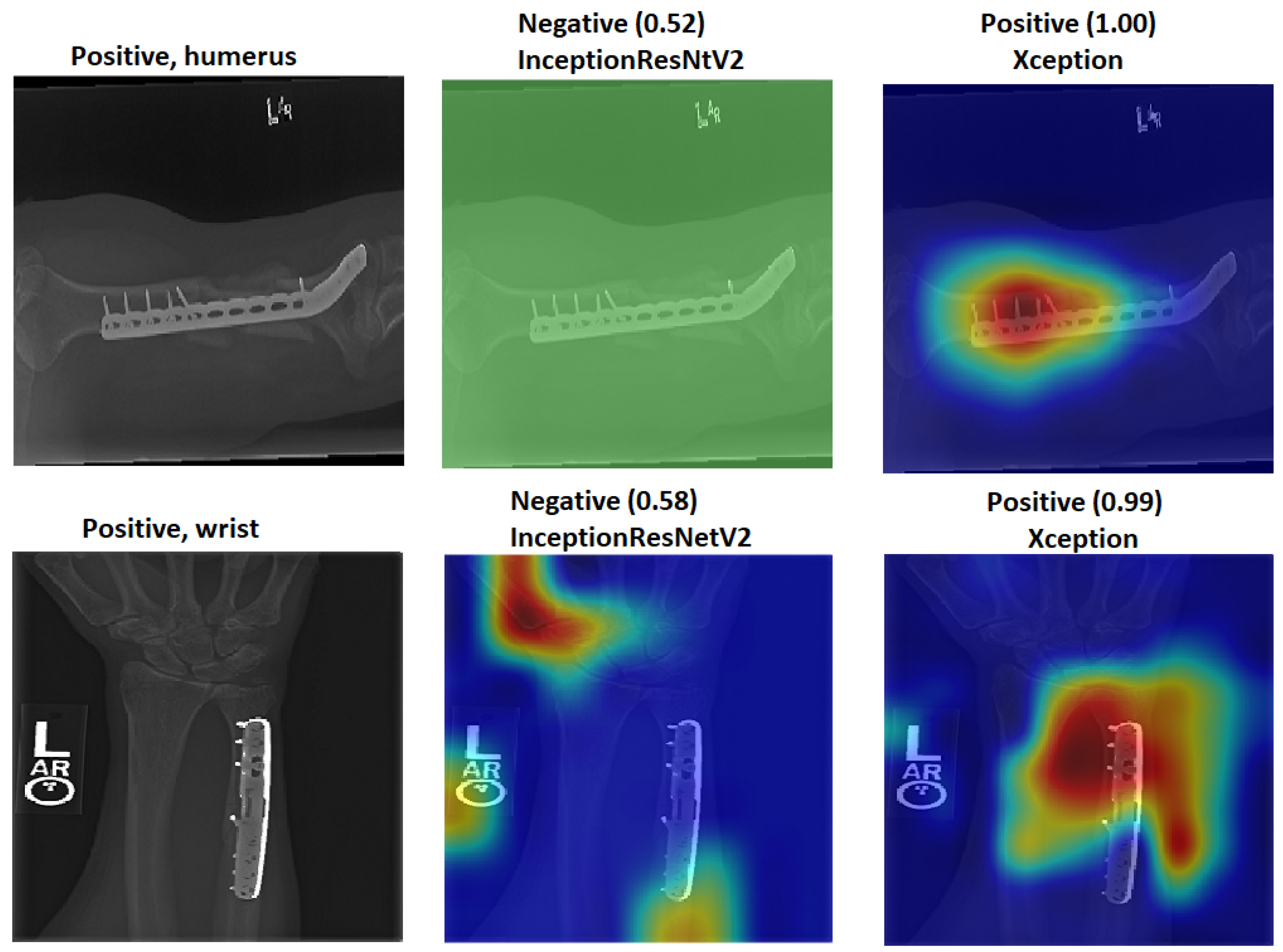
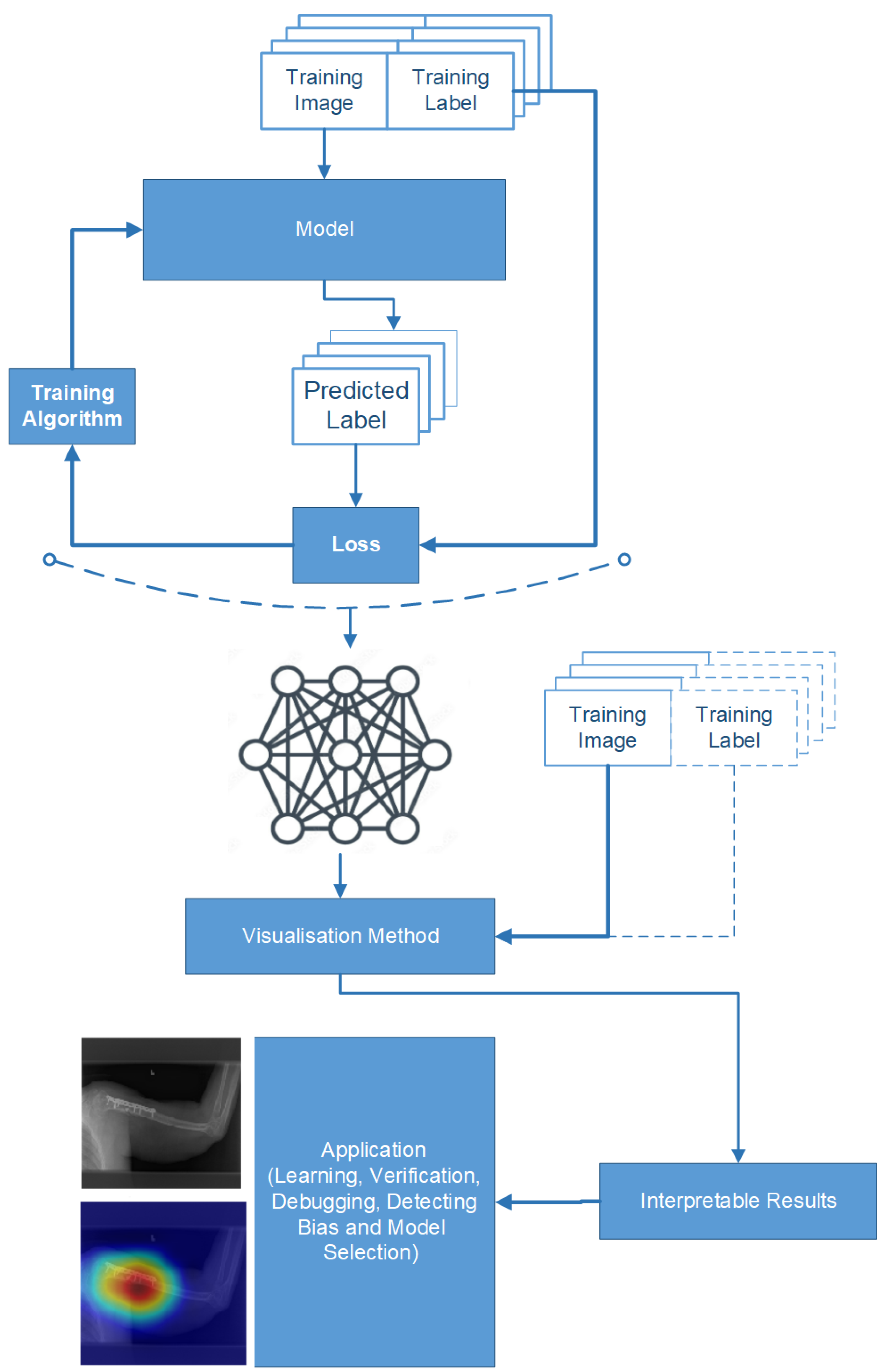
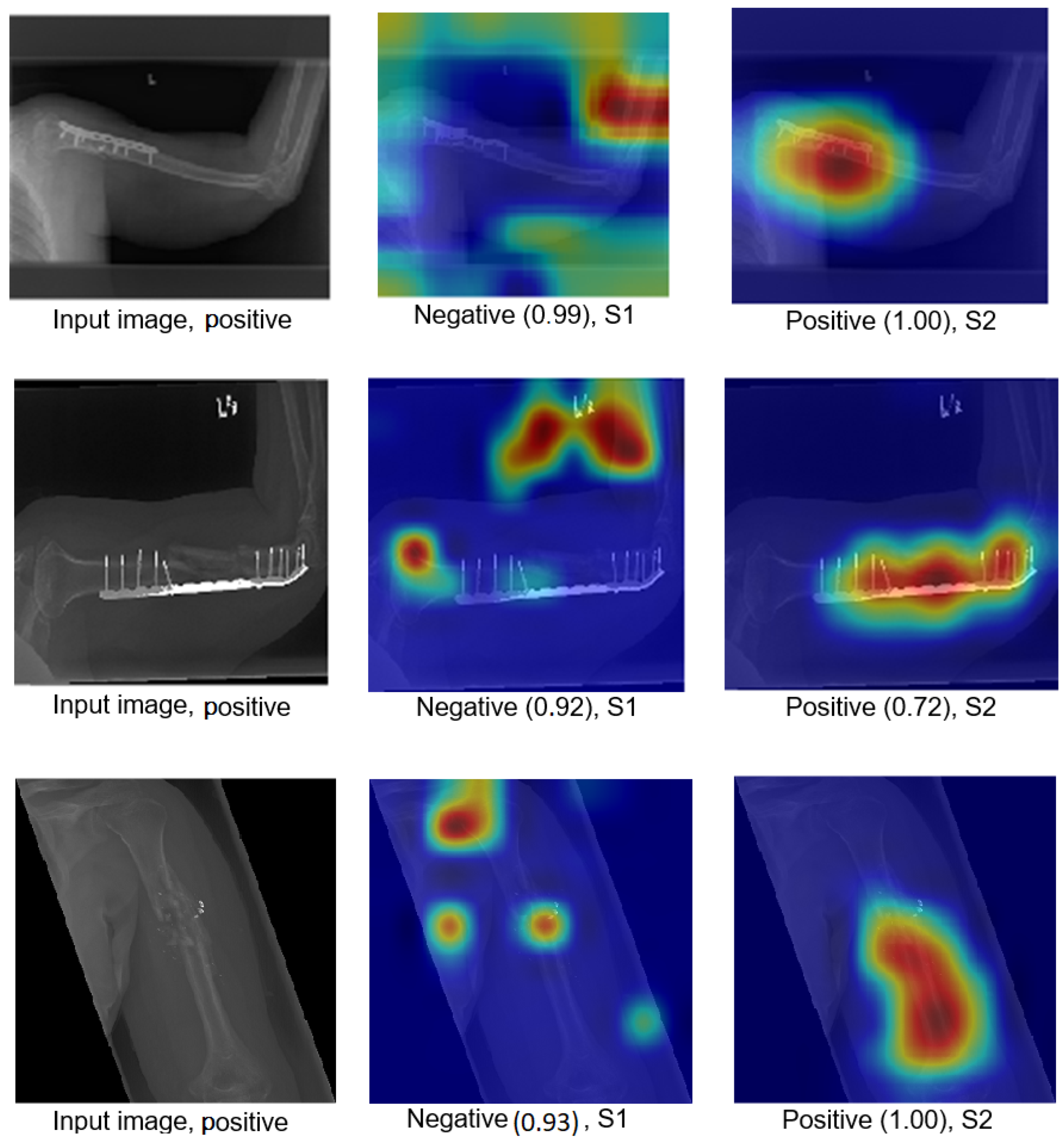
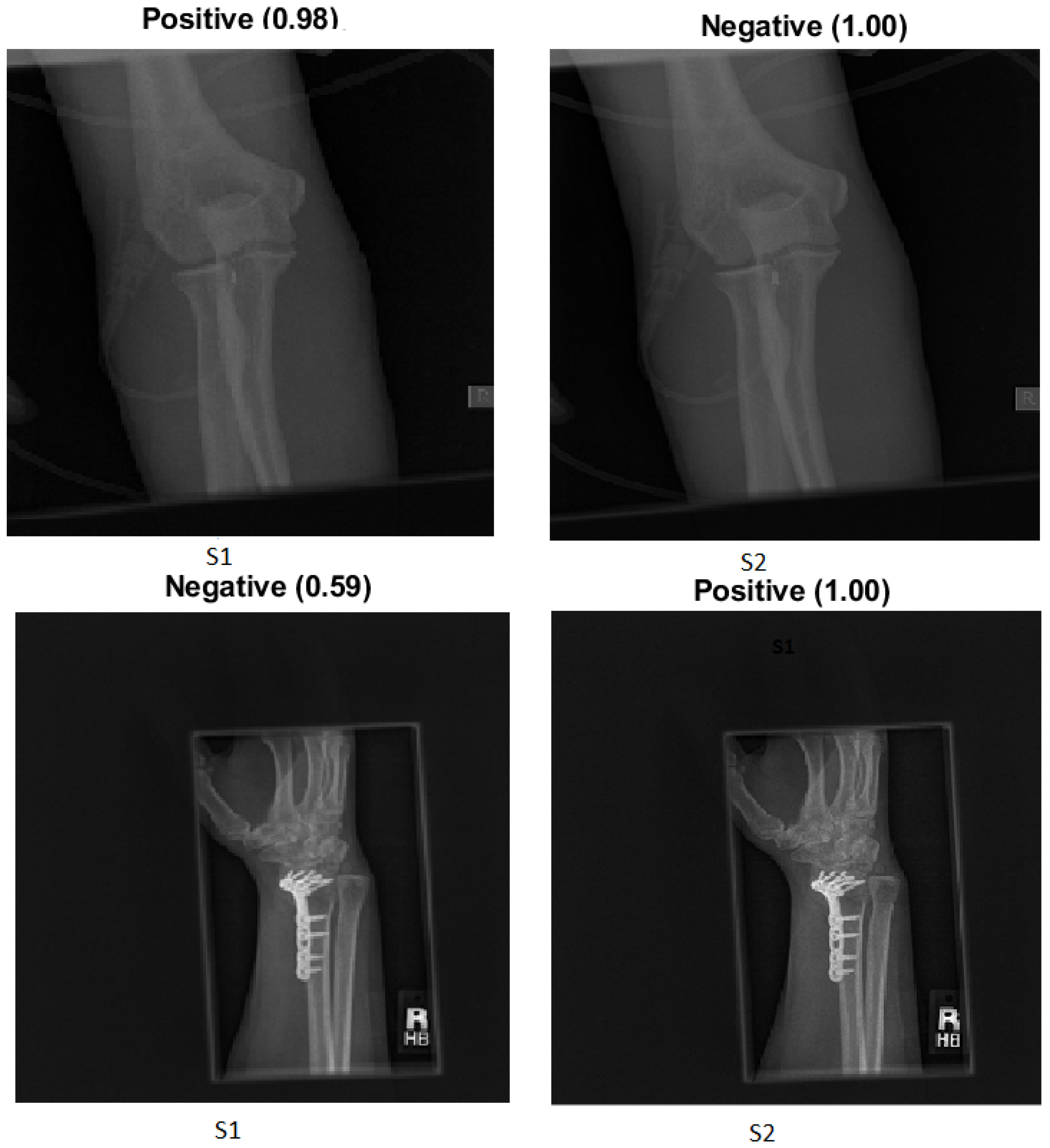
We explain how the decision was made to adopt two visualisation tools, i.e., gradient-weighted class activation mapping (Grad-CAM) and local printable model-agnostic explanations (LIME), to verify the robustness of the proposed method.
We show that the proposed TL approach and feature-fusion technique can be applied to various medical image applications, thus expanding their utility and potential impact, as demonstrated by adopting a computed tomography (CT) case that showed significant improvements in the results.
2. Related Work
This section provides an overview of the latest techniques used in the field. One such technique is the use of CNNs, which have demonstrated remarkable success in computer vision tasks and have become crucial for image classification. As mentioned above, the availability of large datasets and the time-consuming nature of training classifiers pose significant challenges to achieving optimal training results. Various techniques have been proposed to increase the size of the datasets, and one such strategy is active learning (AL). AL involves iteratively selecting the most-informative samples from an unlabelled dataset for annotation and model training. The primary goal of AL is to maximise the model’s performance while minimising the amount of labelled data required for training. This is particularly beneficial in scenarios where obtaining labelled data can be costly or time-consuming. The AL process relies on initially training the model on a small labelled dataset and then using a query strategy to determine which unlabelled samples should be selected for labelling. The query strategy is crucial, as it selects samples based on their potential impact on the model’s performance. There are several common query strategies used, including uncertainty sampling, query-by-committee, and information-density-based methods. After selecting and labelling these informative samples, the samples are incorporated into the training set to update the model using the newly labelled data. The training process for AL is often repeated over time until the model reaches a desired performance level or satisfies other specific criteria relevant to the desired application. This iterative approach helps the model learn from diverse and informative examples, which gradually improves its performance with fewer labelled samples. A key limitation of AL algorithms is that they are based on labelling one sample at a time. This means that, after each sample is labelled, the model needs to be retrained, which can be computationally expensive and time-consuming [
32]. Researchers have thus been working on optimising this process to reduce the retraining burden.
For example, Wen et al. [
33] conducted a study on using AL for nucleus segmentation in pathology images in which they investigated how AL performance improves for three different algorithm families: support vector machines (SVMs), random forest (RF), and CNNs. By employing AL, the researchers aimed to enhance the efficiency and accuracy of nucleus segmentation in medical imaging. Moreover, synthetic data generation is a powerful technique used to address data scarcity or privacy concerns by creating additional data artificially. In various domains, obtaining a sufficient amount of labelled data to train ML models can be challenging. Synthetic data generation offers a solution by generating new data points that are similar to the existing data, but are not direct replicas, thus expanding the dataset [
3]. GANs have emerged as highly successful and versatile approaches that can find applications across various domains, including image generation, text generation, and video synthesis. GANs possess the remarkable ability to generate high-quality, diverse, and realistic synthetic data, making them invaluable for a wide range of tasks, including data augmentation, data synthesis, and creative applications [
34]. Adar et al. [
35] demonstrated the successful application of GANs for enhancing the classification performance for liver lesion classification by employing data augmentation techniques. In their study, conducted in 2018, the authors used GANs to increase the amount and diversity of training data, specifically for liver lesion classification. The authors observed a significant improvement in sensitivity and specificity compared to using traditional data-augmentation methods. Specifically, the classification performance increased from 78.6% sensitivity and 88.4% specificity (when using traditional enhancements) to 85.7% sensitivity and 92.4% specificity (when using GAN-generated data). This improvement can be attributed to the GAN’s ability to provide a more-diverse and -representative training dataset, allowing the classifier to better generalise and make more-accurate predictions on real-world liver lesion data.
Furthermore, Yi et al. [
36] extensively explored and discussed the application of GAN image synthesis in various critical medical imaging domains. They highlighted the significant impact of GANs on improving medical image generation, analysis, and diagnostics across a range of applications. In the domain of brain magnetic resonance imaging (MRI), GANs have proven to be particularly valuable. Calimeri et al. [
37] and Bermudez et al. [
38] successfully used GAN-based image synthesis to generate realistic brain MRI scans. This synthetic data augmentation has led to the improved training of brain image analysis models and better performance in tasks such as segmentation and disease classification. For lung cancer diagnostics, Chuquicusma et al. [
39] demonstrated the effectiveness of GANs in generating synthetic lung nodules and lesions. This data synthesis enabled the development and validation of robust and accurate lung-cancer-detection models, even when dealing with limited real-world data. High-resolution skin imaging is another domain where GANs have shown promise. Baur et al. [
40] used GANs to synthesise high-resolution melanoma images. This approach enhanced the quality and diversity of the dataset used for training skin-cancer-detection models, leading to improved diagnostic accuracy and the early detection of skin cancer. While data augmentation is a powerful tool for improving model performance, its lack of interpretability can be a concern, especially in sensitive or critical applications. Ensuring transparency and explainability is essential for building trust and confidence in AI models, enabling users to comprehend why certain augmentations lead to improved performance. By incorporating human-understandable augmentation strategies and leveraging model interpretability techniques, researchers and practitioners can strike a balance between performance enhancement and interpretability, thereby making AI systems more trustworthy and responsible [
41].
Additionally, Tahmina et al. [
42] applied data augmentation in their study to detect humerus fractures. They used preprocessed images to increase the quality of the dataset, and the performance of their study was 78%.
Despite the promise of data augmentation, there are challenges that must be considered. Selecting appropriate augmentation techniques and parameters requires careful consideration. In addition, achieving the correct balance between augmenting and maintaining the integrity of the medical data is crucial to ensuring that the synthetic examples remain consistent with the real-world distribution of medical images. In this context, TL and pre-training are two alternative strategies for learning the low-level properties in CNNs. TL has proven to be an effective technique to train CNNs with limited data, thus improving the performance of DL models. TL enables one to leverage the knowledge and features learned from pre-existing models and apply them to new tasks or domains, thus reducing the need for extensive data collection and training time. By capitalising on pre-trained models, TL enables the efficient and effective training of CNNs even with smaller datasets. CNNs have been effectively trained using TL techniques in which the weights of pre-trained CNNs are used to classify other target images [
20]. TL can be broadly categorised into two types: fine-tuned TL and feature extraction TL. The feature extraction TL approach employs a well-trained CNN model that was trained on a large dataset. The convolutional layers of the pre-trained model are frozen, whereas the fully connected layers are discarded. The frozen convolutional layers act as fixed feature extractors, which capture meaningful representations from the input images. These extracted features are then fed into a new classifier, which can be implemented using new fully connected layers or a supervised ML approach. During this type of TL, only the parameters of the new classifier are trained using the pre-learned features of the pre-trained CNN model [
43].
This approach transfers knowledge from the pre-trained CNN model, which has learned rich representations from a large dataset, to the new medical task at hand. By using the extracted features and training only the classifier, feature-extracting TL enables efficient training with limited data, thus reducing the need for extensive computational resources and training time.
In contrast, the fine-tuning TL approach involves replacing the classifier layers while using the pre-trained CNN model that was trained on a large dataset as a base. In this approach, the convolutional and classifier layers are fine-tuned during the training process. The weights of the convolutional layers are initialised with the pre-trained weights from the CNN model, while the classifier layers start with random weights. The entire network is trained through this training process, allowing it to adapt and learn task-specific representations [
44].
The fine-tuning TL approach is beneficial when the target task requires more-specialised knowledge and the available target dataset is more extensive. The model can learn task-specific features and improve its performance by updating the weights of the convolutional and classifier layers.
Both feature-extracting TL and fine-tuning TL have their advantages and are, thus, used based on the specific requirements of the task at hand. Feature-extracting TL is particularly useful when limited training data are available, as it leverages the pre-trained model’s learned features. Fine-tuning TL, however, can enhance performance by allowing the model to learn task-specific representations by updating both the convolutional and classifier layers. For this reason, this study used the fine-tuning TL type.
Various studies applied DL models to detect abnormalities in X-ray images, such as Ortiz et al. [
45], who investigated three AI models to detect pneumonia in chest X-ray images; the authors used the feature-extraction technique with three different ML classifiers, and the accuracy of this study was 83.00% and with an 89% sensitivityfor radiomics, an accuracy of 89.9% with a 93.6% sensitivity for fractal dimension, and 91.3% accuracy with a 90.5% sensitivity for superpixel-based histon. Moreover, Canayaz et al. [
46] implemented feature fusion by combining AlexNet and VGG19 models to classify COVID-19, pneumonia, and normal X-ray images; the authors’ approach achieved 99.38% accuracy. In addition, Rajinikanth et al. [
47] used InceptionV3, which was pre-trained, to detect pneumonia in chest X-ray images. The authors used deep feature extraction and feature reduction with the Firefly Algorithm and multi-class classification using five-fold cross-validation; the results of the K-nearest neighbour (KNN) classifier demonstrated an accuracy of 85.18%. Furthermore, Rajinikanth et al. [
48] also applied one-fold and two-fold training by using UNet lung section segmentation.
Indeed, DL models have been implemented in various studies to improve the detection of abnormalities in musculoskeletal images. Rajpurkar et al. [
19] conducted a study using a dataset called MURA, consisting of 40,005 musculoskeletal images. Their research employed a DenseNet169 CNN architecture, as described in Huang et al. [
49], whereby each layer was linked to all other layers in a feedforward fashion, thus achieving a deep network design. The model classified the images as abnormal if the prediction probability was greater than 0.5. The performance of the model was evaluated using two metrics: sensitivity and specificity. The sensitivity, which measures the ability of the model to identify true positives correctly, was 81.5%. The specificity, which measures the model’s ability to identify true negatives correctly, was 88.7%. These metrics indicate the model’s ability to detect normal and abnormal cases accurately. The model’s overall performance was also assessed using the area under the receiver operator characteristic (AUROC) metric, which considers the trade-off between sensitivity and specificity. The model achieved an AUROC of 92.9%, indicating its solid discriminative power. When diagnosing abnormalities in fingers and wrists, the model’s performance was roughly equivalent to that of the best radiologists. Despite the model’s agreement with the gold standard being similar to that of other radiologists, it was still relatively low. However, when diagnosing abnormalities in elbows, forearms, hands, humerus, and shoulders, the model’s performance was worse than that of the best radiologists [
19].
To investigate this, Chada [
50] conducted a study to evaluate the performance of three state-of-the-art CNN architectures, namely DenseNet169, DenseNet201, and InceptionResNetV2, on the MURA dataset. The researchers fine-tuned these CNN models using the Adam optimiser with a learning rate of 0.001. The evaluation was performed separately for humerus and finger images.
For the humerus task, the best performance was observed with the DenseNet201 model, which achieved a Cohen’s Kappa score of 76.4%. This indicated a substantial agreement between the predictions of the model and the ground-truth labels. For the images of fingers, however, the InceptionResNetV2 model demonstrated the best performance, obtaining a Cohen’s Kappa score (which assesses the agreement between the model’s predictions and the ground-truth beyond chance) of 55.5%. These results highlight the effectiveness of these CNN architectures in detecting abnormalities in musculoskeletal images, with performance variations depending on the specific anatomical areas. However, the performance of finger X-rays was less promising, undoubtedly due to the limitations of high inter-radiologist variation.
Another study focused on classifying proximal humerus fractures using the Neer classification system [
51]. The researchers used a pre-trained ResNet-152 classifier that was fine-tuned for the specific task of classifying fractures. This approach leveraged the pre-trained weights of the ResNet-152 model and trained the classifier layers on the target dataset. Using this TL technique, the model accurately classified 86% of the proximal humerus fractures according to the Neer classification system. Despite the fact that the Neer classification is the most-regularly used technique for proximal humerus fracture classification, the reliability of this study needs to improve. The author in [
51] assessed the diagnostic performance of CNNs with a cropped single-shoulder X-ray image, but this might not be applicable to the relative clinical scenario.
Furthermore, Lindsey et al. [
52] investigated the detection of wrist fractures, comparing the performance of radiologists with and without the assistance of CNN models. The study aimed to assess how the use of CNN models affected radiologists’ diagnostic capabilities. The results indicated a marked increase in radiological performance when aided by CNN models, highlighting the potential of DL models as supportive tools in the field of fracture detection. However, the study had a number of drawbacks, such as the fact that the experiment was a review of the data performed through the web interface that simulated an image archiving and communication system (PACS) used by medical professionals for medical imaging. Furthermore, the accuracy of the physicians’ and the model’s diagnoses in this study was restricted to the determination of what is visible inside a radiograph. Finally, the diagnosed condition’s improvement or deterioration was influenced by factors other than DL accuracy in diagnosis.
Saif et al. [
53] proposed a capsule-network-based approach to classify abnormalities in the musculoskeletal system. They conducted experiments by training their network on images of different sizes, specifically images of 64 × 64, 128 × 128, and 224 × 224 px. The goal was to determine the optimal image size to achieve an accurate classification, and the results indicated that, when using 224 × 224 px images, the network achieved the highest training accuracy (96%) for wrist radiography images. However, it is possible that the network’s performance in some studies was influenced by overfitting, a situation where the model becomes excessively tailored to the training data, which leads to the poor generalisation for unseen data.
However, in 2019, Varma et al. [
54] introduced the MURA dataset, which included a private dataset of 93,455 lower-extremity radiographs with images of the foot, ankle, knee, and hip. This dataset was explicitly curated for abnormality detection using less-adequate extremity radiographs. To evaluate the performance of different CNN architectures, Varma et al. trained ResNet-50, DenseNet-161, and ResNet-101 on a subset of their private dataset. Despite the structural differences between these architectures, the authors found that performance did not vary significantly. However, the authors then proposed a comparison explicitly focused on the DenseNet-161 architecture, which was trained on both the ImageNet and MURA datasets, to assess the impact of TL on model performance. This comparison aimed to investigate the effect of TL, where a CNN model is initially pre-trained on a large-scale dataset (such as ImageNet) and then fine-tuned on a specific task or domain (in this case, the MURA dataset). Using pre-trained weights and pre-learned representations from the large-scale dataset, TL can potentially improve the performance of CNN models for the target task. However, the limitation of this study comes from the fact that it reviewed data with datasets from a single institution; thus, the performance of the authors’ models may differ in the real world when different images are used.
Furthermore, Kandel et al. [
55] conducted a study using the MURA dataset to examine the performance of six CNN architectures, namely, VGG, Xception, ResNet, GoogLeNet, InceptionResNet, and DenseNet, to detect bone abnormalities. They compared models trained from scratch with pre-trained models using ImageNet and then fine-tuned them on the MURA dataset. The study’s results highlighted that TL has the potential to enhance model performance while reducing the susceptibility to overfitting. Among the five state-of-the-art CNNs evaluated for the MURA dataset, the humerus datasets achieved the highest precision (81.66%). Although the authors used the TL approach, the training-from-scratch approach’s poor performance could have been due to the number of images in the dataset, as well as the hyperparameter selection. The CNNs considered are distinguished by their incorporation of a significant number of trainable parameters (such as weights), and the number of images used to train these networks is insufficient to develop an effective model. Hyperparameters indicate the significance of the learning rate. Although the authors used a lower value of the learning rate in the fine-tuning technique to avoid significantly modifying the original weights of the designs, the training-from-scratch strategy may demand a higher value of the learning rate.
Feature fusion of DL techniques also was implemented by Bhan et al. [
56] to classify fracture or non-fracture in the MURA dataset; the five pre-trained models were DenseNet-169, MobileNetV2 ResNet-50, ResNeXt-50 and VGG16, and then, these pre-trained models were combined in this study. The results of the feature-fusion approach were that the humerus achieved an 87.85% accuracy and a 75.72% Cohen’s Kappa, while the accuracy was 83.13% and a 66.25% Cohen’s Kappa for the shoulder. In the same study, the performance of the wrist classification was 86.65% accuracy and a 72.59% Cohen’s Kappa.
This literature review focused on the significant challenge caused by the limited availability of annotated data in the medical domain. The scarcity of annotated medical datasets prevents the full potential and effectiveness of DL algorithms. This challenge has motivated the main objective of this article, which was to explore strategies that can achieve greater performance with minimal data in the field of medical DL.