Evaluation of CIN2/3 Lesion Regression in GynTect® DNA Methylation-Marker-Negative Patients in a Longitudinal Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. Cytology, HPV Testing and Methylation Analyses
2.3. Sample Size Calculation and Statistical Analysis
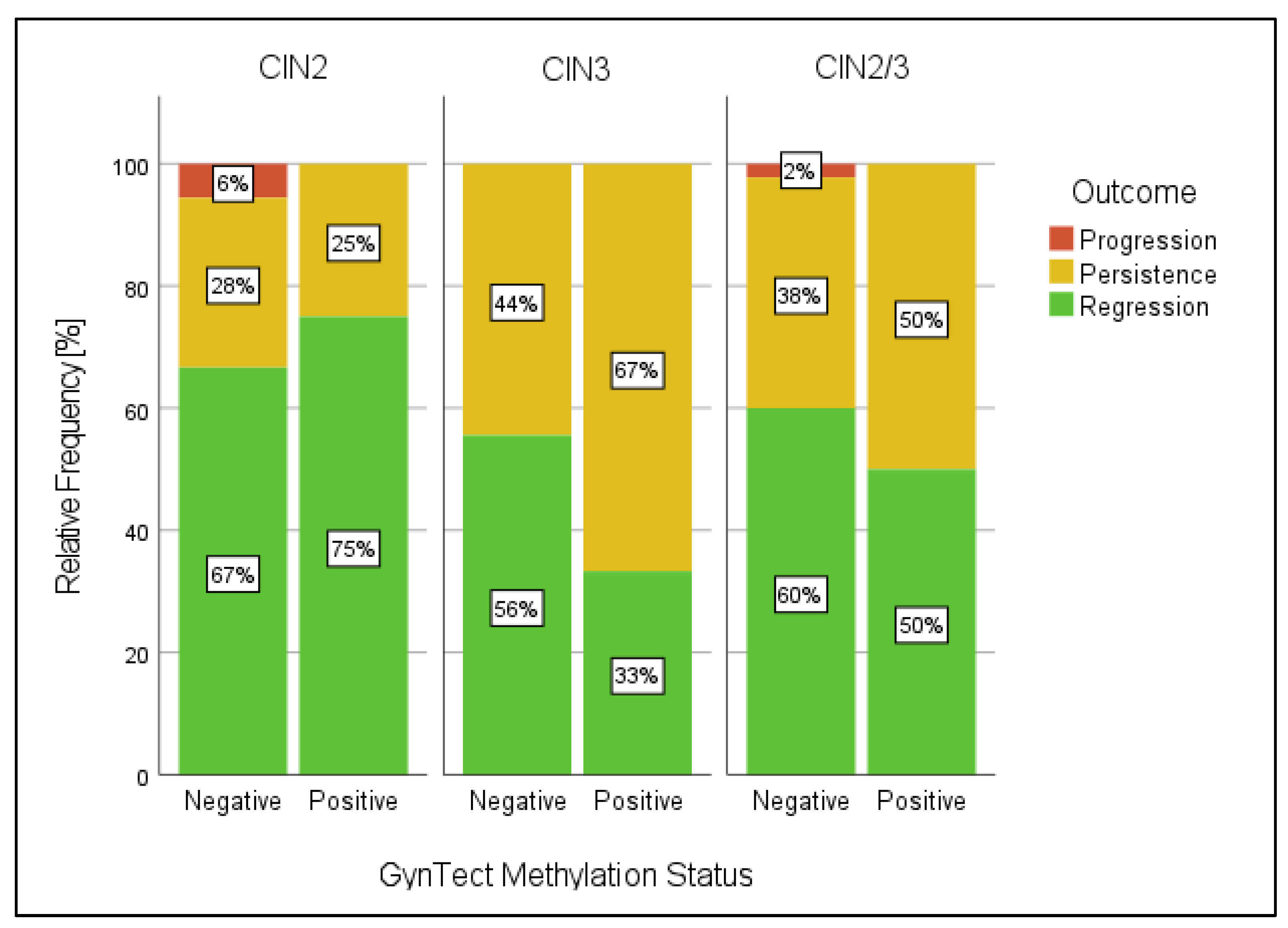
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wentzensen, N.; Arbyn, M.; Berkhof, J.; Bower, M.; Canfell, K.; Einstein, M.; Farley, C.; Monsonego, J.; Franceschi, S. Eurogin 2016 Roadmap: How HPV knowledge is changing screening practice. Int. J. Cancer 2017, 140, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Simon, M.; de Sanjosé, S.; A Clarke, M.; Poljak, M.; Rezhake, R.; Berkhof, J.; Nyaga, V.; Gultekin, M.; Canfell, K.; et al. Accuracy and effectiveness of HPV mRNA testing in cervical cancer screening: A systematic review and meta-analysis. Lancet Oncol. 2022, 23, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Ikenberg, H.; Bergeron, C.; Schmidt, D.; Griesser, H.; Alameda, F.; Angeloni, C.; Bogers, J.; Dachez, R.; Denton, K.; Hariri, J.; et al. Screening for Cervical Cancer Precursors With p16/Ki-67 Dual-Stained Cytology: Results of the PALMS Study. J. Natl. Cancer Inst. 2013, 105, 1550–1557. [Google Scholar] [CrossRef]
- Wentzensen, N.; Fetterman, B.; Castle, P.E.; Schiffman, M.; Wood, S.N.; Stiemerling, E.; Tokugawa, D.; Bodelon, C.; Poitras, N.; Lorey, T.; et al. p16/Ki-67 Dual Stain Cytology for Detection of Cervical Precancer in HPV-Positive Women. J. Natl. Cancer Inst. 2015, 107, djv257. [Google Scholar] [CrossRef]
- Stanczuk, G.A.; Baxter, G.J.; Currie, H.; Forson, W.; Lawrence, J.R.; Cuschieri, K.; Wilson, A.; Patterson, L.; Govan, L.; Black, J.; et al. Defining Optimal Triage Strategies for hrHPV Screen–Positive Women—An Evaluation of HPV 16/18 Genotyping, Cytology, and p16/Ki-67 Cytoimmunochemistry. Cancer Epidemiol. Biomarkers Prev. 2017, 26, 1629–1635. [Google Scholar] [CrossRef]
- Zhang, L.; Tan, W.; Yang, H.; Zhang, S.; Dai, Y. Detection of Host Cell Gene/HPV DNA Methylation Markers: A Promising Triage Approach for Cervical Cancer. Front. Oncol. 2022, 12, 831949. [Google Scholar] [CrossRef]
- Taryma-Leśniak, O.; Sokolowska, K.E.; Wojdacz, T.K. Current status of development of methylation biomarkers for in vitro diagnostic IVD applications. Clin. Epigenet. 2020, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, R.D.M.; Snijders, P.J.F.; Heideman, D.A.M.; Meijer, C.J.L.M. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat. Rev. Cancer 2014, 14, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Eichelkraut, K.; Schmidt, D.; Zeiser, I.; Hilal, Z.; Tettenborn, Z.; Hansel, A.; Ikenberg, H. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer 2018, 18, 1197. [Google Scholar] [CrossRef]
- Loopik, D.L.M.; Doucette, S.M.; Bekkers, R.L.; Bentley, J.R.M. Regression and Progression Predictors of CIN2 in Women Younger Than 25 Years. J. Low. Genit. Tract Dis. 2016, 20, 213–217. [Google Scholar] [CrossRef]
- Lee, M.H.; Finlayson, S.J.; Gukova, K.; Hanley, G.; Miller, D.; Sadownik, L.A. Outcomes of Conservative Management of High Grade Squamous Intraepithelial Lesions in Young Women. J. Low. Genit. Tract Dis. 2018, 22, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Tainio, K.; Athanasiou, A.; Tikkinen, K.; Aaltonen, R.; Cárdenas, J.; Hernándes, C.; Glazer-Livson, S.; Jakobsson, M.; Joronen, K.; Kiviharju, M.; et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: Systematic review and meta-analysis. BMJ 2018, 360, k499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, C.-X. Spontaneous Regression of Cervical Intraepithelial Neoplasia 2: A Meta-analysis. Gynecol. Obstet. Investig. 2019, 84, 562–567. [Google Scholar] [CrossRef]
- Massad, L.S.; Einstein, M.H.; Huh, W.K.; Katki, H.A.; Kinney, W.K.; Schiffman, M.; Solomon, D.; Wentzensen, N.; Lawson, H.W. 2012 Updated Consensus Guidelines for the Management of Abnormal Cervical Cancer Screening Tests and Cancer Precursors. J. Low. Genit. Tract Dis. 2013, 17, S1–S27. [Google Scholar] [CrossRef]
- Grigore, M.; Cruickshank, M.E.; Nieminen, P.; Tjalma, W.; Moss, E.; Redman, C. National guidelines for management of cervical squamous intraepithelial lesion: A survey of European Federation for colposcopy members. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021, 256, 46–50. [Google Scholar] [CrossRef]
- Kremer, W.W.; Dick, S.; Heideman, D.A.; Steenbergen, R.D.; Bleeker, M.C.; Verhoeve, H.R.; van Baal, W.M.; van Trommel, N.; Kenter, G.G.; Meijer, C.J.; et al. Clinical Regression of High-Grade Cervical Intraepithelial Neoplasia Is Associated with Absence of FAM19A4/miR124-2 DNA Methylation (CONCERVE Study). J. Clin. Oncol. 2022, 40, 3037–3046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, X.; Hu, S.; Chen, S.; Zhao, S.; Dong, L.; Carvalho, A.L.; Muwonge, R.; Zhao, F.; Basu, P. Triage performance and predictive value of the human gene methylation panel among women positive on self-collected HPV test: Results from a prospective cohort study. Int. J. Cancer 2022, 151, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Hansel, A.; Steinbach, D.; Greinke, C.; Schmitz, M.; Eiselt, J.; Scheungraber, C.; Gajda, M.; Hoyer, H.; Runnebaum, I.B.; Dürst, M. A Promising DNA Methylation Signature for the Triage of High-Risk Human Papillomavirus DNA-Positive Women. PLoS ONE 2014, 9, e91905. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Wunsch, K.; Hoyer, H.; Scheungraber, C.; Runnebaum, I.B.; Hansel, A.; Dürst, M. Performance of a methylation specific real-time PCR assay as a triage test for HPV-positive women. Clin. Epigenetics 2017, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- S3-Leitlinie. Prävention des Zervixkarzinoms. AWMF Registernummer: 015/027OL. Available online: http://www.leitlinienprogramm-onkologie.de/leitlinien/zervixkarzinom-praevention/ (accessed on 5 June 2023).
- Dippmann, C.; Schmitz, M.; Wunsch, K.; Schütze, S.; Beer, K.; Greinke, C.; Ikenberg, H.; Hoyer, H.; Runnebaum, I.B.; Hansel, A.; et al. Triage of hrHPV-positive women: Comparison of two commercial methylation-specific PCR assays. Clin. Epigenet. 2020, 12, 171. [Google Scholar] [CrossRef]
- A’Hern, R.P. Sample size tables for exact single-stage phase II designs. Stat. Med. 2001, 20, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Koeneman, M.M.; Hendriks, N.; Kooreman, L.F.; Winkens, B.; Kruitwagen, R.F.; Kruse, A.J. Prognostic factors for spontaneous regression of high-risk human papillomavirus-positive cervical intra-epithelial neoplasia grade 2. Int. J. Gynecol. Cancer 2019, 29, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; MacIntyre, D.A.; Ntritsos, G.; Smith, A.; Tsilidis, K.K.; Marchesi, J.R.; Bennett, P.R.; Moscicki, A.-B.; Kyrgiou, M. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat. Commun. 2020, 11, 1999. [Google Scholar] [CrossRef] [PubMed]
- Salvadó, A.; Miralpeix, E.; Solé-Sedeno, J.M.; Kanjou, N.; Lloveras, B.; Duran, X.; Mancebo, G. Predictor factors for conservative management of cervical intraepithelial neoplasia grade 2: Cytology and HPV genotyping. Gynecol. Oncol. 2021, 162, 569–574. [Google Scholar] [CrossRef]
- Halle, M.K.; Munk, A.C.; Engesæter, B.; Akbari, S.; Frafjord, A.; Hoivik, E.A.; Forsse, D.; Fasmer, K.E.; Woie, K.; Haldorsen, I.S.; et al. A Gene Signature Identifying CIN3 Regression and Cervical Cancer Survival. Cancers 2021, 13, 5737. [Google Scholar] [CrossRef]
- Guan, P.; Howell-Jones, R.; Li, N.; Bruni, L.; de Sanjosé, S.; Franceschi, S.; Clifford, G.M. Human papillomavirus types in 115,789 HPV-positive women: A meta-analysis from cervical infection to cancer. Int. J. Cancer 2012, 131, 2349–2359. [Google Scholar] [CrossRef]
- Khan, M.J.; Castle, P.E.; Lorincz, A.T.; Wacholder, S.; Sherman, M.; Scott, D.R.; Rush, B.B.; Glass, A.G.; Schiffman, M. The Elevated 10-Year Risk of Cervical Precancer and Cancer in Women with Human Papillomavirus (HPV) Type 16 or 18 and the Possible Utility of Type-Specific HPV Testing in Clinical Practice. J. Natl. Cancer Inst. 2005, 97, 1072–1079. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoyer, H.; Stolte, C.; Böhmer, G.; Hampl, M.; Hagemann, I.; Maier, E.; Denecke, A.; Hirchenhain, C.; Patzke, J.; Jentschke, M.; et al. Evaluation of CIN2/3 Lesion Regression in GynTect® DNA Methylation-Marker-Negative Patients in a Longitudinal Study. Cancers 2023, 15, 3951. https://doi.org/10.3390/cancers15153951
Hoyer H, Stolte C, Böhmer G, Hampl M, Hagemann I, Maier E, Denecke A, Hirchenhain C, Patzke J, Jentschke M, et al. Evaluation of CIN2/3 Lesion Regression in GynTect® DNA Methylation-Marker-Negative Patients in a Longitudinal Study. Cancers. 2023; 15(15):3951. https://doi.org/10.3390/cancers15153951
Chicago/Turabian StyleHoyer, Heike, Claudia Stolte, Gerd Böhmer, Monika Hampl, Ingke Hagemann, Elisabeth Maier, Agnieszka Denecke, Christine Hirchenhain, Jan Patzke, Matthias Jentschke, and et al. 2023. "Evaluation of CIN2/3 Lesion Regression in GynTect® DNA Methylation-Marker-Negative Patients in a Longitudinal Study" Cancers 15, no. 15: 3951. https://doi.org/10.3390/cancers15153951
APA StyleHoyer, H., Stolte, C., Böhmer, G., Hampl, M., Hagemann, I., Maier, E., Denecke, A., Hirchenhain, C., Patzke, J., Jentschke, M., Gerick, A., Heller, T., Hippe, J., Wunsch, K., Schmitz, M., & Dürst, M. (2023). Evaluation of CIN2/3 Lesion Regression in GynTect® DNA Methylation-Marker-Negative Patients in a Longitudinal Study. Cancers, 15(15), 3951. https://doi.org/10.3390/cancers15153951









