Functional In Vivo Screening Identifies microRNAs Regulating Metastatic Dissemination of Prostate Cancer Cells to Bone Marrow
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Cell Lines
2.3. RNA Extraction, Reverse Transcription and qRT-PCR
2.4. Transduction of Cells with miRZiP Library
2.5. In Vivo PCa Model of Metastatic Migration
2.6. DNA Extraction and PCR
2.7. DNA Sequencing
2.8. Immunohistochemical Analysis
2.9. RNA Immunoprecipitation Sequencing (RIP-Seq)
2.10. RNA Sequencing
2.11. In Vitro Functional Analyses
2.12. Data Processing and Statistical Analysis
3. Results
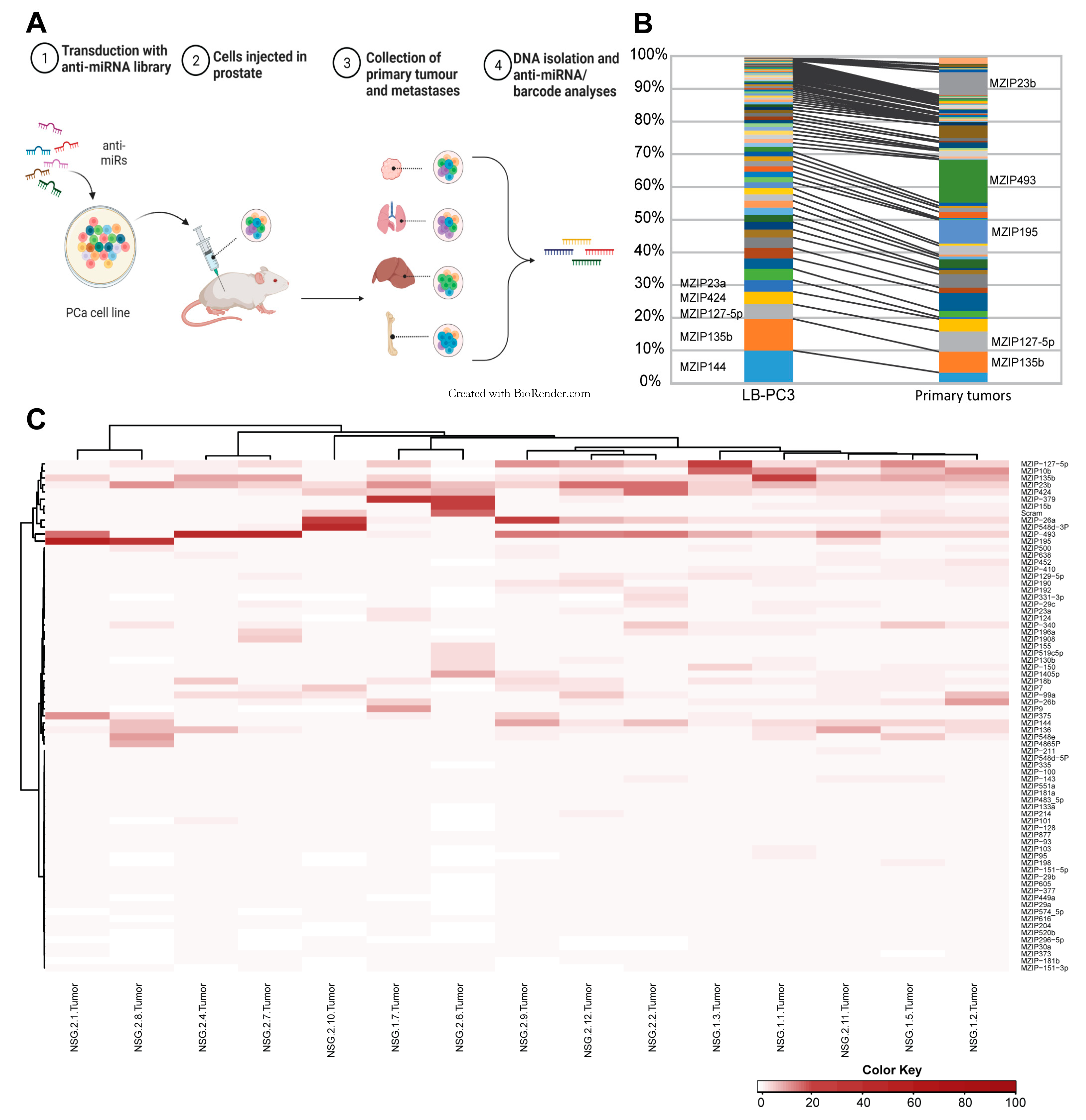
3.1. Transduction of PCa Cells with Anti-miR Library
3.2. In Vivo Functional Library Screening
3.3. Metastatic Dissemination
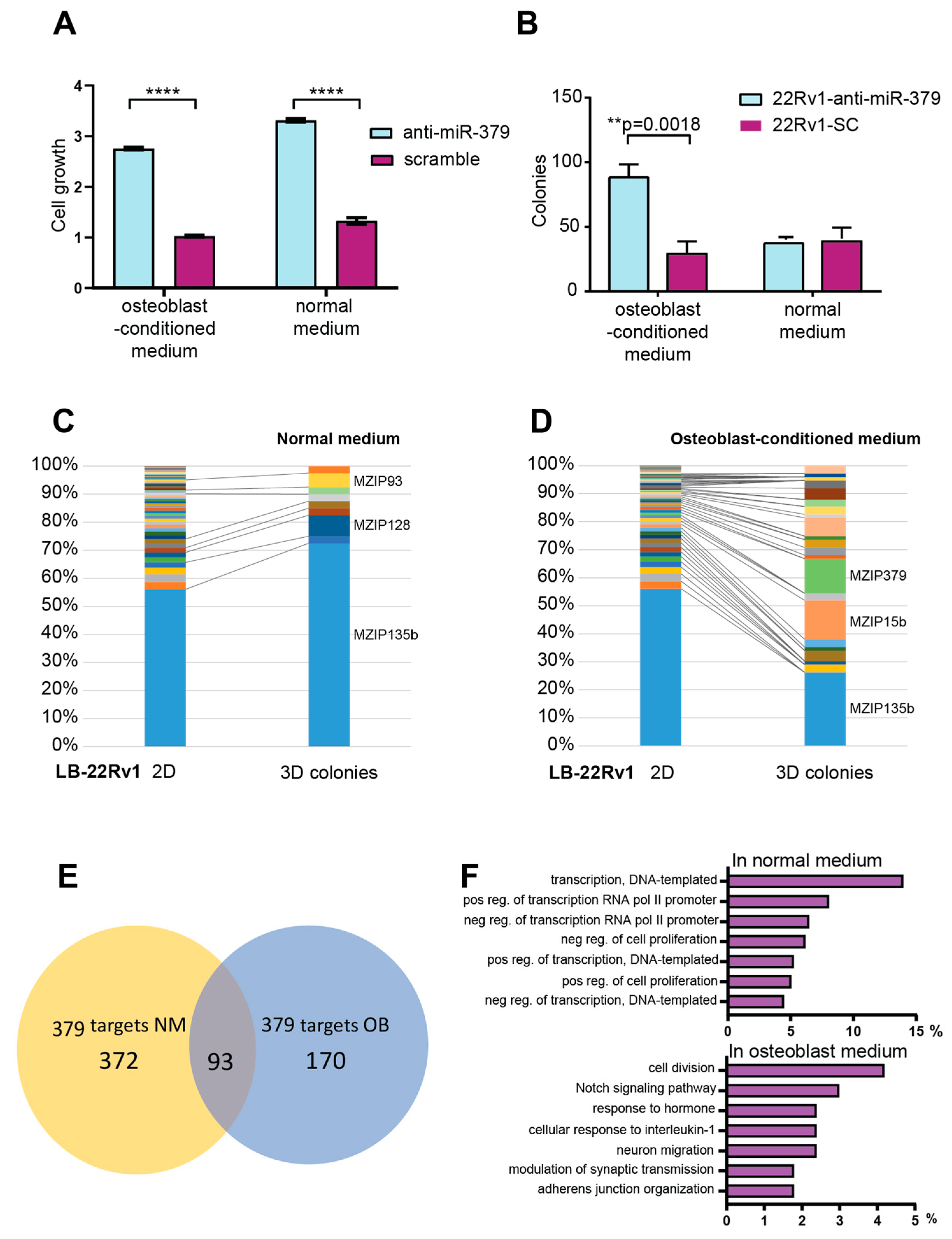
3.4. In Vitro Functional Analyses
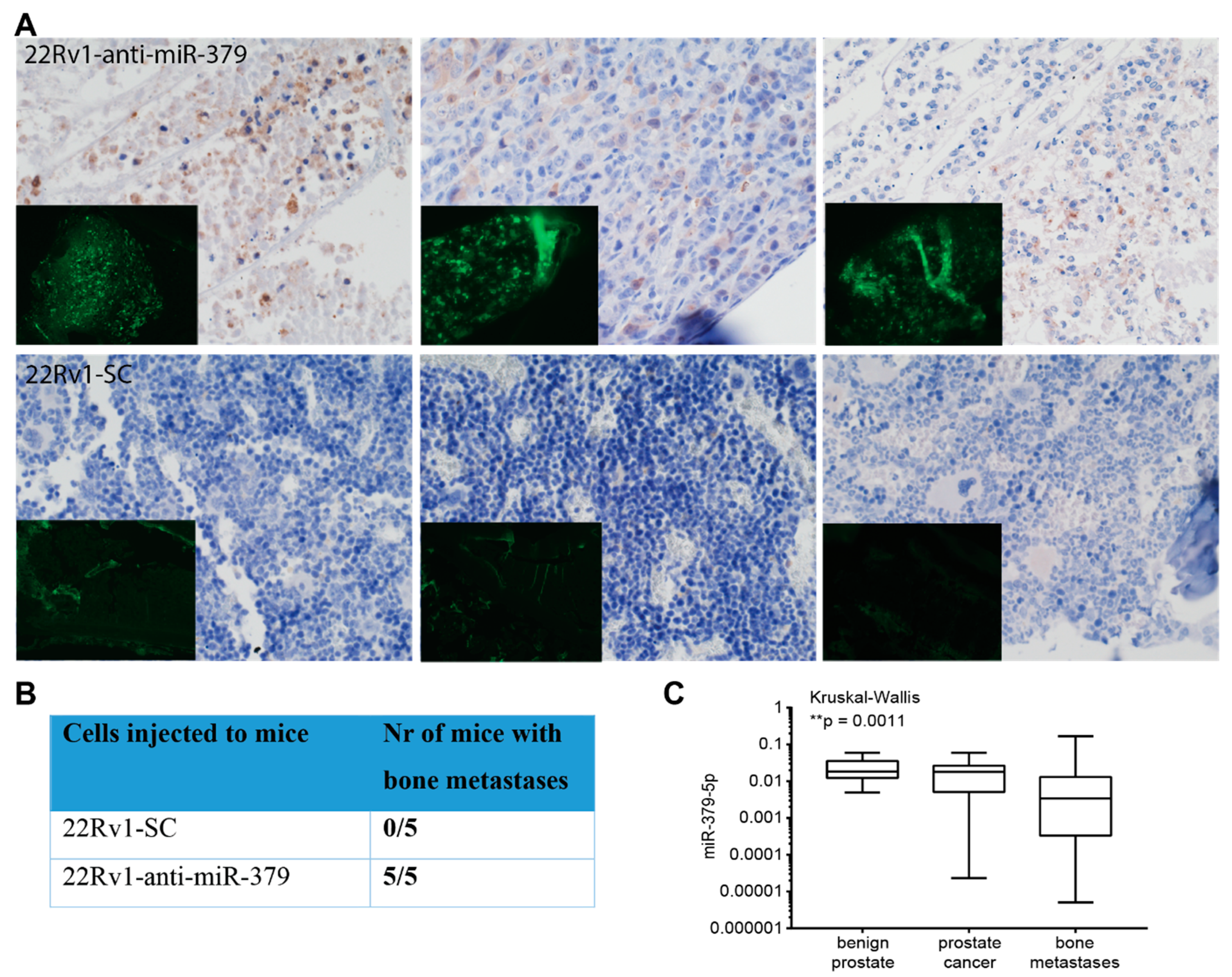
3.5. Metastatic Dissemination to Bone
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| PCa | Prostate cancer |
| OB-CM | Osteoblast-conditioned medium |
| GFP | green fluorescent protein |
| IP | immunoprecipitation |
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. 2018. Available online: http://gco.iarc.fr/today/home (accessed on 24 July 2023).
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer statistics, 2008. CA Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef] [PubMed]
- Cornford, P.; Bellmunt, J.; Bolla, M.; Briers, E.; De Santis, M.; Gross, T.; Henry, A.M.; Joniau, S.; Lam, T.B.; Mason, M.D.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part II: Treatment of Relapsing, Metastatic, and Castration-Resistant Prostate Cancer. Eur. Urol. 2017, 71, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.J.; Saylor, P.J.; Smith, M.R. Treatment and prevention of bone complications from prostate cancer. Bone 2011, 48, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [Green Version]
- Siemens, H.; Jackstadt, R.; Hunten, S.; Kaller, M.; Menssen, A.; Gotz, U.; Hermeking, H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011, 10, 4256–4271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef]
- Larne, O.; Martens-Uzunova, E.; Hagman, Z.; Edsjo, A.; Lippolis, G.; den Berg, M.S.; Bjartell, A.; Jenster, G.; Ceder, Y. miQ—A novel microRNA based diagnostic and prognostic tool for prostate cancer. Int. J. Cancer 2013, 132, 2867–2875. [Google Scholar] [CrossRef]
- Hagman, Z.; Haflidadottir, B.S.; Ansari, M.; Persson, M.; Bjartell, A.; Edsjo, A.; Ceder, Y. The tumour suppressor miR-34c targets MET in prostate cancer cells. Br. J. Cancer 2013, 109, 1271–1278. [Google Scholar] [CrossRef] [Green Version]
- Hagman, Z.; Haflidadottir, B.S.; Ceder, J.A.; Larne, O.; Bjartell, A.; Lilja, H.; Edsjo, A.; Ceder, Y. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer 2013, 108, 1668–1676. [Google Scholar] [CrossRef] [Green Version]
- Hörnberg, E.; Ylitalo, E.B.; Crnalic, S.; Antti, H.; Stattin, P.; Widmark, A.; Bergh, A.; Wikström, P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS ONE 2011, 6, e19059. [Google Scholar] [CrossRef] [Green Version]
- Ylitalo, E.B.; Thysell, E.; Jernberg, E.; Lundholm, M.; Crnalic, S.; Egevad, L.; Stattin, P.; Widmark, A.; Bergh, A.; Wikström, P. Subgroups of Castration-resistant Prostate Cancer Bone Metastases Defined Through an Inverse Relationship Between Androgen Receptor Activity and Immune Response. Eur Urol. 2017, 71, 776–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Campbell, J.P.; Merkel, A.R.; Masood-Campbell, S.K.; Elefteriou, F.; Sterling, J.A. Models of bone metastasis. J. Vis. Exp. 2012, 67, e4260. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Sahu, A.; Asangani, I.A.; Cao, Q.; Patel, L.; Vergara, I.A.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larne, O.; Hagman, Z.; Lilja, H.; Bjartell, A.; Edsjö, A.; Ceder, Y. miR-145 suppress the androgen receptor in prostate cancer cells and correlates to prostate cancer prognosis. Carcinogenesis 2015, 36, 858–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saal, L.H.; Vallon-Christersson, J.; Hakkinen, J.; Hegardt, C.; Grabau, D.; Winter, C.; Brueffer, C.; Tang, M.H.; Reutersward, C.; Schulz, R.; et al. The Sweden Cancerome Analysis Network—Breast (SCAN-B) Initiative: A large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015, 7, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, G.; Haflidadottir, B.S.; Jaremo, H.; Persson, M.; Catela Ivkovic, T.; Wikstrom, P.; Ceder, Y. Regulation of cell-cell adhesion in prostate cancer cells by microRNA-96 through upregulation of E-Cadherin and EpCAM. Carcinogenesis 2020, 41, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Haflidadottir, B.S.; Larne, O.; Martin, M.; Persson, M.; Edsjo, A.; Bjartell, A.; Ceder, Y. Upregulation of miR-96 enhances cellular proliferation of prostate cancer cells through FOXO1. PLoS ONE 2013, 8, e72400. [Google Scholar] [CrossRef] [Green Version]
- D’Agostino, R.B.; Belanger, A.; D’Agostino, R.B., Jr. A suggestion for using powerful and informative test of normality. Am. Stat. 1990, 44, 316–321. [Google Scholar]
- GraphPad Prism; 6.01 edn.; GraphPad Software Inc.: La Jolla, CA, USA, 2012.
- Li, J.; Meng, S.; Xu, M.; Wang, S.; He, L.; Xu, X.; Wang, X.; Xie, L. Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels. Oncotarget 2018, 9, 3752–3764. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Chen, Q.B.; Han, Z.D.; Zhang, Y.Q.; He, H.C.; Chen, J.H.; Chen, Y.R.; Yang, S.B.; Wu, Y.D.; Zeng, Y.R.; et al. miR-195 Inhibits Tumor Progression by Targeting RPS6KB1 in Human Prostate Cancer. Clin. Cancer Res. 2015, 21, 4922–4934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Enokida, H.; Chiyomaru, T.; Kinoshita, T.; Nakagawa, M.; Naya, Y.; Ichikawa, T.; Seki, N. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget 2014, 5, 7748–7759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aakula, A.; Leivonen, S.K.; Hintsanen, P.; Aittokallio, T.; Ceder, Y.; Borresen-Dale, A.L.; Perala, M.; Ostling, P.; Kallioniemi, O. MicroRNA-135b regulates ERalpha, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol. Oncol. 2015, 9, 1287–1300. [Google Scholar] [CrossRef] [Green Version]
- Ostling, P.; Leivonen, S.K.; Aakula, A.; Kohonen, P.; Makela, R.; Hagman, Z.; Edsjo, A.; Kangaspeska, S.; Edgren, H.; Nicorici, D.; et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011, 71, 1956–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, M.A.; Ishteiwy, R.A.; Magani, F.; Udayakumar, T.; Reiner, T.; Yates, T.J.; Miller, P.; Perez-Stable, C.; Rai, P.; Verdun, R.; et al. The microRNA-23b/-27b cluster suppresses prostate cancer metastasis via Huntingtin-interacting protein 1-related. Oncogene 2016, 35, 4752–4761. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, D.; Lv, J.; Wang, S.; Zhang, Q. miR-410-3p promotes prostate cancer progression via regulating PTEN/AKT/mTOR signaling pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2459–2465. [Google Scholar] [CrossRef]
- Guo, J.; Wang, M.; Liu, X. MicroRNA-195 suppresses tumor cell proliferation and metastasis by directly targeting BCOX1 in prostate carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 91. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Liu, X.; Liu, Y.; Yang, J.; Lv, G.; Dong, S. MicroRNA-335 and -543 suppress bone metastasis in prostate cancer via targeting endothelial nitric oxide synthase. Int. J. Mol. Med. 2015, 36, 1417–1425. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Du, W.; Zhang, H.; Wang, Y.; Li, K.; Meng, Y.; Wang, J. Transcriptional downregulation of miR-127-3p by CTCF promotes prostate cancer bone metastasis by targeting PSMB5. FEBS Lett. 2020, 594, 466–476. [Google Scholar] [CrossRef]
- Hao, G.J.; Hao, H.J.; Ding, Y.H.; Wen, H.; Li, X.F.; Wang, Q.R.; Zhang, B.B. Suppression of EIF4G2 by miR-379 potentiates the cisplatin chemosensitivity in nonsmall cell lung cancer cells. FEBS Lett. 2017, 591, 636–645. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Niu, X.; Tao, J.; Li, P.; Lu, Q.; Xu, A.; Chen, W.; Wang, Z. MicroRNA-379-5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol. Rep. 2017, 37, 3502–3508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.S.; Li, H.S.; Huang, J.Q.; Dong, S.H.; Huang, Z.J.; Yi, W.; Zhan, G.F.; Feng, J.T.; Sun, J.C.; Huang, X.H. MicroRNA-379-5p inhibits tumor invasion and metastasis by targeting FAK/AKT signaling in hepatocellular carcinoma. Cancer Lett. 2016, 375, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Qin, S.; Cao, F.; Ding, S.; Li, M. MicroRNA-379 inhibits metastasis and epithelial-mesenchymal transition via targeting FAK/AKT signaling in gastric cancer. Int. J. Oncol. 2017, 51, 867–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, G.; Edsjo, A.; Bjartell, A.; Ceder, Y. Quantification of microRNA editing using two-tailed RT-qPCR for improved biomarker discovery. RNA 2021, 27, 1412–1424. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivkovic, T.C.; Cornella, H.; Voss, G.; Ku, A.; Persson, M.; Rigo, R.; Gruvberger-Saal, S.K.; Saal, L.H.; Ceder, Y. Functional In Vivo Screening Identifies microRNAs Regulating Metastatic Dissemination of Prostate Cancer Cells to Bone Marrow. Cancers 2023, 15, 3892. https://doi.org/10.3390/cancers15153892
Ivkovic TC, Cornella H, Voss G, Ku A, Persson M, Rigo R, Gruvberger-Saal SK, Saal LH, Ceder Y. Functional In Vivo Screening Identifies microRNAs Regulating Metastatic Dissemination of Prostate Cancer Cells to Bone Marrow. Cancers. 2023; 15(15):3892. https://doi.org/10.3390/cancers15153892
Chicago/Turabian StyleIvkovic, Tina Catela, Helena Cornella, Gjendine Voss, Anson Ku, Margareta Persson, Robert Rigo, Sofia K. Gruvberger-Saal, Lao H. Saal, and Yvonne Ceder. 2023. "Functional In Vivo Screening Identifies microRNAs Regulating Metastatic Dissemination of Prostate Cancer Cells to Bone Marrow" Cancers 15, no. 15: 3892. https://doi.org/10.3390/cancers15153892
APA StyleIvkovic, T. C., Cornella, H., Voss, G., Ku, A., Persson, M., Rigo, R., Gruvberger-Saal, S. K., Saal, L. H., & Ceder, Y. (2023). Functional In Vivo Screening Identifies microRNAs Regulating Metastatic Dissemination of Prostate Cancer Cells to Bone Marrow. Cancers, 15(15), 3892. https://doi.org/10.3390/cancers15153892





