Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review
Abstract
:Simple Summary
Abstract
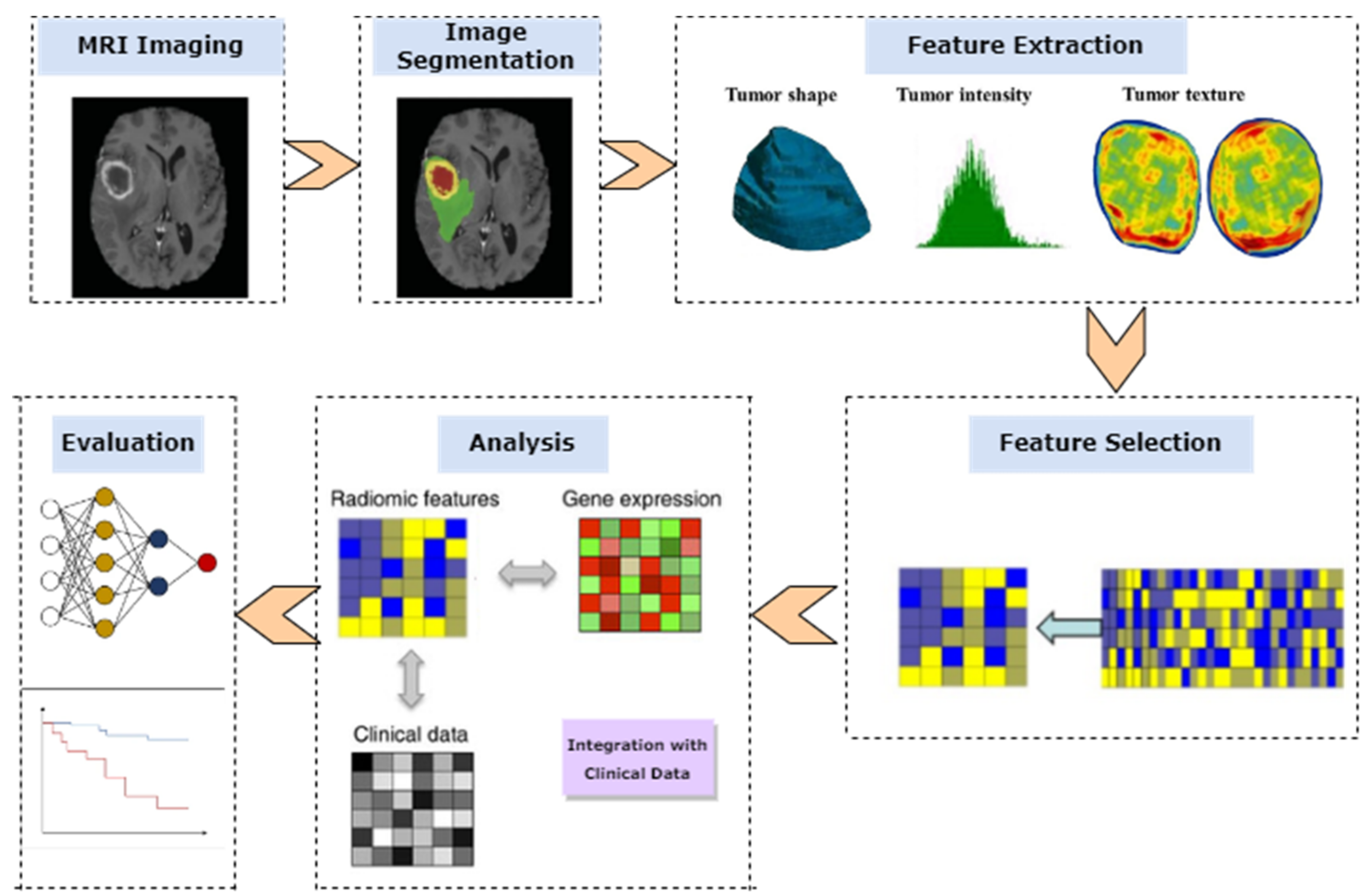
1. Introduction
2. Materials and Methods
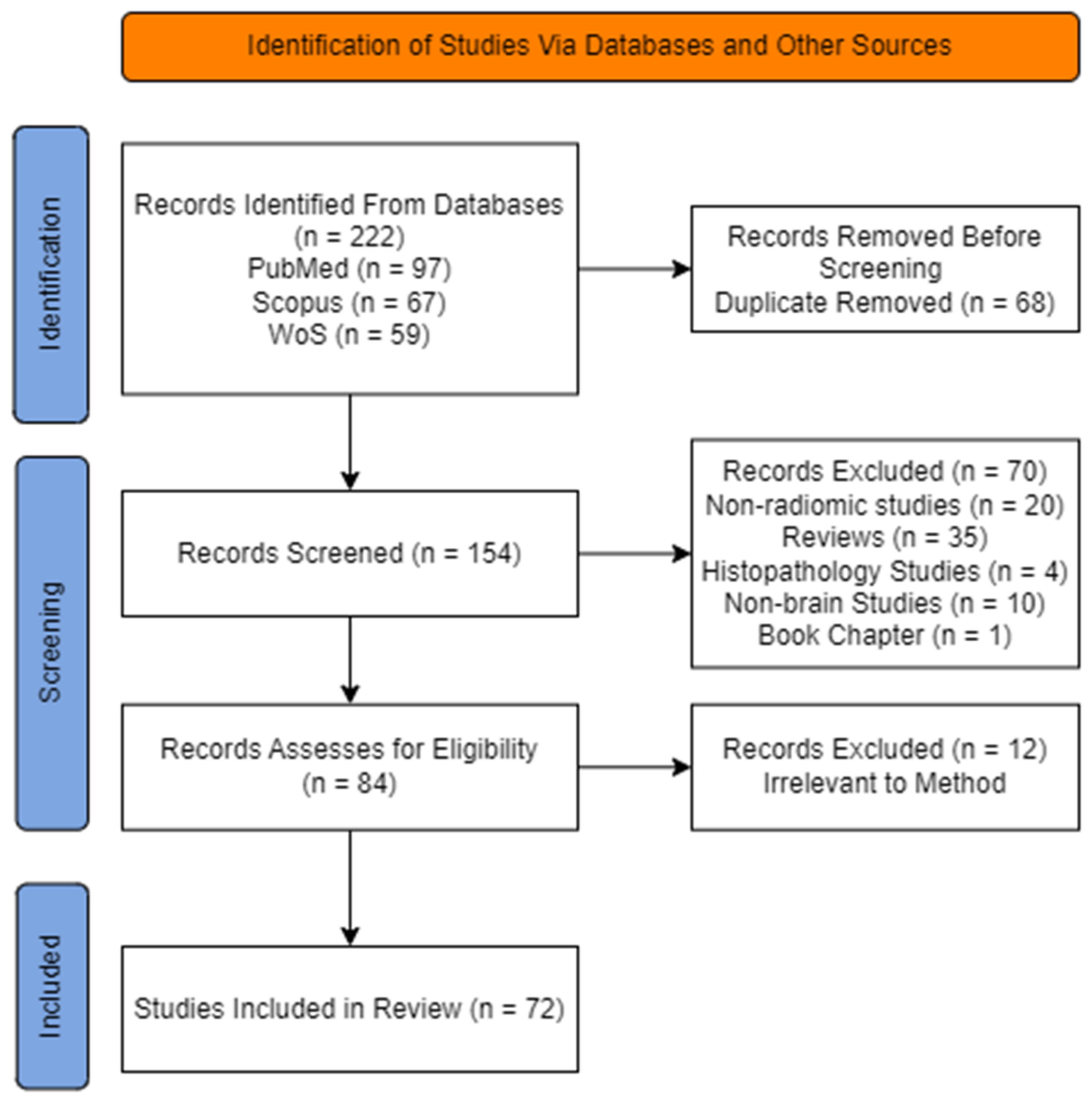
2.1. Search Strategy and Selection Criteria
2.2. Planning and Performance of the Review
2.3. Studies Corresponding Publication Year
2.4. Characteristics of Studies
| Reference | Application Field | Diseases | NP (Type) | MRI Sequence | Region for Feature Extraction | Software Used | NF | FS | CM | VM | Performance | RQS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prasanna et al., 2017 [10] | Prognosis | GBM | 65(36 long-term survival, 29 short-term survival) | T2W, Gd-T1W, FLAIR | ET, NCR, PTR | MATLAB | 134 | 12 (mRMR) | RF | 3-Fold CV | CI = 0.68~0.78 | 56% |
| Shofty et al., 2018 [25] | Diagnosis | LGG | 47 (26 oligodendroglia, 21 astrocytomas) | T1WGd, T2W, FLAIR | Pre-defined Lesion area of tumor | MATLAB | 152 | PCA | SVM, KNN, Ensemble classifier | 5-Fold CV | AUC = 0.87 | 69% |
| Akbari et al., 2018 [26] | Diagnosis and prognosis | GBM | 129 (74 male, 55 female) | T1WGd, T1W, T2W, T2-FLAIR, DTI | ET, non-ET, ED | CaPTk | 436 | Yes | SVM | 10-Fold CV | AUC = 0.92 | 75% |
| Cho et al., 2018 [27] | Prognosis and survival | Glioma | 285 (210 HGG, 75 LGG) | T1W, T2W, T1ce, FLAIR | ET, non-ET, ED | PyRadiomics, MATLAB | 468 (3 Types) | yes (Top 5) | SVM, RF | 5-Fold CV | AUC = 0.903 | 75% |
| Rathore et al. 2018 [28] | Prognosis | GBM | 31 | T1W, T2W, T1ce, FLAIR, DTI | ED, ET, NET | CaPTk | n/a | n/a | SVM | LOO CV | AUC =0.91 | 72% |
| Binder et al., 2018 [29] | Survival | GBM | 260 | T1W, T2W, T1ce, FLAIR | ET, non-ET, ED | CaPTk | 1650 | yes (p > 0.05) | Multivariate classification framework | 5-Fold CV | ------- | 78% |
| Abidin et al., 2019 [30] | Diagnosis | METs and Glioma | 52 | T1ce, T2 FLAIR | Tumor | Amira | 630 | no | AdaBoost | 10-Fold CV | AUC = 0.84 | 64% |
| Talamonti et al., 2019 [31] | Survival | Medulloblastoma | 70 | T1W TSE MDC, T2W TSE, T2W FLAIR | Necrosis, solid tumor, and oedema | PyRadiomics | ---- | yes | SVM | LOO-CV | ------ | 72% |
| Hajianfar et al., 2019 [32] | Diagnosis | GBM | 82 | T1W, T2W, T1ce, FLAIR | NCR, WT, ET, ED | R, Python | 7000 | Top 20 | Ada-Boost, DT | 10-Fold CV | AUC = 0.74 | 81% |
| Hamerla et al., 2019 [33] | Diagnosis | Meningioma | 147 | T1W, T2W, T1ce, FLAIR | Peritumoral ED | PyRadiomics | 12,733 | 16 | SVM, RF, NLP, XGBoost | 10-Fold CV | AUC = 0.97 | 81% |
| Kniep et al., 2019 [34] | Diagnosis | METs | 189 | T1ce, T1W, FLAIR | Multiple metastases | Python | 1423 | 59 | RF | 5-Fold CV | AUC = 0.90 | 72% |
| Jeong et al., 2019 [35] | Diagnosis | GBM | 25 (13 HGG, 12 LGG) | T2W FLAIR, T1W | Solid tumor | MATLAB | 1689 | 7 (types) of delta- and radiomic features | RF | LOO CV | AUC = 0.938 | 69% |
| Wei et al., 2019 [36] | Diagnosis | GBM | 105 | T1ce, T2-FLAIR & ADC | Tumor & PED | R | 3051 | 100 | LR | No CV | AUC = 0.926 | 78% |
| Wening et al., 2019 [37] | Survival | GBM | 211 | Multimodal | ED, ET, NEC | PyRadiomics | 9871 | 95 | LR | No CV | ACC = 0.56 (long, mid, and short-term survival) | 67% |
| Kim et al., 2019 [38] | Prognosis | GBM | 83 | T1W, T2W, T1ce, FLAIR, DTI, DSC | NER | ANTsR | 6472 | Top 10 (LASSO) | GLM | 10-Fold CV | CI = 0.87 | 72% |
| Prasanna et al., 2019 [39] | Survival | Glioma | 241 | T1c, T2w, and FLAIR | ET, WT, TC | MATLAB | 234 | yes (Top 2) | CNN, RF | 3-Fold CV | ST- 0.57 MT-0.63 LT-0.43 | 56% |
| Qian et al., 2019 [40] | Diagnosis | GBM & METs | 412 (GBM 242, 170 METs) | T1W, T1c, T2w | Tumor & peritumoral region | PyRadiomics | 1303 | 12 | SVM, LASSO, MLP, ADaBoost | 5-Fold CV | AUC = 0.95 | 86% |
| Carré et al., 2019 [41] | Diagnosis | GBM | 243 (108 grade II and III gliomas, 135 grade IV GBM) | T1w-gd and T2w-flair | OED, NCR, ET | PyRadiomics | 1462 | 91 (18 first-order and 73 second- order) | RF, NB, LR, SVM, NN | 5-Fold CV | ACC = 0.82 (95% CI 0.80–0.8 5, p = 0.005) | 72% |
| Shofty et al., 2020 [42] | Diagnosis | METs | 53 | Multi-modal | Brain lesions | MATLAB | 195 | 50 (PCA) | SVM | 5-Fold CV | AUC = 0.78 | 75% |
| Sudre et al., 2020 [43] | Diagnosis | Glioma | 333 (101 LGG, 232 HGG | T2 W, FLAIR | Tumor | NiftyReg | Several | 29 (Shape, histogram, Haralick) | RF | 2-Fold CV | AUC = 0.80 | 67% |
| Crisi et al., 2020 [44] | Prognosis | GBM | 59 | T1-GRE, T2- GRE, T2FLAIR | ET, NEC | LIFEx | 92 | 14 | NB, DT, MLP | 10-Fold CV | AUC = 0.84 | 47% |
| Wei et al. 2020, [45] | Diagnosis | IHPC, meningioma | 292 (IHPC = 155 meningiomas = 137) | T1WI, CE- T1WI, and T2WI | TC and PED | PyRadiomics | 473 | 64 | Recursive feature elimination, RF | 3-Fold CV | AUC = 0.913 (Tr), 0.914 (val) | 86% |
| Beig et al., 2020 [46] | Survival | GBM | 203 | Gd-T1W, T2W, FLAIR | NCR, PED, ET | MATLAB | 936 | 25 | Cox regression | 5-Fold CV | ----- | 81% |
| Lohmann et al., 2020 [47] | Early progression | GBM | 34 | PET | Tumor | PyRadiomics | 944 | 4 (shape, Histogram, GLSZM) | RF | 5-Fold CV | AUC = 0.79 | 58% |
| Correa et al., 2020 [48] | Diagnosis | METs | 37 | post-Gd T1w, T2w, and FLAIR | Lesion and lesion habitat | -------- | 4740 (Haralick, Gabor, Laws, CoLlAGe) | top 3 (Laws) | RF | 3-Fold CV | AUC = 0.97 | 67% |
| Kumar et al., 2020 [49] | Prognosis | Glioma | 285 (210 HGG, 75 LGG) | T1, T1c, T2 FLAIR | NET, NCR, ED, ET | PyRadiomics | 1158 | 580 | RF | 5-fold CV | AUC = 0.97 | 58% |
| Verma et al., 2020 [50] | Survival | GBM | 156 | Gd-T1W, T2W, FLAIR | ET, NET, NCR | R studio | 3024 (Haralick, Laws, CoLlAGe) | ------ | LASSO | 10-fold CV | CI = 0.80 | 56% |
| Choi et al., 2020 [51] | Survival | GBM | 144 | T1W, T2W, T1ce, FLAIR | PED | PyRadiomics | 478 | 7 | Cox-Lasso | 10-fold CV | --- | 75% |
| Yousaf et al., 2020 [52] | Survival | GBM | 335 (259 HGG, 76 LGG) | T1W, T2W, T1ce and FLAIR | Tumor | MATLAB | 30,632 | 14 | RF | 10-fold CV | ---- | 53% |
| Zhang et al., 2020 [53] | Diagnosis and prognosis | GBM | 104 | T1C, T1, T2, FLAIR | ET, NCR, ED | MATLAB | 180 | ------ | SVM | No CV | ACC = 87.88% | 56% |
| Choi et al., 2020 [54] | Diagnosis | GBM | 136 | T2W | Tumor & PED | PyRadiomics | 107 | 9 | Random Forest | No CV | AUC = 0.758 | 83% |
| Sakai et al., 2020 [55] | Diagnosis | Glioma | 100 (22 IDH1 mutant, 78 wildtypes | FLAIR, DWI | Tumor | Olea sphere | 92 | ---- | XGBoost | 5-fold CV | AUC = 0.97 | 67% |
| Demire et al. 2021 [56] | Diagnosis | GBM & METs | 60 (35 GBM, 25 METs) | T1WI, T2WI, FLAIR, postcontrast T1WI | NEC, NET, ET, Oedema | Third- party | 856 | ----- | SVM, RF, NB | 5-Fold CV | AUC = 0.97 | 50% |
| Tixier et al., 2021 [57] | Survival | GBM | 234 | T1W | Gd -ET, NEC, NET, TC | Python | 88 | 57 | Lasso | 5-Fold CV | AUC = 0.75 | 61% |
| Russo et al., 2021 [58] | Diagnosis | Glioma | 56 | PET | Tumor | LIFEx | 44 | ----- | NN, RF, SVM | 5-Fold CV | AUC = 0.78 | 50% |
| Yan et al., 2021 [59] | Diagnosis | GBM | 41 | T1ce, T1W, T2W, FLAIR | Tumor | CaPTk | 841 | 153 | RF | No CV | ACC = 81% | 64% |
| Ye et al., 2021 [60] | Diagnosis and prognosis | GBM | 285 (210 HGG, 75 LGG) | T1W, T2W, T2 FLAIR | GD-ET, PED | PyRadiomics | 94 | Top 30 | RF, KNN, SVM, MLP, CNN | No CV | AUC = 0.65 (short-, mid-, and long-term survival) | 67% |
| Joo et al., 2021 [61] | Diagnosis | Meningioma | 454 | T2W, T1ce | Tumor & PED | MATLAB | 3222 | Top 6 | RF | 10-Fold CV | AUC = 0.76 | 56% |
| Pasquini et al., 2021 [62] | Diagnosis | High-grade glioma | 156 | T1W, T2W, FLAIR, PWI, DWI | WT, CET, NEC, NET | MATLAB | 1871 | Top 15 | RF | 10-Fold CV | AUC = 74.2% | 56% |
| Cao et al., 2021 [63] | Prognosis | Lower-grade glioma | 102 (60 men, 42 women) | T1W, T2W, FLAIR, DWI | WT, NEC | MATLAB | 56 | Top 10 | RF | No CV | AUC = 0.879 | 53% |
| Patel et al., 2021 [64] | Prognosis | GBM | 76 | CE-T1W, T2W, DWI | Whole Brain | PyRadiomics | 307 | 6 | RF, NB | 10-Fold CV | AUC = 0.8 | 70% |
| Soltani et al., 2021 [65] | Diagnosis and prognosis | GBM | 211 | T1, T1CE, T2, and T2- FLAIR | ED, ET, NEC | PyRadiomics | 3910 | 67 | ANN, KNN, RF | No CV | ACC = 0.57 (short-, mid-, and long-term survival) | 56% |
| Wagner et at., 2021 [66] | Prognosis | LGG | 115 | T2-FLAIR, Gd-T1W | Segmented tumor | PyRadiomics | 851 | 10 | RF | 4-fold CV | AUC = 0.75 | 58% |
| Le et al., 2021 [67] | Diagnosis and prognosis | Glioma | 120 | T2-FLAIR, Gd-T1W | ET, NET, ED | CaPTk | 704 | 13 | XGBoost | LOO-CV | AUC = 0.85 | 61% |
| Kumar et al., 2021, [68] | Diagnosis | Glioma | 369 (293 HGG, 76 LGG) | T2 FLAIR, T1W, postcontrast T1W and | NET, NCR, ED, ET | Python | 428 | ---- | LR, SVM, KNN, ERT | 5-fold CV | AUC = 0.95 | 67% |
| Cepeda et al., 2021 [69] | Survival | GBM | 203 | T1CE, T1, T2, FLAIR | Tumor, peritumoral | MATLAB | 15,720 | ---- | Naive Bayes | No CV | AUC = 0.769 | 61% |
| Maliket al., 2021 [70] | Diagnosis (clinical study) | LGG & GBM | 78 (42 GBM, 36 LGG) | T1ce, T2-FLAIR, DWI | PED, TC | PyRadiomics | 3822 | 9 (RFE) | SVM, KNN, LDA, AdaBoost | LOO CV | AUC = 0.96 | 67% |
| Samani et al., 2021 [71] | Diagnosis | GBM & METs | 106 (66 GBM, 40 METs) | DTI | PTR | PyRadiomics | All first-order features | Top 2% (PCA) | SVM, CNN | 5-fold CV | ACC = 85% | 61% |
| Xiao et al., 2021 [72] | Diagnosis | GBM & Brain Abscess | 118 (86 GBM, 32 brain abscess) | T1W, T2W, T1ce, FLAIR | NCR, PED, TC | PyRadiomics | 1004 | 43 (PCA) | RF, LR | 5-fold CV | AUC = 0.89 | 56% |
| Gutta et al., 2021 [73] | Diagnosis | Glioma | 237 | T1CE, T1W, T2W, T2- FLAIR | ET, NET & ED | PyRadiomics | 1284 | 45 | SVM, RF | No CV | ACC = 87% | 67% |
| Zhang et al., 2021 [74] | Diagnosis | Glioma | 162 | Gd-T1W, T1W, T2W, T2-FLAIR | TC, ED | PyRadiomics | 1102 | Top 10 | autoML | 4-fold CV | AUC = 0.951 | 58% |
| Xu et al., 2021 [75] | Prognosis | GBM | 236 | T1, T1-Gd, T2W, T2- FLAIR | ET, ED, NET, NCR | PyRadiomics | 1320 | 45 | Cox regression | 5-fold CV | C-index = 0.64 | 61% |
| Meißner et al., 2022 [76] | Survival | METs | 59 | T1CE, T2W | Tumor | PyRadiomics | 1316 | 100 | SVM | 10-fold CV | AUC = 0.92 | 67% |
| Shaheen et al., 2022 [77] | Survival | Glioma | 178 | T1W, T2W, T1ce, FLAIR | PTE, NEC, ENC | PyRadiomics | 89 | 50 | SVM | --- | AUC = 0.73 | 61% |
| Deng et al., 2022 [78] | Survival | Glioma | 84 | T2W, T1ce, FLAIR | Tumor, NCR, ED | PyRadiomics | 1316 | 12 | RF | ---- | AUC = 0.879 | 61% |
| Liu et al., 2022 [79] | Prognosis | GBM | 200 | T1CE, T2 | Tumor and peritumoral region | PyRadiomics | 8412 | Top 20 | RF, SVM | 10-fold CV | AUC = 0.91 | 61% |
| Do et al., 2022 [80] | Prognosis | GBM | 53 | T1W, T1Gd, T2, T2-FLAIR | NCR, PED, ET | Python | 704 | 22 | RF, SVM, XGBoost | 5-fold CV | AUC = 0.93 | 50% |
| Chiu et al., 2022 [81] | Diagnosis | GBM | 54 | T1Gd, T2W, T2-FLAIR, T1CE | NCR, ET, PED | Python | 1316 | ---- | RF | No CV | AUC = 0.96 | 53% |
| Chen et al., 2022 [82] | Diagnosis | Meningioma | 819 | T1W, T2W, T1CE | Solid tumor, NCR | Python | 2942 | top 9 | RF | No CV | AUC = 0.95 | 56% |
| Xu et al., 2022 [83] | Prognosis | Glioma | 74 | T1W, T2W- FLAIR, T1CE | Solid tumor | PyRadiomics | 112 | 7 | Stack, KNN, LR, RF, SVM, NB | 5-fold CV | AUC = 0.76 | 67% |
| Kumar et al., 2022 [84] | Diagnosis | Glioma | 285 (210 HGG, 75 LGG) | T2W, T1ce, FLAIR | NCR, ET, NET, PED | PyRadiomics | 321 | 42 | RF, DT, SVM, LR | 5-fold CV | AUC = 0.975 | 86% |
| Verma et al., 2022 [85] | Survival | GBM | 150 | Gadolinium—T1w, T2w, FLAIR | ET, NCR | MATLAB | 3792 | 316 | ---- | 5-fold CV | AUC = 0.78 | 75% |
| Wang et al., 2022 [86] | Diagnosis | METs | 228 | T1ce | Solid tumor and NCR | Python | 960 | 548 (LASSO) | SVM | 5-fold CV | AUC = 0.928 | 53% |
| Yang et al., 2022 [87] | Diagnosis | GBM | 187 | T1W, T2W, T1ce, FLAIR | Tumor and PED | PyRadiomics | 190 | Yes (LASSO) | Cox regression | 10-fold CV | CI = 0.658 | 69% |
| Liu et al., 2022 [88] | Diagnosis | GBM, MET, and lymphoma | 324 (134 GBM 82 Lymphoma 108 MET) | T2W, T1ce | WT, PED | PyRadiomics | 8412 | Top 20 (LASSO) | RF, linear, AdaBoost | 10-fold CV | AUC = 0.91 | 62% |
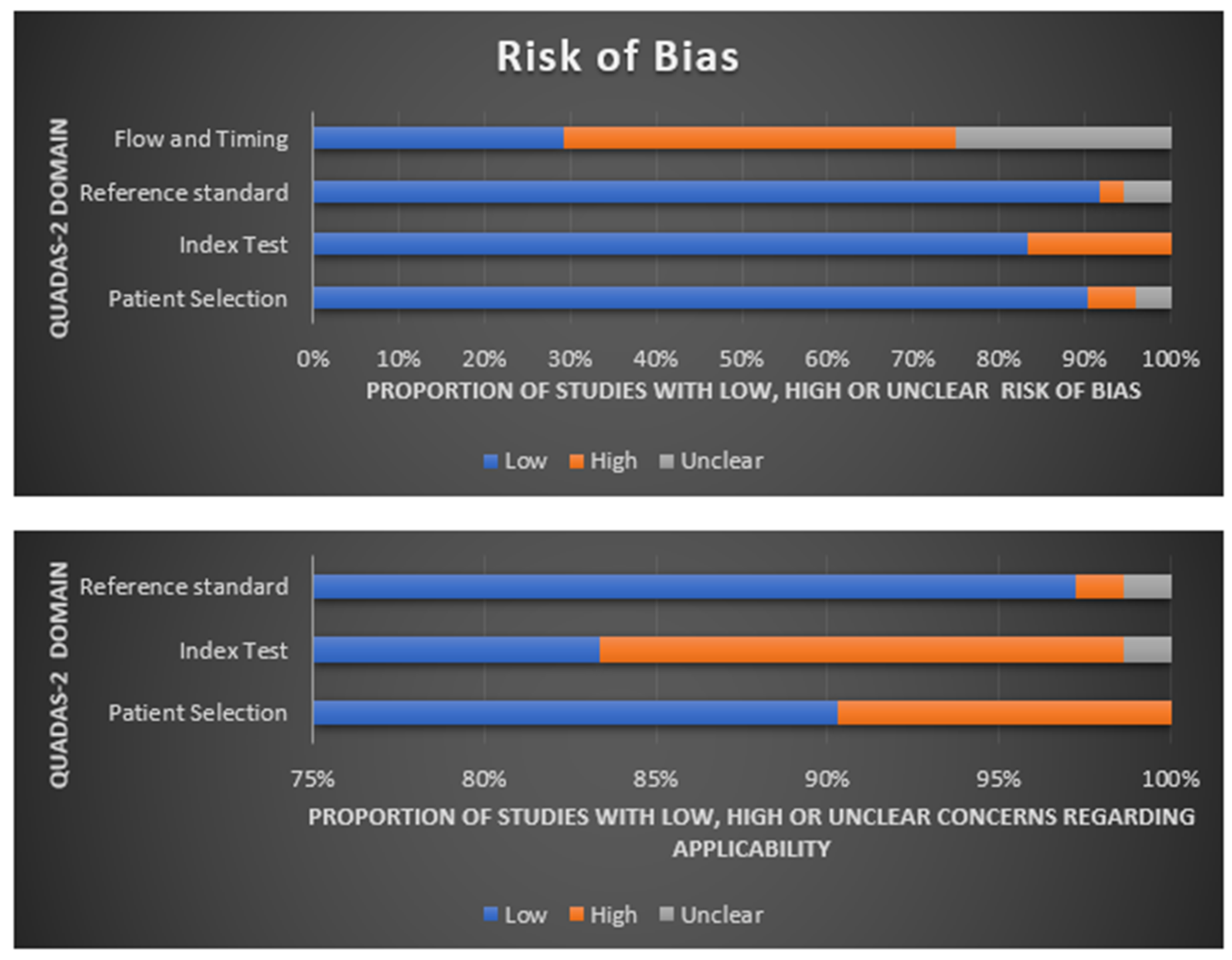
2.5. Quality Assessment
3. Results
3.1. Radiomics for Glioma Grading and Differential Diagnosis
3.2. Radiomics for Non-Glial Tumors
3.3. Radiomics for Survival Prediction
3.4. Radiomics for Brain-Habitat Analysis
3.5. Radiomics for Genetic-Mutation-Status Prediction
3.6. Frequently Selected Radiomics Features
3.7. Evaluation Metrics
4. Discussion
4.1. Promises of Radiomics and Machine Learning for Brain Tumor Analysis
4.2. Research Gaps and Future Challenges
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| AdaBoost | Adaptive Boosting |
| CI | Concordance Index |
| CNN | Convolutional Neural Network |
| DT | Decision Tree |
| DTI | Diffusion Tensor Imaging |
| DWI | Diffusion-Weighted Imaging |
| ED | Edema |
| ERT | Extremely Randomized Trees |
| ET | Enhancing Tumor |
| GBM | Glioblastoma |
| GLM | Generalized Linear Model |
| HGG | High-Grade Glioma |
| LR | Logistic Regression |
| KNN | K- Nearest Neighbour Algorithm |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| LDA | Linear Discriminant Analysis |
| LGG | Low-Grade Glioma |
| LOO | Leave One Out |
| METs | Metastasis |
| MLP | Multi-Layer Perceptron |
| mRMR | Minimum Redundancy Maximum Relevance |
| NB | Naïve Bayes |
| IHPC | Intracranial hemangiopericytoma |
| NCR | Necrosis |
| NER | Non-Enhancing Region |
| NET | Non-Enhancing Tumor |
| NN | Neural Network |
| PCA | Principal Component Analysis |
| PED | Peritumoral Edema |
| PTR | Peritumoral Region |
| RF | Random Forest |
| RFE | Recursive Feature Elimination |
| SVM | Support Vector Machine |
| T1ce | Contrast-Enhanced T1 Imaging |
| XGBoost | eXtreme Gradient Boosting |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-Oncology 2015, 17 (Suppl. 4), iv1–iv62. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Chaudhury, B.; Hall, L.O.; Goldgof, D.B.; Gillies, R.J.; Gatenby, R.A. Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J. Magn. Reson. Imaging 2017, 46, 115–123. [Google Scholar] [CrossRef]
- Tykocki, T.; Eltayeb, M. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 2018, 54, 7–13. [Google Scholar] [CrossRef]
- Delgado-López, P.; Corrales-García, E. Survival in glioblastoma: A review on the impact of treatment modalities. Clin. Transl. Oncol. 2016, 18, 1062–1071. [Google Scholar] [CrossRef]
- Zaccagna, F.; Grist, J.T.; Quartuccio, N.; Riemer, F.; Fraioli, F.; Caracò, C.; Halsey, R.; Aldalilah, Y.; Cunningham, C.H.; Massoud, T.F. Imaging and treatment of brain tumors through molecular targeting: Recent clinical advances. Eur. J. Radiol. 2021, 142, 109842. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [Green Version]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150. [Google Scholar] [CrossRef] [Green Version]
- Prasanna, P.; Patel, J.; Partovi, S.; Madabhushi, A.; Tiwari, P. Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur. Radiol. 2017, 27, 4188–4197. [Google Scholar] [CrossRef]
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in Brain Tumor: Image Assessment, Quantitative Feature Descriptors, and Machine-Learning Approaches. Am. J. Neuroradiol. 2018, 39, 208–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beig, N.; Bera, K.; Tiwari, P. Introduction to radiomics and radiogenomics in neuro-oncology: Implications and challenges. Neurooncol. Adv. 2020, 2, iv3–iv14. [Google Scholar] [CrossRef] [PubMed]
- Di Ieva, A. Fractal analysis of microvascular networks in malignant brain tumors. Clin. Neuropathol. 2012, 31, 342–351. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“How-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, S.; Chen, H.; Luo, L. Brain Tumor Segmentation and Survival Prediction Using Multimodal MRI Scans With Deep Learning. Front. Neurosci. 2019, 13, 810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Segato, A.; Marzullo, A.; Calimeri, F.; De Momi, E. Artificial intelligence for brain diseases: A systematic review. APL Bioeng. 2020, 4, 41503. [Google Scholar] [CrossRef] [PubMed]
- Di Ieva, A.; Russo, C.; Liu, S.; Jian, A.; Bai, M.Y.; Qian, Y.; Magnussen, J.S. Application of deep learning for automatic segmentation of brain tumors on magnetic resonance imaging: A heuristic approach in the clinical scenario. Neuroradiology 2021, 63, 1253–1262. [Google Scholar] [CrossRef]
- Jose, L.; Liu, S.; Russo, C.; Cong, C.; Song, Y.; Rodriguez, M.; Di Ieva, A. Artificial Intelligence-Assisted Classification of Gliomas Using Whole-Slide Images. Arch. Pathol. Lab. Med. 2022. [Google Scholar] [CrossRef]
- Liu, S.; Shah, Z.; Sav, A.; Russo, C.; Berkovsky, S.; Qian, Y.; Coiera, E.; Di Ieva, A. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep. 2020, 10, 7733. [Google Scholar] [CrossRef]
- Gao, Y.; Xiao, X.; Han, B.; Li, G.; Ning, X.; Wang, D.; Cai, W.; Kikinis, R.; Berkovsky, S.; Di Ieva, A.; et al. Deep Learning Methodology for Differentiating Glioma Recurrence From Radiation Necrosis Using Multimodal Magnetic Resonance Imaging: Algorithm Development and Validation. JMIR Med. Inform. 2020, 8, e19805. [Google Scholar] [CrossRef]
- Jian, A.; Liu, S.; Di Ieva, A. Artificial Intelligence for Survival Prediction in Brain Tumors on Neuroimaging. Neurosurgery 2022, 91, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Macyszyn, L.; Da, X.; Bilello, M.; Wolf, R.L.; Martinez-Lage, M.; Biros, G.; Alonso-Basanta, M.; O’Rourke, D.M.; Davatzikos, C. Imaging Surrogates of Infiltration Obtained Via Multiparametric Imaging Pattern Analysis Predict Subsequent Location of Recurrence of Glioblastoma. Neurosurgery 2016, 78, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shofty, B.; Artzi, M.; Ben Bashat, D.; Liberman, G.; Haim, O.; Kashanian, A.; Bokstein, F.; Blumenthal, D.T.; Ram, Z.; Shahar, T. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Bakas, S.; Pisapia, J.M.; Nasrallah, M.P.; Rozycki, M.; Martinez-Lage, M.; Morrissette, J.J.D.; Dahmane, N.; O’Rourke, D.M.; Davatzikos, C. In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncology 2018, 20, 1068–1079. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.H.; Lee, S.H.; Kim, J.; Park, H. Classification of the glioma grading using radiomics analysis. PeerJ 2018, 6, e5982. [Google Scholar] [CrossRef] [Green Version]
- Rathore, S.; Akbari, H.; Doshi, J.; Shukla, G.; Rozycki, M.; Bilello, M.; Lustig, R.; Davatzikos, C. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: Implications for personalized radiotherapy planning. J. Med. Imaging 2018, 5, 21219. [Google Scholar] [CrossRef]
- Binder, Z.A.; Thorne, A.H.; Bakas, S.; Wileyto, E.P.; Bilello, M.; Akbari, H.; Rathore, S.; Ha, S.M.; Zhang, L.; Ferguson, C.J.; et al. Epidermal Growth Factor Receptor Extracellular Domain Mutations in Glioblastoma Present Opportunities for Clinical Imaging and Therapeutic Development. Cancer Cell 2018, 34, 163–177.e7. [Google Scholar] [CrossRef] [Green Version]
- Abidin, A.Z.; Dar, I.; D’Souza, A.M.; Lin, E.P.; Wismüller, A. Investigating a quantitative radiomics approach for brain tumor classification. In Proceedings of the Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 16–21 February 2019. [Google Scholar]
- Talamonti, C.; Piffer, S.; Greto, D.; Mangoni, M.; Ciccarone, A.; Dicarolo, P.; Fantacci, M.E.; Fusi, F.; Oliva, P.; Palumbo, L.; et al. Radiomic and dosiomic profiling of paediatric medulloblastoma tumours treated with intensity modulated radiation therapy. Commun. Comput. Inf. Sci. 2019, 1089, 56–64. [Google Scholar]
- Hajianfar, G.; Shiri, I.; Maleki, H.; Oveisi, N.; Haghparast, A.; Abdollahi, H.; Oveisi, M. Noninvasive O6 Methylguanine-DNA Methyltransferase Status Prediction in Glioblastoma Multiforme Cancer Using Magnetic Resonance Imaging Radiomics Features: Univariate and Multivariate Radiogenomics Analysis. World Neurosurg. 2019, 132, e140–e161. [Google Scholar] [CrossRef] [Green Version]
- Hamerla, G.; Meyer, H.J.; Schob, S.; Ginat, D.T.; Altman, A.; Lim, T.; Gihr, G.A.; Horvath-Rizea, D.; Hoffmann, K.T.; Surov, A. Comparison of machine learning classifiers for differentiation of grade 1 from higher gradings in meningioma: A multicenter radiomics study. Magn. Reson. Imaging 2019, 63, 244–249. [Google Scholar] [CrossRef]
- Kniep, H.C.; Madesta, F.; Schneider, T.; Hanning, U.; Schönfeld, M.H.; Schön, G.; Fiehler, J.; Gauer, T.; Werner, R.; Gellissen, S. Radiomics of brain MRI: Utility in prediction of metastatic tumor type. Radiology 2019, 290, 479–487. [Google Scholar] [CrossRef]
- Jeong, J.J.; Ji, B.; Lei, Y.; Wang, L.; Liu, T.; Ali, A.; Curran, W.J.; Mao, H.; Yang, X. Machine-learning based classification of glioblastoma using dynamic susceptibility enhanced MR image. In Proceedings of the Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 16–21 February 2019. [Google Scholar]
- Wei, J.; Yang, G.; Hao, X.; Gu, D.; Tan, Y.; Wang, X.; Dong, D.; Zhang, S.; Wang, L.; Zhang, H.; et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur. Radiol. 2019, 29, 877–888. [Google Scholar] [CrossRef] [Green Version]
- Weninger, L.; Haarburger, C.; Merhof, D. Robustness of Radiomics for Survival Prediction of Brain Tumor Patients Depending on Resection Status. Front. Comput. Neurosci. 2019, 13, 73. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Yoon, M.J.; Park, J.E.; Choi, E.J.; Lee, J.; Kim, H.S. Radiomics in peritumoral non-enhancing regions: Fractional anisotropy and cerebral blood volume improve prediction of local progression and overall survival in patients with glioblastoma. Neuroradiology 2019, 61, 1261–1272. [Google Scholar] [CrossRef]
- Prasanna, P.; Karnawat, A.; Ismail, M.; Madabhushi, A.; Tiwari, P. Radiomics-based convolutional neural network for brain tumor segmentation on multiparametric magnetic resonance imaging. J. Med. Imaging 2019, 6, 24005. [Google Scholar] [CrossRef]
- Qian, Z.; Li, Y.; Wang, Y.; Li, L.; Li, R.; Wang, K.; Li, S.; Tang, K.; Zhang, C.; Fan, X.; et al. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett. 2019, 451, 128–135. [Google Scholar] [CrossRef]
- Carré, A.; Klausner, G.; Edjlali, M.; Lerousseau, M.; Briend-Diop, J.; Sun, R.; Ammari, S.; Reuzé, S.; Alvarez Andres, E.; Estienne, T.; et al. Standardization of brain MR images across machines and protocols: Bridging the gap for MRI-based radiomics. Sci. Rep. 2020, 10, 12340. [Google Scholar] [CrossRef]
- Shofty, B.; Artzi, M.; Shtrozberg, S.; Fanizzi, C.; DiMeco, F.; Haim, O.; Peleg Hason, S.; Ram, Z.; Bashat, D.B.; Grossman, R. Virtual biopsy using MRI radiomics for prediction of BRAF status in melanoma brain metastasis. Sci. Rep. 2020, 10, 6623. [Google Scholar] [CrossRef] [Green Version]
- Sudre, C.H.; Panovska-Griffiths, J.; Sanverdi, E.; Brandner, S.; Katsaros, V.K.; Stranjalis, G.; Pizzini, F.B.; Ghimenton, C.; Surlan-Popovic, K.; Avsenik, J.; et al. Machine learning assisted DSC-MRI radiomics as a tool for glioma classification by grade and mutation status. BMC Med. Inform. Decis. Mak. 2020, 20, 149. [Google Scholar] [CrossRef]
- Crisi, G.; Filice, S. Predicting MGMT Promoter Methylation of Glioblastoma from Dynamic Susceptibility Contrast Perfusion: A Radiomic Approach. J. Neuroimaging 2020, 30, 458–462. [Google Scholar] [CrossRef]
- Wei, J.W.; Li, L.W.; Han, Y.Q.; Gu, D.; Chen, Q.; Wang, J.M.; Li, R.T.; Zhan, J.; Tian, J.; Zhou, D.B. Accurate Preoperative Distinction of Intracranial Hemangiopericytoma From Meningioma Using a Multihabitat and Multisequence-Based Radiomics Diagnostic Technique. Front. Oncol. 2020, 10, 534. [Google Scholar] [CrossRef]
- Beig, N.; Bera, K.; Prasanna, P.; Antunes, J.; Correa, R.; Singh, S.; Saeed Bamashmos, A.; Ismail, M.; Braman, N.; Verma, R.; et al. Radiogenomic-Based Survival Risk Stratification of Tumor Habitat on Gd-T1w MRI Is Associated with Biological Processes in Glioblastoma. Clin. Cancer Res. 2020, 26, 1866–1876. [Google Scholar] [CrossRef] [Green Version]
- Lohmann, P.; Elahmadawy, M.A.; Gutsche, R.; Werner, J.M.; Bauer, E.K.; Ceccon, G.; Kocher, M.; Lerche, C.W.; Rapp, M.; Fink, G.R.; et al. FET PET Radiomics for Differentiating Pseudoprogression from Early Tumor Progression in Glioma Patients Post-Chemoradiation. Cancers 2020, 12, 3835. [Google Scholar] [CrossRef]
- Correa, R.; Lei, Q.; Chen, J.; Zeng, J.; Yu, J.; Tiwari, P. Lesion-habitat radiomics to distinguish radiation necrosis from tumor recurrence on post-treatment MRI in metastatic brain tumors. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 16–19 February 2020. [Google Scholar]
- Kumar, R.; Gupta, A.; Arora, H.S.; Pandian, G.N.; Raman, B. CGHF: A Computational Decision Support System for Glioma Classification Using Hybrid Radiomics- and Stationary Wavelet-Based Features. IEEE Access 2020, 8, 79440–79458. [Google Scholar] [CrossRef]
- Verma, R.; Correa, R.; Hill, V.B.; Statsevych, V.; Bera, K.; Beig, N.; Mahammedi, A.; Madabhushi, A.; Ahluwalia, M.; Tiwari, P. Tumor Habitat-derived Radiomic Features at Pretreatment MRI That Are Prognostic for Progression-free Survival in Glioblastoma Are Associated with Key Morphologic Attributes at Histopathologic Examination: A Feasibility Study. Radiol. Artif. Intell. 2020, 2, e190168. [Google Scholar] [CrossRef]
- Choi, S.W.; Cho, H.H.; Koo, H.; Cho, K.R.; Nenning, K.H.; Langs, G.; Furtner, J.; Baumann, B.; Woehrer, A.; Cho, H.J.; et al. Multi-habitat radiomics unravels distinct phenotypic subtypes of glioblastoma with clinical and genomic significance. Cancers 2020, 12, 1707. [Google Scholar] [CrossRef]
- Yousaf, S.; Anwar, S.M.; RaviPrakash, H.; Bagci, U. Brain Tumor Survival Prediction Using Radiomics Features. In Proceedings of the Third International Workshop, MLCN 2020, and Second International Workshop, Lima, Peru, 4–8 October 2020; Volume 12449, pp. 284–293. [Google Scholar]
- Zhang, X.; Lu, D.; Gao, P.; Tian, Q.; Lu, H.; Xu, X.; He, X.; Liu, Y. Survival-relevant high-risk subregion identification for glioblastoma patients: The MRI-based multiple instance learning approach. Eur. Radiol. 2020, 30, 5602–5610. [Google Scholar] [CrossRef]
- Choi, Y.; Nam, Y.; Lee, Y.S.; Kim, J.; Ahn, K.J.; Jang, J.; Shin, N.Y.; Kim, B.S.; Jeon, S.S. IDH1 mutation prediction using MR-based radiomics in glioblastoma: Comparison between manual and fully automated deep learning-based approach of tumor segmentation. Eur. J. Radiol. 2020, 128, 109031. [Google Scholar] [CrossRef]
- Sakai, Y.; Yang, C.; Kihira, S.; Tsankova, N.; Khan, F.; Hormigo, A.; Lai, A.; Cloughesy, T.; Nael, K. MRI radiomic features to predict idh1 mutation status in gliomas: A machine learning approach using gradient tree boosting. Int. J. Mol. Sci. 2020, 21, 8004. [Google Scholar] [CrossRef]
- Demirel, E.; Gökaslan, C.O.; Dilek, O.; Ozdemir, C.; Boyacı, M.G.; Korkmaz, S. Differential diagnosis of glioblastoma and solitary brain metastasis—The success of artificial intelligence models created with radiomics data obtained by automatic segmentation from conventional MRI sequences. Ceska Slov. Neurol. Neurochir. 2021, 84, 541–546. [Google Scholar] [CrossRef]
- Tixier, F.; Jaouen, V.; Hognon, C.; Gallinato, O.; Colin, T.; Visvikis, D. Evaluation of conventional and deep learning based image harmonization methods in radiomics studies. Phys. Med. Biol. 2021, 66, 245009. [Google Scholar] [CrossRef]
- Russo, G.; Stefano, A.; Alongi, P.; Comelli, A.; Catalfamo, B.; Mantarro, C.; Longo, C.; Altieri, R.; Certo, F.; Cosentino, S.; et al. Feasibility on the use of radiomics features of 11[C]-MET PET/CT in central nervous system tumours: Preliminary results on potential grading discrimination using a machine learning model. Curr. Oncol. 2021, 28, 5318–5331. [Google Scholar] [CrossRef]
- Yan, J.L.; Toh, C.H.; Ko, L.; Wei, K.C.; Chen, P.Y. A Neural Network Approach to Identify Glioblastoma Progression Phenotype from Multimodal MRI. Cancers 2021, 13, 2006. [Google Scholar] [CrossRef]
- Ye, J.M.; Huang, H.; Jiang, W.W.; Xu, X.M.; Xie, C.; Lu, B.; Wang, X.C.; Lai, X.B. Tumor Grade and Overall Survival Prediction of Gliomas Using Radiomics. Sci. Program. 2021, 2021, 9913466. [Google Scholar] [CrossRef]
- Joo, L.; Park, J.E.; Park, S.Y.; Nam, S.J.; Kim, Y.H.; Kim, J.H.; Kim, H.S. Extensive peritumoral edema and brain-to-tumor interface MRI features enable prediction of brain invasion in meningioma: Development and validation. Neuro-Oncology 2021, 23, 324–333. [Google Scholar] [CrossRef]
- Pasquini, L.; Napolitano, A.; Lucignani, M.; Tagliente, E.; Dellepiane, F.; Rossi-Espagnet, M.C.; Ritrovato, M.; Vidiri, A.; Villani, V.; Ranazzi, G.; et al. AI and High-Grade Glioma for Diagnosis and Outcome Prediction: Do All Machine Learning Models Perform Equally Well? Front. Oncol. 2021, 11, 601425. [Google Scholar] [CrossRef]
- Cao, M.; Suo, S.; Zhang, X.; Wang, X.; Xu, J.; Yang, W.; Zhou, Y. Qualitative and Quantitative MRI Analysis in IDH1 Genotype Prediction of Lower-Grade Gliomas: A Machine Learning Approach. BioMed Res. Int. 2021, 2021, 1235314. [Google Scholar] [CrossRef]
- Patel, M.; Zhan, J.; Natarajan, K.; Flintham, R.; Davies, N.; Sanghera, P.; Grist, J.; Duddalwar, V.; Peet, A.; Sawlani, V. Machine learning-based radiomic evaluation of treatment response prediction in glioblastoma. Clin. Radiol. 2021, 76, 628.e17–628.e27. [Google Scholar] [CrossRef]
- Soltani, M.; Bonakdar, A.; Shakourifar, N.; Babaie, R.; Raahemifar, K. Efficacy of Location-Based Features for Survival Prediction of Patients with Glioblastoma Depending on Resection Status. Front. Oncol. 2021, 11, 661123. [Google Scholar] [CrossRef]
- Wagner, M.W.; Hainc, N.; Khalvati, F.; Namdar, K.; Figueiredo, L.; Sheng, M.; Laughlin, S.; Shroff, M.M.; Bouffet, E.; Tabori, U.; et al. Radiomics of pediatric low-grade gliomas: Toward a pretherapeutic differentiation of BRAF-mutated and BRAF-fused tumors. Am. J. Neuroradiol. 2021, 42, 759–765. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Hung, T.N.K.; Do, D.T.; Lam, L.H.T.; Dang, L.H.; Huynh, T.T. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 2021, 132, 104320. [Google Scholar] [CrossRef]
- Kumar, R.; Gupta, A.; Arora, H.S.; Raman, B. GRGE: Detection of Gliomas Using Radiomics, GA Features and Extremely Randomized Trees. In Proceedings of the 2021 International Conference on Information Networking (ICOIN), Jeju Island, Republic of Korea, 13–16 January 2021; pp. 379–384. [Google Scholar]
- Cepeda, S.; Pérez-Nuñez, A.; García-García, S.; García-Pérez, D.; Arrese, I.; Jiménez-Roldán, L.; García-Galindo, M.; González, P.; Velasco-Casares, M.; Zamora, T.; et al. Predicting Short-Term Survival after Gross Total or Near Total Resection in Glioblastomas by Machine Learning-Based Radiomic Analysis of Preoperative MRI. Cancers 2021, 13, 5047. [Google Scholar] [CrossRef]
- Malik, N.; Geraghty, B.; Dasgupta, A.; Maralani, P.J.; Sandhu, M.; Detsky, J.; Tseng, C.L.; Soliman, H.; Myrehaug, S.; Husain, Z.; et al. MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J. Neurooncol. 2021, 155, 181–191. [Google Scholar] [CrossRef]
- Samani, Z.R.; Parker, D.; Wolf, R.; Hodges, W.; Brem, S.; Verma, R. Distinct tumor signatures using deep learning-based characterization of the peritumoral microenvironment in glioblastomas and brain metastases. Sci. Rep. 2021, 11, 14469. [Google Scholar] [CrossRef]
- Xiao, D.; Wang, J.; Wang, X.; Fu, P.; Zhao, H.; Yan, P.; Jiang, X. Distinguishing brain abscess from necrotic glioblastoma using MRI-based intranodular radiomic features and peritumoral edema/tumor volume ratio. J. Integr. Neurosci. 2021, 20, 623–634. [Google Scholar] [CrossRef]
- Gutta, S.; Acharya, J.; Shiroishi, M.S.; Hwang, D.; Nayak, K.S. Improved glioma grading using deep convolutional neural networks. Am. J. Neuroradiol. 2021, 42, 233–239. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Su, X.; Yang, X.; Wang, W.; Wan, X.; Tan, Q.; Chen, N.; Yue, Q.; Gong, Q. Automated machine learning to predict the co-occurrence of isocitrate dehydrogenase mutations and O6-methylguanine-DNA methyltransferase promoter methylation in patients with gliomas. J. Magn. Reson. Imaging 2021, 54, 197–205. [Google Scholar] [CrossRef]
- Xu, X.; Samaras, D.; Prasanna, P. Radiologically Defined Tumor-habitat Adjacency as a Prognostic Biomarker in Glioblastoma. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 3998–4001. [Google Scholar]
- Meißner, A.K.; Gutsche, R.; Galldiks, N.; Kocher, M.; Jünger, S.T.; Eich, M.L.; Montesinos-Rongen, M.; Brunn, A.; Deckert, M.; Wendl, C.; et al. Radiomics for the noninvasive prediction of the BRAF mutation status in patients with melanoma brain metastases. Neuro-Oncology 2022, 24, 1331–1340. [Google Scholar] [CrossRef]
- Shaheen, A.; Bukhari, S.T.; Nadeem, M.; Burigat, S.; Bagci, U.; Mohy-Ud-Din, H. Overall Survival Prediction of Glioma Patients With Multiregional Radiomics. Front. Neurosci. 2022, 16, 911065. [Google Scholar] [CrossRef]
- Deng, D.B.; Liao, Y.T.; Zhou, J.F.; Cheng, L.N.; He, P.; Wu, S.N.; Wang, W.S.; Zhou, Q. Non-Invasive Prediction of Survival Time of Midline Glioma Patients Using Machine Learning on Multiparametric MRI Radiomics Features. Front. Neurol. 2022, 13, 866274. [Google Scholar] [CrossRef]
- Liu, D.; Chen, J.; Ge, H.; Hu, X.; Yang, K.; Liu, Y.; Hu, G.; Luo, B.; Yan, Z.; Song, K.; et al. Differentiation of malignant brain tumor types using intratumoral and peritumoral radiomic features. Front. Oncol. 2022, 12, 848846. [Google Scholar] [CrossRef]
- Do, D.T.; Yang, M.R.; Lam, L.H.T.; Le, N.Q.K.; Wu, Y.W. Improving MGMT methylation status prediction of glioblastoma through optimizing radiomics features using genetic algorithm-based machine learning approach. Sci. Rep. 2022, 12, 13412. [Google Scholar] [CrossRef]
- Chiu, F.Y.; Yen, Y. Efficient Radiomics-Based Classification of Multi-Parametric MR Images to Identify Volumetric Habitats and Signatures in Glioblastoma: A Machine Learning Approach. Cancers 2022, 14, 1475. [Google Scholar] [CrossRef]
- Chen, H.; Li, S.; Zhang, Y.; Liu, L.; Lv, X.; Yi, Y.; Ruan, G.; Ke, C.; Feng, Y. Deep learning-based automatic segmentation of meningioma from multiparametric MRI for preoperative meningioma differentiation using radiomic features: A multicentre study. Eur. Radiol. 2022, 32, 7248–7259. [Google Scholar] [CrossRef]
- Xu, J.Q.; Ren, Y.; Zhao, X.Y.; Wang, X.Q.; Yu, X.C.; Yao, Z.W.; Zhou, Y.; Feng, X.Y.; Zhou, X.J.; Wang, H. Incorporating multiple magnetic resonance diffusion models to differentiate low- and high-grade adult gliomas: A machine learning approach. Quant. Imaging Med. Surg. 2022, 12, 5171–5183. [Google Scholar] [CrossRef]
- Kumar, R.; Gupta, A.; Arora, H.S.; Raman, B. IBRDM: An Intelligent Framework for Brain Tumor Classification Using Radiomics- and DWT-based Fusion of MRI Sequences. ACM Trans. Internet Technol. 2022, 22, 9. [Google Scholar] [CrossRef]
- Verma, R.; Hill, V.B.; Statsevych, V.; Bera, K.; Correa, R.; Leo, P.; Ahluwalia, M.; Madabhushi, A.; Tiwari, P. Stable and Discriminatory Radiomic Features from the Tumor and Its Habitat Associated with Progression-Free Survival in Glioblastoma: A Multi-Institutional Study. Am. J. Neuroradiol. 2022, 43, 1115–1123. [Google Scholar] [CrossRef]
- Wang, Y.; Lang, J.; Zuo, J.Z.; Dong, Y.; Hu, Z.; Xu, X.; Zhang, Y.; Wang, Q.; Yang, L.; Wong, S.T.C.; et al. The radiomic-clinical model using the SHAP method for assessing the treatment response of whole-brain radiotherapy: A multicentric study. Eur. Radiol. 2022, 32, 8737–8747. [Google Scholar] [CrossRef]
- Yang, Y.; Han, Y.; Zhao, S.; Xiao, G.; Guo, L.; Zhang, X.; Cui, G. Spatial heterogeneity of edema region uncovers survival-relevant habitat of Glioblastoma. Eur. J. Radiol. 2022, 154, 110423. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Zhang, P.; Bi, Y.; Yang, C.; Wu, M.; He, D.; Huang, S.; Yang, K.; Qi, S.; Wang, J. MRI radiomic features of peritumoral edema may predict the recurrence sites of glioblastoma multiforme. Front. Oncol. 2022, 12, 1042498. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. An ensemble learning approach for brain cancer detection exploiting radiomic features. Comput. Methods Programs Biomed. 2020, 185, 105134. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro-Oncology 2021, 23, 304–313. [Google Scholar] [CrossRef]
- Verduin, M.; Primakov, S.; Compter, I.; Woodruff, H.C.; van Kuijk, S.M.J.; Ramaekers, B.L.T.; te Dorsthorst, M.; Revenich, E.G.M.; ter Laan, M.; Pegge, S.A.H.; et al. Prognostic and Predictive Value of Integrated Qualitative and Quantitative Magnetic Resonance Imaging Analysis in Glioblastoma. Cancers 2021, 13, 722. [Google Scholar] [CrossRef]
- Su, X.; Chen, N.; Sun, H.; Liu, Y.; Yang, X.; Wang, W.; Zhang, S.; Tan, Q.; Su, J.; Gong, Q.; et al. Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro-Oncology 2020, 22, 393–401. [Google Scholar] [CrossRef]
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef]
- Montemurro, N.; Pahwa, B.; Tayal, A.; Shukla, A.; De Jesus Encarnacion, M.; Ramirez, I.; Nurmukhametov, R.; Chavda, V.; De Carlo, A. Macrophages in Recurrent Glioblastoma as a Prognostic Factor in the Synergistic System of the Tumor Microenvironment. Neurol. Int. 2023, 15, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.; Choudhury, A.; Keough, M.B.; Seo, K.; Ni, L.; Kakaizada, S.; Lee, A.; Aabedi, A.; Popova, G.; Lipkin, B.; et al. Glioblastoma remodelling of human neural circuits decreases survival. Nature 2023, 617, 599–607. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabassum, M.; Suman, A.A.; Suero Molina, E.; Pan, E.; Di Ieva, A.; Liu, S. Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review. Cancers 2023, 15, 3845. https://doi.org/10.3390/cancers15153845
Tabassum M, Suman AA, Suero Molina E, Pan E, Di Ieva A, Liu S. Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review. Cancers. 2023; 15(15):3845. https://doi.org/10.3390/cancers15153845
Chicago/Turabian StyleTabassum, Mehnaz, Abdulla Al Suman, Eric Suero Molina, Elizabeth Pan, Antonio Di Ieva, and Sidong Liu. 2023. "Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review" Cancers 15, no. 15: 3845. https://doi.org/10.3390/cancers15153845
APA StyleTabassum, M., Suman, A. A., Suero Molina, E., Pan, E., Di Ieva, A., & Liu, S. (2023). Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review. Cancers, 15(15), 3845. https://doi.org/10.3390/cancers15153845






