Vessel Delineation Using U-Net: A Sparse Labeled Deep Learning Approach for Semantic Segmentation of Histological Images
Abstract
Simple Summary
Abstract
1. Introduction
- Organ Segmentation: Segmentation of various organs in medical images, such as liver, kidney, pancreas, gall bladder, and lung [21,22,23]. This provides important support for diagnosing, surgical planning, and disease monitoring. However, these methods are based on CT or MRI data and are focused on localization of a target structure not on a precise segmentation [23].
- Cell Segmentation: Segmentation of various cell types in microscopy images [24], such as detection of metastases in histopathology images [25], segmentation of cells’ nuclei [26,27], and segmentation of blood cells in microscopic images [28,29]. These methods are focused on localization rather than on an accurate segmentation and are applied to very homogeneous images.
- Tumor Segmentation: Tumor segmentation in different types of medical images, such as ultrasound [30], computed tomography (CT) [31,32,33,34,35], and magnetic resonance imaging (MRI) scans [36,37]. Accurate segmentation of tumors is crucial for diagnosing, treatment planning, and monitoring of cancer patients. Conventionally, the image types for tumor segmentation are gained from ultrasound, CT, and MRI, showing characteristics that cannot directly be transferred to histological images.
- Vessel Segmentation: Segmentation of blood vessels in various medical images, such as magnetic resonance angiography (MRA) scans [38], retinal images [39,40,41], and brain scans [42], which is important for the diagnosis and treatment of various diseases, such as diabetic retinopathy and stroke. Vessel segmentation is applied to various image modalities, yet the vessels are mostly homogeneous in structure. Typically, segmented images express two classes. Thus, for histological images, a scalable multiclass model that can be extended to classes specific to the desired application must be built.
2. Materials and Methods
2.1. Sample Acquisition
2.2. Data Preparation
- Vessel walls are fine but continuous structures: Walls are always labeled completely, indicating that there are no gaps, even with low image intensity.
- Vessel lumens are separated from the tissue by a vessel wall: Vessel lumens are also labeled by drawing a line on the inside of the enclosing wall. Also, a line on the outside of the wall as tissue is labeled to indicate the separation from tissue and lumen by a wall. The remaining lumen is labeled by lines crossing the whole area of the lumen.
- Remaining blood cells within the vessel lumen should be labeled as lumen: The labeled lines within the lumen go across the remaining cells to identify them as part of the lumen.
- The tissue is a large continuous structure making up most part of the image: Tissue is labeled by long lines across the heterogeneous areas of tissue. These lines indicate its continuousness and penalize frequent changes in the segmentation.
- The background is also large and continuous and located at the images’ edges: The background is also labeled with long lines across its area. A separating line at the tissue border indicates the transition from tissue to background.
- Contrast: random increase or decrease in contrast by up to 90%;
- Brightness: random increase or decrease in the brightness by up to 30%;
- Zoom: random scaling between 50% and 200% of the original size, where x and y directions are scaled independently;
- Rotation: random angle between −90° and 90°;
- Flipping: no flipping, horizontal or vertical flipping.
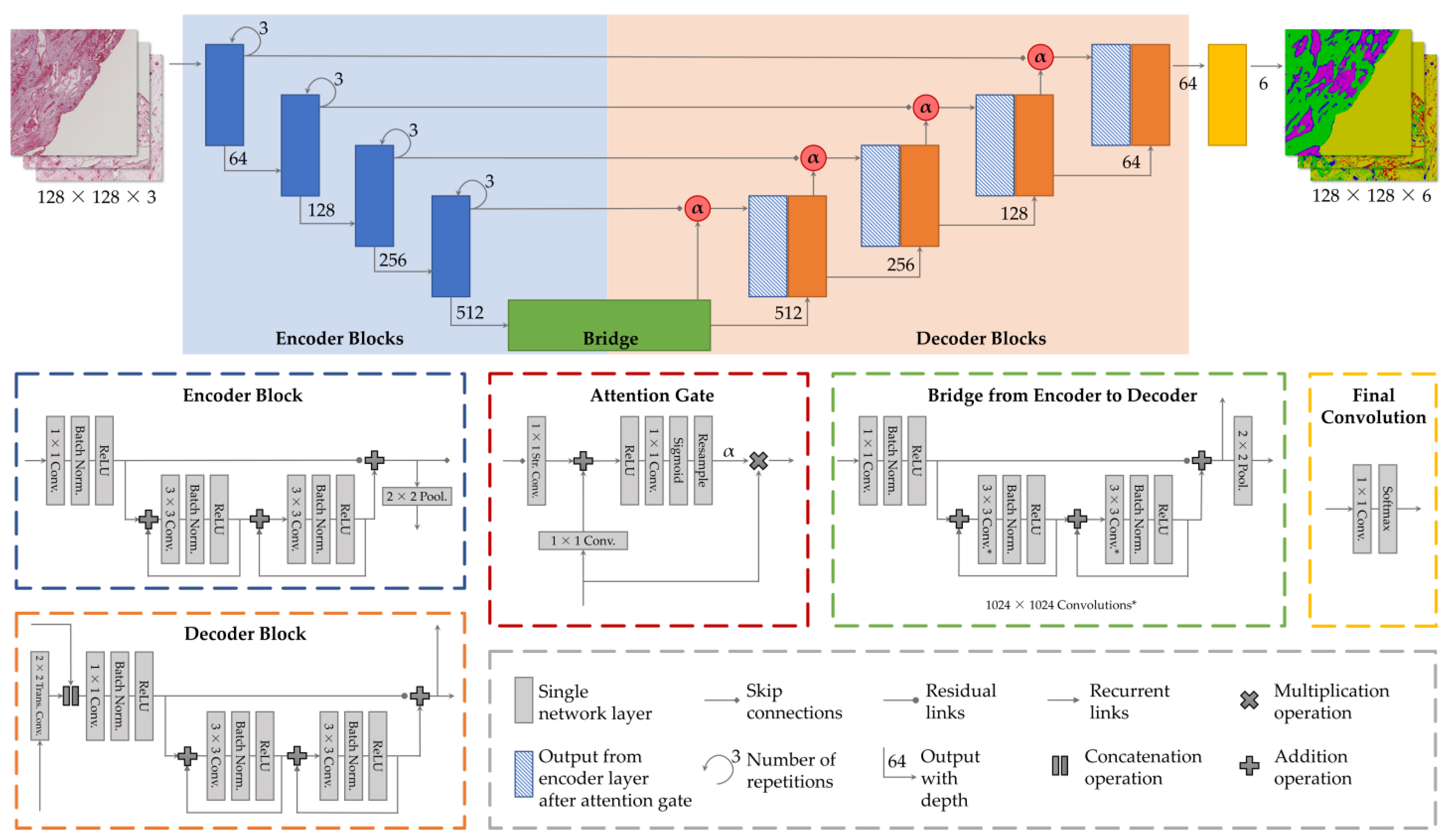
2.3. Network Architecture
2.3.1. Attention Gates
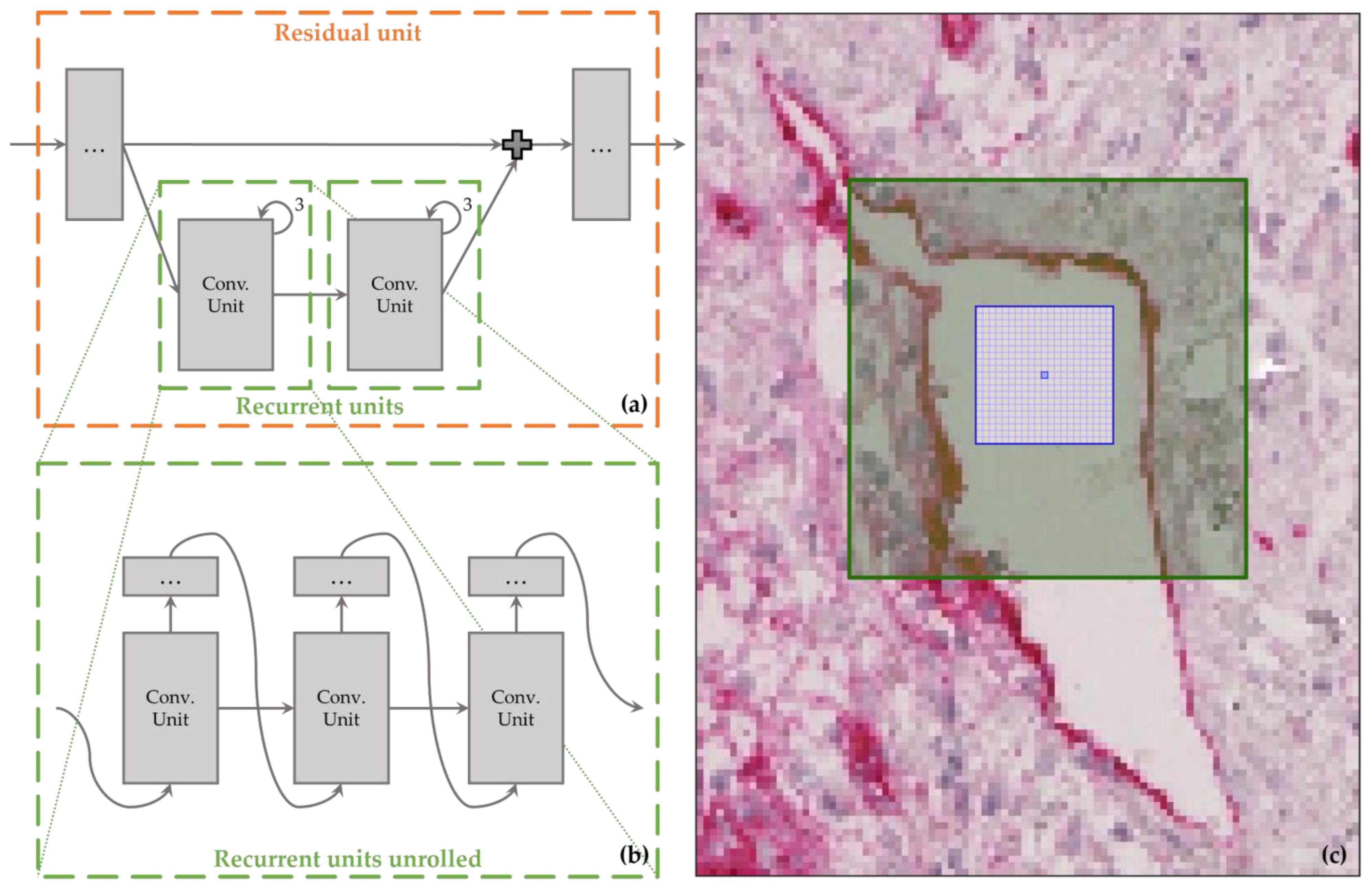
2.3.2. Recurrent Links
2.3.3. Residual Links
2.3.4. Loss Function
2.3.5. Hyperparameters
- Batch size: 16 (for all architectures);
- Dropout: 0.25 (for Recurrent U-Net) and 0.125 (for all other architectures);
- Learning rate: 1 × 10−4 (for all architectures).
2.4. Used Software and Hardware
3. Results
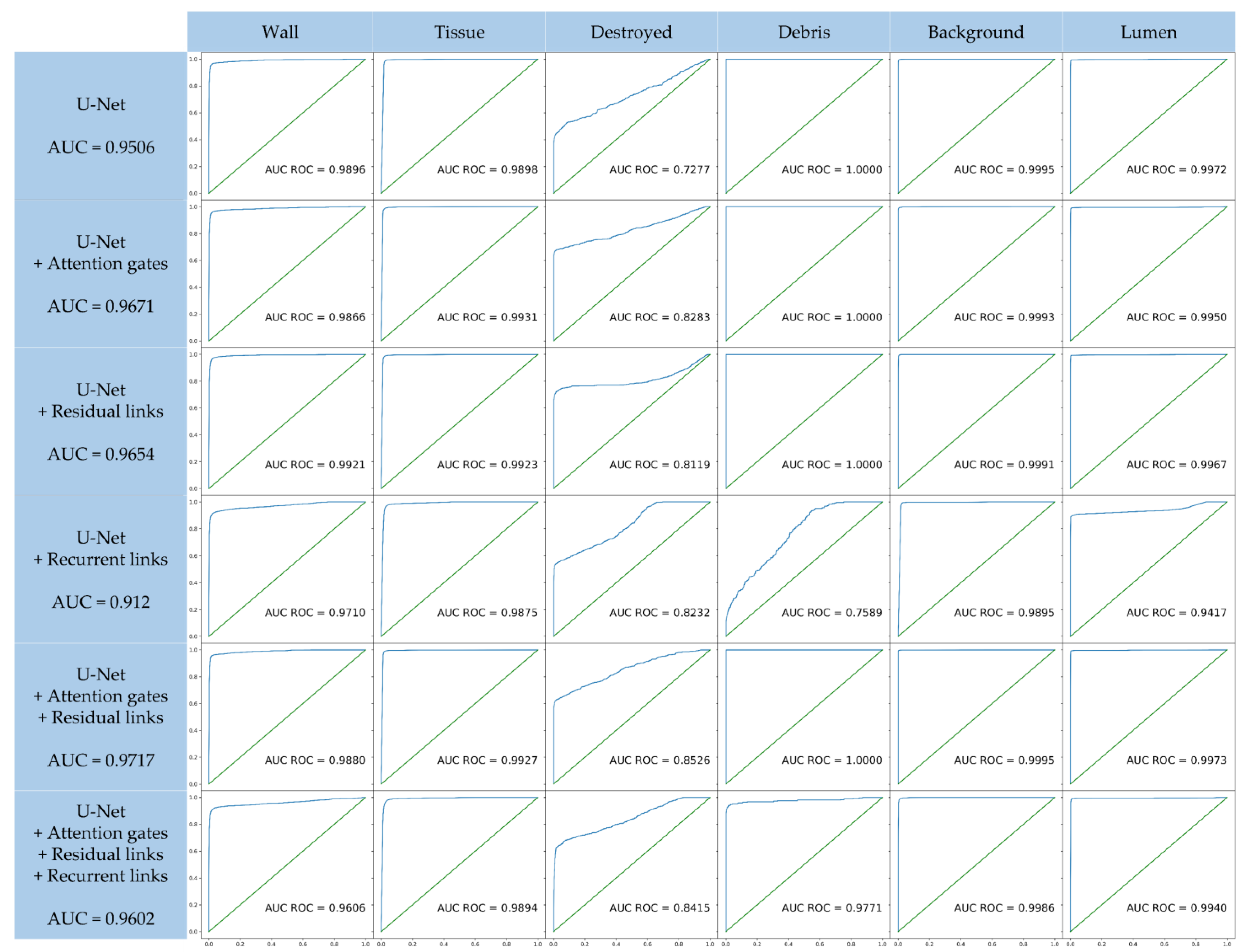
3.1. Quantitative Ablation Study
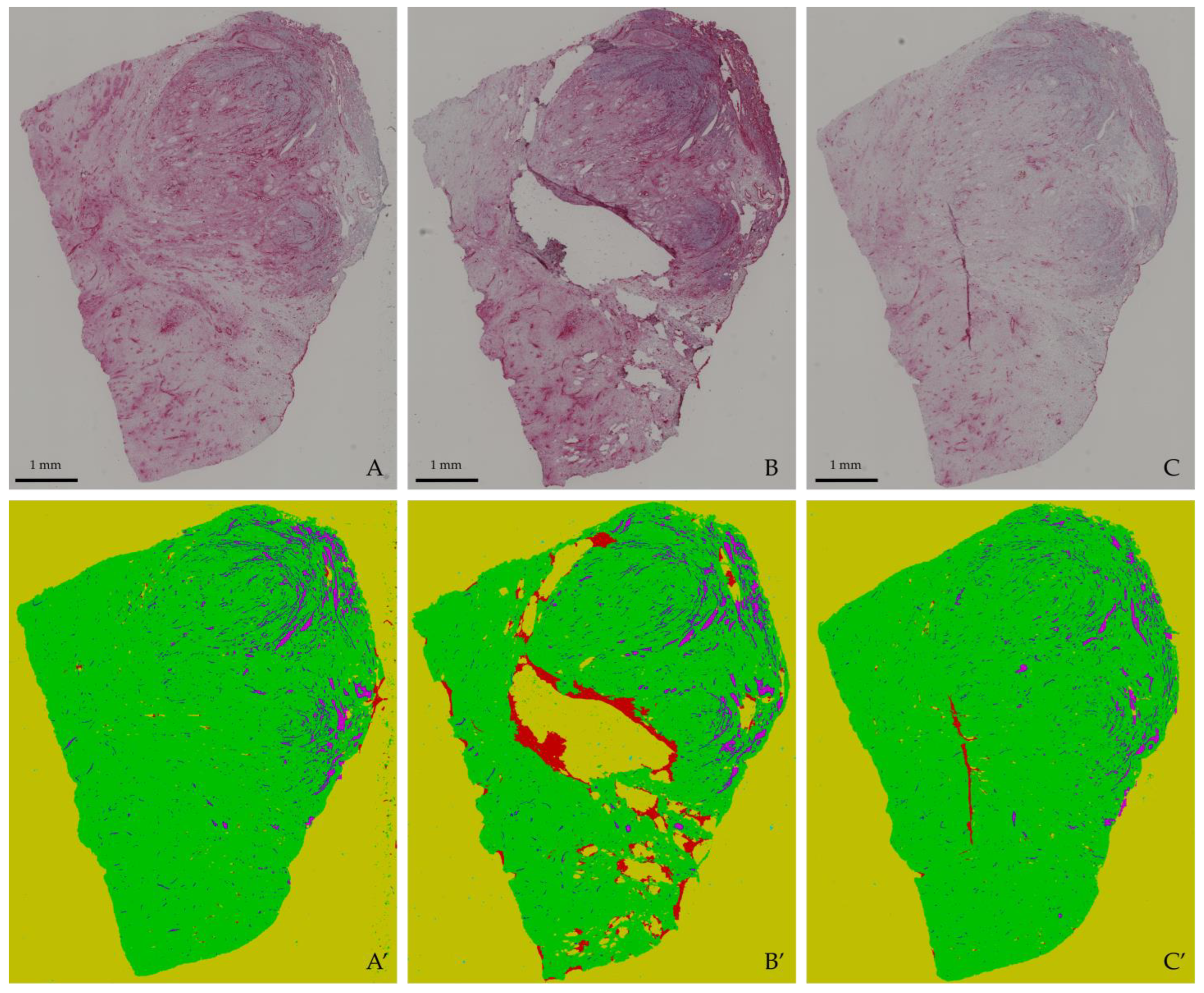
3.2. Qualitative Segmentation Analysis
3.3. Selection of the Best Architecture
4. Discussion and Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Yousef, R.; Gupta, G.; Yousef, N.; Khari, M. A holistic overview of deep learning approach in medical imaging. Multimed. Syst. 2022, 28, 881–914. [Google Scholar] [CrossRef] [PubMed]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2022, 11, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Ding, Y.; Qadri, S.F.; Yang, J. Convolutional-neural-network-based feature extraction for liver segmentation from CT images. In Proceedings of the Eleventh International Conference on Digital Image Processing (ICDIP 2019), Guangzhou, China, 10–13 May 2019; Jiang, X., Hwang, J.-N., Eds.; SPIE: Bellingham, WA, USA, 2019; p. 159, ISBN 9781510630758. [Google Scholar]
- Qadri, S.F.; Lin, H.; Shen, L.; Ahmad, M.; Qadri, S.; Khan, S.; Khan, M.; Zareen, S.S.; Akbar, M.A.; Bin Heyat, M.B.; et al. CT-Based Automatic Spine Segmentation Using Patch-Based Deep Learning. Int. J. Intell. Syst. 2023, 2023, 2345835. [Google Scholar] [CrossRef]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 2015, 18, 539–553. [Google Scholar] [CrossRef]
- Sung, K.E.; Beebe, D.J. Microfluidic 3D models of cancer. Adv. Drug Deliv. Rev. 2014, 79, 68–78. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model ®evolution: Unveiling Tumor-Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef]
- Gerardo-Nava, J.L.; Jansen, J.; Günther, D.; Klasen, L.; Thiebes, A.L.; Niessing, B.; Bergerbit, C.; Meyer, A.A.; Linkhorst, J.; Barth, M.; et al. Transformative Materials to Create 3D Functional Human Tissue Models In Vitro in a Reproducible Manner. Adv Healthc. Mater. 2023, 2301030. [Google Scholar] [CrossRef]
- Ben Hamida, A.; Devanne, M.; Weber, J.; Truntzer, C.; Derangère, V.; Ghiringhelli, F.; Forestier, G.; Wemmert, C. Deep learning for colon cancer histopathological images analysis. Comput. Biol. Med. 2021, 136, 104730. [Google Scholar] [CrossRef]
- Mo, Y.; Wu, Y.; Yang, X.; Liu, F.; Liao, Y. Review the state-of-the-art technologies of semantic segmentation based on deep learning. Neurocomputing 2022, 493, 626–646. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Bae, H.; Puranik, A.S.; Gauvin, R.; Edalat, F.; Carrillo-Conde, B.; Peppas, N.A.; Khademhosseini, A. Building vascular networks. Sci. Transl. Med. 2012, 4, 160ps23. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.; Brady, E.; Zheng, Y.; Moore, E.; Stevens, K.R. Engineering the multiscale complexity of vascular networks. Nat. Rev. Mater. 2022, 7, 702–716. [Google Scholar] [CrossRef]
- Chen, E.P.; Toksoy, Z.; Davis, B.A.; Geibel, J.P. 3D Bioprinting of Vascularized Tissues for in vitro and in vivo Applications. Front. Bioeng. Biotechnol. 2021, 9, 664188. [Google Scholar] [CrossRef]
- Lindemann, M.C.; Luttke, T.; Nottrodt, N.; Schmitz-Rode, T.; Slabu, I. FEM based simulation of magnetic drug targeting in a multibranched vessel model. Comput. Methods Programs Biomed. 2021, 210, 106354. [Google Scholar] [CrossRef]
- Helms, F.; Haverich, A.; Wilhelmi, M.; Böer, U. Establishment of a Modular Hemodynamic Simulator for Accurate In Vitro Simulation of Physiological and Pathological Pressure Waveforms in Native and Bioartificial Blood Vessels. Cardiovasc. Eng. Technol. 2022, 13, 291–306. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional In Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Kakeya, H.; Okada, T.; Oshiro, Y. 3D U-JAPA-Net: Mixture of Convolutional Networks for Abdominal Multi-organ CT Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 426–433. ISBN 978-3-030-00936-6. [Google Scholar]
- Seo, H.; Huang, C.; Bassenne, M.; Xiao, R.; Xing, L. Modified U-Net (mU-Net) With Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans. Med. Imaging 2020, 39, 1316–1325. [Google Scholar] [CrossRef]
- Li, C.; Tan, Y.; Chen, W.; Luo, X.; He, Y.; Gao, Y.; Li, F. ANU-Net: Attention-based nested U-Net to exploit full resolution features for medical image segmentation. Comput. Graph. 2020, 90, 11–20. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.W.; Jia, S.; Ashraf, A.B.; Hu, P. Integrative Data Augmentation with U-Net Segmentation Masks Improves Detection of Lymph Node Metastases in Breast Cancer Patients. Cancers 2020, 12, 2934. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Nuclei Segmentation with Recurrent Residual Convolutional Neural Networks based U-Net (R2U-Net). In Proceedings of the NAECON 2018—IEEE National Aerospace and Electronics Conference, Dayton, OH, USA, 23–26 July 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 228–233, ISBN 978-1-5386-6557-2. [Google Scholar]
- Long, F. Microscopy cell nuclei segmentation with enhanced U-Net. BMC Bioinform. 2020, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Benazzouz, M.; Benomar, M.L.; Moualek, Y. Modified U-Net for cytological medical image segmentation. Int. J. Imaging Syst. Tech. 2022, 32, 1761–1773. [Google Scholar] [CrossRef]
- Zhang, M.; Li, X.; Xu, M.; Li, Q. Automated Semantic Segmentation of Red Blood Cells for Sickle Cell Disease. IEEE J. Biomed. Health Inform. 2020, 24, 3095–3102. [Google Scholar] [CrossRef]
- Li, X.; Guo, Y.; Jiang, F.; Xu, L.; Shen, F.; Jin, Z.; Wang, Y. Multi-Task Refined Boundary-Supervision U-Net (MRBSU-Net) for Gastrointestinal Stromal Tumor Segmentation in Endoscopic Ultrasound (EUS) Images. IEEE Access 2020, 8, 5805–5816. [Google Scholar] [CrossRef]
- Li, S.; Tso, G.K.; He, K. Bottleneck feature supervised U-Net for pixel-wise liver and tumor segmentation. Expert Syst. Appl. 2020, 145, 113131. [Google Scholar] [CrossRef]
- Zhang, Y.; Lei, B.; Fu, C.; Du, J.; Zhu, X.; Han, X.; Du, L.; Gao, W.; Wang, T.; Ma, G. HBNet: Hybrid Blocks Network for Segmentation of Gastric Tumor from Ordinary CT Images. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4, ISBN 978-1-5386-9330-8. [Google Scholar]
- Wang, L.; Wang, B.; Xu, Z. Tumor Segmentation Based on Deeply Supervised Multi-Scale U-Net. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 746–749, ISBN 978-1-7281-1867-3. [Google Scholar]
- Liu, Y.-C.; Shahid, M.; Sarapugdi, W.; Lin, Y.-X.; Chen, J.-C.; Hua, K.-L. Cascaded atrous dual attention U-Net for tumor segmentation. Multimed. Tools Appl. 2021, 80, 30007–30031. [Google Scholar] [CrossRef]
- Pang, S.; Du, A.; Orgun, M.A.; Wang, Y.; Yu, Z. Tumor attention networks: Better feature selection, better tumor segmentation. Neural Netw. 2021, 140, 203–222. [Google Scholar] [CrossRef]
- Hasan, S.M.K.; Linte, C.A. A Modified U-Net Convolutional Network Featuring a Nearest-neighbor Re-sampling-based Elastic-Transformation for Brain Tissue Characterization and Segmentation. In Proceedings of the 2018 IEEE Western New York Image and Signal Processing Workshop (WNYISPW), Rochester, NY, USA, 5 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Shi, W.; Pang, E.; Wu, Q.; Lin, F. Brain Tumor Segmentation Using Dense Channels 2D U-net and Multiple Feature Extraction Network. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 273–283. ISBN 978-3-030-46639-8. [Google Scholar]
- Angermann, C.; Haltmeier, M. Random 2.5D U-net for Fully 3D Segmentation. In Machine Learning and Medical Engineering for Cardiovascular Health and Intravascular Imaging and Computer Assisted Stenting; Liao, H., Balocco, S., Wang, G., Zhang, F., Liu, Y., Ding, Z., Duong, L., Phellan, R., Zahnd, G., Breininger, K., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 158–166. ISBN 978-3-030-33326-3. [Google Scholar]
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl. Based Syst. 2019, 178, 149–162. [Google Scholar] [CrossRef]
- Wu, Y.; Xia, Y.; Song, Y.; Zhang, D.; Liu, D.; Zhang, C.; Cai, W. Vessel-Net: Retinal Vessel Segmentation Under Multi-path Supervision. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 264–272. ISBN 978-3-030-32238-0. [Google Scholar]
- Adarsh, R.; Amarnageswarao, G.; Pandeeswari, R.; Deivalakshmi, S. Dense Residual Convolutional Auto Encoder for Retinal Blood Vessels Segmentation. In Proceedings of the 2020 6th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 6–7 March 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 280–284, ISBN 978-1-7281-5196-0. [Google Scholar]
- Zhang, M.; Zhang, C.; Wu, X.; Cao, X.; Young, G.S.; Chen, H.; Xu, X. A neural network approach to segment brain blood vessels in digital subtraction angiography. Comput. Methods Programs Biomed. 2020, 185, 105159. [Google Scholar] [CrossRef]
- Palzer, J.; Mues, B.; Goerg, R.; Aberle, M.; Rensen, S.S.; Olde Damink, S.W.M.; Vaes, R.D.W.; Cramer, T.; Schmitz-Rode, T.; Neumann, U.P.; et al. Magnetic Fluid Hyperthermia as Treatment Option for Pancreatic Cancer Cells and Pancreatic Cancer Organoids. Int. J. Nanomed. 2021, 16, 2965–2981. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; ISBN 978-1-4614-7137-0. [Google Scholar]
- Shakeel, F.; Sabhitha, A.S.; Sharma, S. Exploratory review on class imbalance problem: An overview. In Proceedings of the 2017 8th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Delhi, India, 3–5 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Bria, A.; Marrocco, C.; Tortorella, F. Addressing class imbalance in deep learning for small lesion detection on medical images. Comput. Biol. Med. 2020, 120, 103735. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Gupta, A.; Girshick, R. Training Region-Based Object Detectors with Online Hard Example Mining. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 761–769, ISBN 978-1-4673-8851-1. [Google Scholar]
- Dong, Q.; Gong, S.; Zhu, X. Class Rectification Hard Mining for Imbalanced Deep Learning. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1869–1878, ISBN 978-1-5386-1032-9. [Google Scholar]
- Dai, J.; Qi, H.; Xiong, Y.; Li, Y.; Zhang, G.; Hu, H.; Wei, Y. Deformable Convolutional Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 764–773, ISBN 978-1-5386-1032-9. [Google Scholar]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Advances in Neural Information Processing Systems 30 (NIPS 2017); Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: New York, NY, USA, 2017; pp. 6000–6010. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging 2019, 6, 14006. [Google Scholar] [CrossRef]
- Tomar, N.K.; Jha, D.; Riegler, M.A.; Johansen, H.D.; Johansen, D.; Rittscher, J.; Halvorsen, P.; Ali, S. FANet: A Feedback Attention Network for Improved Biomedical Image Segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–14. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, F.; Gao, J.; Cao, S. Multi-Path Recurrent U-Net Segmentation of Retinal Fundus Image. Appl. Sci. 2020, 10, 3777. [Google Scholar] [CrossRef]
- Araujo, A.; Norris, W.; Sim, J. Computing Receptive Fields of Convolutional Neural Networks. Distill 2019, 4, e21. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778, ISBN 978-1-4673-8851-1. [Google Scholar]
- Li, D.; Dharmawan, D.A.; Ng, B.P.; Rahardja, S. Residual U-Net for Retinal Vessel Segmentation. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1425–1429, ISBN 978-1-5386-6249-6. [Google Scholar]
- Yu, W.; Fang, B.; Liu, Y.; Gao, M.; Zheng, S.; Wang, Y. Liver Vessels Segmentation Based on 3d Residual U-NET. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 250–254, ISBN 978-1-5386-6249-6. [Google Scholar]
- Zhang, J.; Lv, X.; Zhang, H.; Liu, B. AResU-Net: Attention Residual U-Net for Brain Tumor Segmentation. Symmetry 2020, 12, 721. [Google Scholar] [CrossRef]
- Pan, L.-S.; Li, C.-W.; Su, S.-F.; Tay, S.-Y.; Tran, Q.-V.; Chan, W.P. Coronary artery segmentation under class imbalance using a U-Net based architecture on computed tomography angiography images. Sci. Rep. 2021, 11, 14493. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 1–54. [Google Scholar] [CrossRef]
- Wasikowski, M.; Chen, X.w. Combating the Small Sample Class Imbalance Problem Using Feature Selection. IEEE Trans. Knowl. Data Eng. 2010, 22, 1388–1400. [Google Scholar] [CrossRef]
- Qu, W.; Balki, I.; Mendez, M.; Valen, J.; Levman, J.; Tyrrell, P.N. Assessing and mitigating the effects of class imbalance in machine learning with application to X-ray imaging. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 2041–2048. [Google Scholar] [CrossRef] [PubMed]
- Powers, D.M.W. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar] [CrossRef]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar] [CrossRef]
- Wager, S.; Wang, S.; Liang, P.S. Dropout Training as Adaptive Regularization. In Advances in Neural Information Processing Systems; Burges, C.J., Bottou, L., Welling, M., Ghahramani, Z., Weinberger, K.Q., Eds.; Curran Associates, Inc.: New York, NY, USA, 2013. [Google Scholar]
- TensorFlow Developers. TensorFlow; Zenodo: Geneve, Switzerland, 2023. [Google Scholar]
- Chollet, F.; Zhu, Q.S.; Rahman, F.; Lee, T.; Qian, C.; de Marmiesse, G.; Jin, H.; Zabluda, O.; Marks, S.; Watson, M.; et al. Keras. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 28 June 2023).
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Kupfer, B.; Netanyahu, N.S.; Shimshoni, I. An Efficient SIFT-Based Mode-Seeking Algorithm for Sub-Pixel Registration of Remotely Sensed Images. IEEE Geosci. Remote Sens. Lett. 2015, 12, 379–383. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, G.; Deng, J.; Xue, Q.; Hou, W.; He, Y. Image Registration Algorithm for Sequence Pathology Slices of Pulmonary Nodule. In Proceedings of the 2019 8th International Symposium on Next Generation Electronics (ISNE), Zhengzhou, China, 9–10 October 2019; IEEE: Piscataway, NY, USA, 2019; pp. 1–3, ISBN 978-1-7281-2062-1. [Google Scholar]
- Lobachev, O.; Ulrich, C.; Steiniger, B.S.; Wilhelmi, V.; Stachniss, V.; Guthe, M. Feature-based multi-resolution registration of immunostained serial sections. Med. Image Anal. 2017, 35, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Saalfeld, S.; Fetter, R.; Cardona, A.; Tomancak, P. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 2012, 9, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Hermann, J.; Brehmer, K.; Jankowski, V.; Lellig, M.; Hohl, M.; Mahfoud, F.; Speer, T.; Schunk, S.J.; Tschernig, T.; Thiele, H.; et al. Registration of Image Modalities for Analyses of Tissue Samples Using 3D Image Modelling. Proteom. Clin. Appl. 2020, 15, e1900143. [Google Scholar] [CrossRef] [PubMed]
- Paknezhad, M.; Loh, S.Y.M.; Choudhury, Y.; Koh, V.K.C.; Yong, T.T.K.; Tan, H.S.; Kanesvaran, R.; Tan, P.H.; Peng, J.Y.S.; Yu, W.; et al. Regional registration of whole slide image stacks containing major histological artifacts. BMC Bioinform. 2020, 21, 558. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Z.; Yu, Q. Point-Based Registration for Multi-stained Histology Images. In Proceedings of the 2020 IEEE 5th International Conference on Image, Vision and Computing (ICIVC), Beijing, China, 10–12 July 2020; IEEE: Piscataway, NY, USA, 2020; pp. 92–96, ISBN 978-1-7281-6661-2. [Google Scholar]
- Deng, R.; Yang, H.; Jha, A.; Lu, Y.; Chu, P.; Fogo, A.B.; Huo, Y. Map3D: Registration-Based Multi-Object Tracking on 3D Serial Whole Slide Images. IEEE Trans. Med. Imaging 2021, 40, 1924–1933. [Google Scholar] [CrossRef]
- Wang, C.-W.; Chen, H.-C. Improved image alignment method in application to X-ray images and biological images. Bioinformatics 2013, 29, 1879–1887. [Google Scholar] [CrossRef]
- Wang, C.-W.; Ka, S.-M.; Chen, A. Robust image registration of biological microscopic images. Sci. Rep. 2014, 4, 6050. [Google Scholar] [CrossRef]
- Liu, Y.; Tian, J.; Hu, R.; Yang, B.; Liu, S.; Yin, L.; Zheng, W. Improved Feature Point Pair Purification Algorithm Based on SIFT During Endoscope Image Stitching. Front. Neurorobot. 2022, 16, 840594. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Bay, H.; Ess, A.; Tuytelaars, T.; van Gool, L. Speeded-Up Robust Features (SURF). Comput. Vis. Image Underst. 2008, 110, 346–359. [Google Scholar] [CrossRef]
- Schwier, M.; Böhler, T.; Hahn, H.K.; Dahmen, U.; Dirsch, O. Registration of histological whole slide images guided by vessel structures. J. Pathol. Inform. 2013, 4, S10. [Google Scholar] [CrossRef]
- Kugler, M.; Goto, Y.; Kawamura, N.; Kobayashi, H.; Yokota, T.; Iwamoto, C.; Ohuchida, K.; Hashizume, M.; Hontani, H. Accurate 3D Reconstruction of a Whole Pancreatic Cancer Tumor from Pathology Images with Different Stains. In Proceedings of the Computational Pathology and Ophthalmic Medical Image Analysis: First International Workshop, COMPAY 2018, and 5th International Workshop, OMIA 2018, Granada, Spain, 16–20 September 2018; Stoyanov, D., Taylor, Z., Ciompi, F., Xu, Y., Martel, A., Maier-Hein, L., Rajpoot, N., van der Laak, J., Veta, M., McKenna, S., et al., Eds.; Springer: Cham, Switzerland, 2018; pp. 35–43, ISBN 9783030009496. [Google Scholar]
- Kugler, M.; Goto, Y.; Tamura, Y.; Kawamura, N.; Kobayashi, H.; Yokota, T.; Iwamoto, C.; Ohuchida, K.; Hashizume, M.; Shimizu, A.; et al. Robust 3D image reconstruction of pancreatic cancer tumors from histopathological images with different stains and its quantitative performance evaluation. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 2047–2055. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yang, B.; Wang, Y.; Tian, J.; Yin, L.; Zheng, W. 2D/3D Multimode Medical Image Registration Based on Normalized Cross-Correlation. Appl. Sci. 2022, 12, 2828. [Google Scholar] [CrossRef]
- Kouw, W.M.; Loog, M. A Review of Domain Adaptation without Target Labels. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 766–785. [Google Scholar] [CrossRef]
- Guan, H.; Liu, M. Domain Adaptation for Medical Image Analysis: A Survey. IEEE Trans. Biomed. Eng. 2022, 69, 1173–1185. [Google Scholar] [CrossRef]
- Sun, Y.; Dai, D.; Xu, S. Rethinking adversarial domain adaptation: Orthogonal decomposition for unsupervised domain adaptation in medical image segmentation. Med. Image Anal. 2022, 82, 102623. [Google Scholar] [CrossRef]
- Xie, Q.; Li, Y.; He, N.; Ning, M.; Ma, K.; Wang, G.; Lian, Y.; Zheng, Y. Unsupervised Domain Adaptation for Medical Image Segmentation by Disentanglement Learning and Self-Training. IEEE Trans. Med. Imaging 2022, 1. [Google Scholar] [CrossRef]
- Ren, J.; Hacihaliloglu, I.; Singer, E.A.; Foran, D.J.; Qi, X. Unsupervised Domain Adaptation for Classification of Histopathology Whole-Slide Images. Front. Bioeng. Biotechnol. 2019, 7, 102. [Google Scholar] [CrossRef]
- Alirezazadeh, P.; Hejrati, B.; Monsef-Esfahani, A.; Fathi, A. Representation learning-based unsupervised domain adaptation for classification of breast cancer histopathology images. Biocybern. Biomed. Eng. 2018, 38, 671–683. [Google Scholar] [CrossRef]
- Liu, X.; Yoo, C.; Xing, F.; Oh, H.; El Fakhri, G.; Kang, J.-W.; Woo, J. Deep Unsupervised Domain Adaptation: A Review of Recent Advances and Perspectives. SIP 2022, 11. [Google Scholar] [CrossRef]
- Ge, Y.; Chen, Z.-M.; Zhang, G.; Heidari, A.A.; Chen, H.; Teng, S. Unsupervised domain adaptation via style adaptation and boundary enhancement for medical semantic segmentation. Neurocomputing 2023, 550, 126469. [Google Scholar] [CrossRef]
- Feng, W.; Ju, L.; Wang, L.; Song, K.; Zhao, X.; Ge, Z. Unsupervised Domain Adaptation for Medical Image Segmentation by Selective Entropy Constraints and Adaptive Semantic Alignment. AAAI 2023, 37, 623–631. [Google Scholar] [CrossRef]
- Garrone, P.; Biondi-Zoccai, G.; Salvetti, I.; Sina, N.; Sheiban, I.; Stella, P.R.; Agostoni, P. Quantitative coronary angiography in the current era: Principles and applications. J. Interv. Cardiol. 2009, 22, 527–536. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, Z.; Zhang, D.; Hau, W.K.; Zhang, H. Progressive Perception Learning for Main Coronary Segmentation in X-Ray Angiography. IEEE Trans. Med. Imaging 2023, 42, 864–879. [Google Scholar] [CrossRef]
- Feezor, R.J.; Caridi, J.; Hawkins, I.; Seeger, J.M. Angiography. Endovascular Surgery; Elsevier: Amsterdam, The Netherlands, 2011; pp. 209–225. ISBN 9781416062080. [Google Scholar]
- Ghekiere, O.; Salgado, R.; Buls, N.; Leiner, T.; Mancini, I.; Vanhoenacker, P.; Dendale, P.; Nchimi, A. Image quality in coronary CT angiography: Challenges and technical solutions. Br. J. Radiol. 2017, 90, 20160567. [Google Scholar] [CrossRef]
- Abdellatif, T.; Brousmiche, K.-L. Formal Verification of Smart Contracts Based on Users and Blockchain Behaviors Models. In Proceedings of the 2018 9th IFIP International Conference on New Technologies, Mobility and Security (NTMS), Paris, France, 26–28 February 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–5, ISBN 978-1-5386-3662-6. [Google Scholar]
- Krichen, M.; Lahami, M.; Al-Haija, Q.A. Formal Methods for the Verification of Smart Contracts: A Review. In Proceedings of the 2022 15th International Conference on Security of Information and Networks (SIN), Sousse, Tunisia, 11–13 November 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–8, ISBN 978-1-6654-5465-0. [Google Scholar]
- Khan, S.N.; Loukil, F.; Ghedira-Guegan, C.; Benkhelifa, E.; Bani-Hani, A. Blockchain smart contracts: Applications, challenges, and future trends. Peer Peer Netw. Appl. 2021, 14, 2901–2925. [Google Scholar] [CrossRef]
- Almakhour, M.; Sliman, L.; Samhat, A.E.; Mellouk, A. Verification of smart contracts: A survey. Pervasive Mob. Comput. 2020, 67, 101227. [Google Scholar] [CrossRef]
- Bao, Y.; Zhu, X.-Y.; Zhang, W.; Shen, W.; Sun, P.; Zhao, Y. On Verification of Smart Contracts via Model Checking. In Theoretical Aspects of Software Engineering; Aït-Ameur, Y., Crăciun, F., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 92–112. ISBN 978-3-031-10362-9. [Google Scholar]
- Bošnački, D.; Wijs, A. Model checking: Recent improvements and applications. Int. J. Softw. Tools Technol. Transf. 2018, 20, 493–497. [Google Scholar] [CrossRef]
- Ellul, J. Towards Configurable and Efficient Runtime Verification of Blockchain Based Smart Contracts at the Virtual Machine Level. In Leveraging Applications of Formal Methods, Verification and Validation: Applications; Margaria, T., Steffen, B., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 131–145. ISBN 978-3-030-61466-9. [Google Scholar]



| Method/Metric | Precision (Equation (2)) | Recall/ Sensitivity (Equation (3)) | Specificity (Equation (6)) | Dice (Equation (4)) | Trained Parameters | Trained Epochs | Dropout Regularization |
|---|---|---|---|---|---|---|---|
| Basic U-Net | 0.9032 (± 0.0144) | 0.8601 (± 0.068) | 0.9877 (± 0.007) | 0.8432 (± 0.0657) | 31,055,622 | 212 | 0.125 |
| U-Net + Attention gates | 0.9053 (± 0.0069) | 0.8621 (± 0.0041) | 0.9864 (± 0.0008) | 0.8524 (± 0.0022) | 31,778,762 | 245 | 0.125 |
| U-Net + Residual links | 0.8961 (± 0.0048) | 0.8435 (± 0.0129) | 0.9879 (± 0.001) | 0.8197 (± 0.0148) | 32,463,174 | 191 | 0.125 |
| U-Net + Recurrent links | 0.8058 (± 0.0481) | 0.8080 (± 0.0113) | 0.9782 (± 0.0038) | 0.7436 (± 0.0415) | 35,631,750 | 108 | 0.25 |
| U-Net + Attention gates + Residual links | 0.9088 (± 0.0061) | 0.8383 (± 0.019) | 0.9869 (± 0.0005) | 0.8247 (± 0.0235) | 33,186,314 | 217 | 0.125 |
| U-Net + Attention gates, Residual links, Recurrent links | 0.7974 (± 0.0074) | 0.8117 (± 0.024) | 0.9787 (± 0.0005) | 0.7432 (± 0.0187) | 37,762,442 | 86 | 0.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glänzer, L.; Masalkhi, H.E.; Roeth, A.A.; Schmitz-Rode, T.; Slabu, I. Vessel Delineation Using U-Net: A Sparse Labeled Deep Learning Approach for Semantic Segmentation of Histological Images. Cancers 2023, 15, 3773. https://doi.org/10.3390/cancers15153773
Glänzer L, Masalkhi HE, Roeth AA, Schmitz-Rode T, Slabu I. Vessel Delineation Using U-Net: A Sparse Labeled Deep Learning Approach for Semantic Segmentation of Histological Images. Cancers. 2023; 15(15):3773. https://doi.org/10.3390/cancers15153773
Chicago/Turabian StyleGlänzer, Lukas, Husam E. Masalkhi, Anjali A. Roeth, Thomas Schmitz-Rode, and Ioana Slabu. 2023. "Vessel Delineation Using U-Net: A Sparse Labeled Deep Learning Approach for Semantic Segmentation of Histological Images" Cancers 15, no. 15: 3773. https://doi.org/10.3390/cancers15153773
APA StyleGlänzer, L., Masalkhi, H. E., Roeth, A. A., Schmitz-Rode, T., & Slabu, I. (2023). Vessel Delineation Using U-Net: A Sparse Labeled Deep Learning Approach for Semantic Segmentation of Histological Images. Cancers, 15(15), 3773. https://doi.org/10.3390/cancers15153773







