Phenotypic and Dermoscopic Patterns of Familial Melanocytic Lesions: A Pilot Study in a Third-Level Center
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Familial Melanoma
1.2. Dermoscopy in Familial Melanoma
1.3. Objectives
2. Materials and Methods
2.1. Study Crireria
2.2. Genomic Analysis
2.3. Patient Examination
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmed, B.; Qadir, M.I.; Ghafoor, S. Malignant Melanoma: Skin Cancer-Diagnosis, Prevention, and Treatment. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Rastrelli, M.; Tropea, S.; Rossi, C.R.; Alaibac, M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 2014, 28, 1005–1011. [Google Scholar] [PubMed]
- Bolick, N.L.; Geller, A.C. Epidemiology of Melanoma. Hematol. Clin. N. Am. 2020, 35, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Karimkhani, C.; Green, A.C.; Nijsten, T.; Weinstock, M.A.; Dellavalle, R.P.; Naghavi, M.; Fitzmaurice, C. The global burden of melanoma: Results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017, 177, 134–140. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Skin Cancer. Available online: https://www.who.int/news-room/questions-and-answers/item/radiation-ultraviolet-(uv)-radiation-and-skin-cancer (accessed on 5 June 2023).
- Linee Guida AIOM Melanoma, Edizione. 2020. Available online: https://www.aiom.it/wp-content/uploads/2020/10/2020_LG_AIOM_Melanoma.pdf (accessed on 5 June 2023).
- Michielin, O.; Van Akkooi, A.C.J.; Ascierto, P.A.; Dummer, R.; Keilholz, U.; ESMO Guidelines Committee. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1884–1901. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase. No. 11; IARC: Lyon, France, 2013. [Google Scholar]
- Teixido, C.; Castillo, P.; Martinez-Vila, C.; Arance, A.; Alos, L. Molecular Markers and Targets in Melanoma. Cells 2021, 10, 2320. [Google Scholar] [CrossRef]
- Nepote, A.; Avallone, G.; Ribero, S.; Cavallo, F.; Roccuzzo, G.; Mastorino, L.; Conforti, C.; Paruzzo, L.; Poletto, S.; Schianca, F.C.; et al. Current Controversies and Challenges on BRAF V600K-Mutant Cutaneous Melanoma. J. Clin. Med. 2022, 11, 828. [Google Scholar] [CrossRef]
- CCavallo, F.; Roccuzzo, G.; Avallone, G.; Conforti, C.; Zalaudek, I.; Giordano, S.; Rubatto, M.; Fava, P.; Ribero, S.; Quaglino, P. Extensive “halo naevi” phenomenon and regression of melanin during nivolumab treatment in metastatic melanoma: A predictor of a better outcome? Dermatol. Ther. 2022, 35, e15559. [Google Scholar] [CrossRef]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Picconi, O.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: I.I. Sun exposure. Eur. J. Cancer 2005, 41, 45–60. [Google Scholar] [CrossRef]
- Amabile, S.; Roccuzzo, G.; Pala, V.; Tonella, L.; Rubatto, M.; Merli, M.; Fava, P.; Ribero, S.; Fierro, M.T.; Queirolo, P.; et al. Clinical Significance of Distant Metastasis-Free Survival (DMFS) in Melanoma: A Narrative Review from Adjuvant Clinical Trials. J. Clin. Med. 2021, 10, 5475. [Google Scholar] [CrossRef]
- Elwood, J.M.; Jopson, J. Melanoma and sun exposure: An overview of published studies. Int. J. Cancer 1997, 73, 198–203. [Google Scholar] [CrossRef]
- Boniol, M.; Autier, P.; Boyle, P.; Gandini, S. Cutaneous melanoma attributable to sunbed use: Systematic review and meta-analysis. BMJ 2012, 345, e4757. [Google Scholar] [CrossRef] [Green Version]
- Green, A.C.; Williams, G.M.; Logan, V.; Strutton, G.M. Reduced Melanoma After Regular Sunscreen Use: Randomized Trial Follow-Up. J. Clin. Oncol. 2011, 29, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Abeni, D.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer 2005, 41, 28–44. [Google Scholar] [CrossRef] [Green Version]
- Abbas, O.; Miller, D.D.; Bhawan, J. Cutaneous malignant melanoma: Update on diagnostic and prognostic biomarkers. Am. J. Dermatopathol. 2014, 36, 363–379. [Google Scholar] [CrossRef]
- Bafounta, M.L.; Beauchet, A.; Aegerter, P.; Saiag, P. Is dermoscopy (epiluminescence microscopy) useful for the diagnosis of melanoma? Results of a meta-analysis using techniques adapted to the evaluation of diagnostic tests. Arch. Dermatol. 2001, 137, 1361–1363. [Google Scholar] [CrossRef] [Green Version]
- Pampena, R.; Kyrgidis, A.; Lallas, A.; Moscarella, E.; Argenziano, G.; Longo, C. A meta-analysis of nevus-associated melanoma: Prevalence and practical implications. J. Am. Acad. Dermatol. 2017, 779, 938–945.e4. [Google Scholar] [CrossRef]
- Ribero, S.; Glass, D.; Bataille, V. Genetic epidemiology of melanoma. Eur. J. Dermatol. 2016, 26, 335–339. [Google Scholar] [CrossRef]
- FFlorell, S.R.; Boucher, K.M.; Garibotti, G.; Astle, J.; Kerber, R.; Mineau, G.; Wiggins, C.; Noyes, R.D.; Tsodikov, A.; Cannon-Albright, L.A.; et al. Population-based analysis of prognostic factors and survival in familial melanoma. J. Clin. Oncol. 2005, 237, 168–177. [Google Scholar] [CrossRef]
- Leachman, S.A.; Carucci, J.; Kohlmann, W.; Banks, K.C.; Asgari, M.M.; Bergman, W.; Bianchi-Scarrà, G.; Brentnall, T.; Paillerets, B.B.-D.; Bruno, W.; et al. Selection criteria for genetic assessment of patients with familial melanoma. J. Am. Acad. Dermatol. 2009, 61, 677.e1–677.e14. [Google Scholar] [CrossRef] [Green Version]
- Tsao, H.; Chin, L.; Garraway, L.A.; Fisher, D.E. Melanoma: From mutations to medicine. Genes Dev. 2012, 26, 1131–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zocchi, L.; Lontano, A.; Merli, M.; Dika, E.; Nagore, E.; Quaglino, P.; Puig, S.; Ribero, S. Familial Melanoma and Susceptibility Genes: A Review of the Most Common Clinical and Dermoscopic Phenotypic Aspect, Associated Malignancies and Practical Tips for Management. J. Clin. Med. 2021, 10, 3760. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Ma, J.; Cai, G.; Fang, S.; Lee, J.E.; Wei, Q.; Amos, C.I. Natural and orthogonal model for estimating gene-gene interactions applied to cutaneous melanoma. Qual. Life Res. 2014, 133, 559–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccioni, L.; Rachakonda, P.S.; Bermejo, J.L.; Planelles, D.; Requena, C.; Hemminki, K.; Nagore, E.; Kumar, R. Variants at the 9p21 locus and melanoma risk. BMC Cancer 2013, 13, 325. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.M.; Struewing, J.P.; Chidambaram, A.; Fraser, M.C.; Tucker, M.A. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J. Natl. Cancer Inst. 2000, 92, 1006–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desnoo, F.; Hayward, N.K. Cutaneous melanoma susceptibility and progression genes. Cancer Lett. 2005, 230, 153–186. [Google Scholar] [CrossRef]
- Harland, M.; E Cust, A.; Badenas, C.; Chang, Y.-M.; A Holland, E.; Aguilera, P.; Aitken, J.F.; Armstrong, B.K.; Barrett, J.H.; Carrera, C.; et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered. Cancer Clin. Pract. 2014, 12, 20. [Google Scholar] [CrossRef]
- Cust, A.E.; Harland, M.; Makalic, E.; Schmidt, D.; Dowty, J.G.; Aitken, J.F.; Agha-Hamilton, C.; Armstrong, B.K.; Barrett, J.H.; Chan, M.; et al. Melanoma risk for CDKN2A mutation carriers who are relatives of population-based case carriers in Australia and the UK. J. Med Genet. 2011, 48, 266–272. [Google Scholar] [CrossRef]
- Bishop, D.T.; Demenais, F.; Goldstein, A.M.; Bergman, W.; Bishop, J.N.; Paillerets, B.B.; Chompret, A.; Ghiorzo, P.; Gruis, N.; Hansson, J.; et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J. Natl. Cancer Inst. 2002, 94, 894–903. [Google Scholar] [CrossRef] [Green Version]
- Tagliabue, E.; Fargnoli, M.C.; Gandini, S.; Maisonneuve, P.; Cornelius, L.a.; Kayser, M.; Nijsten, T.; Han, J.; Kumar, R.; Gruis, N.A.; et al. MC1R gene variants and non-melanoma skin cancer: A pooled-analysis from the M-SKIP project. Br. J. Cancer 2015, 113, 354–363. [Google Scholar] [CrossRef] [Green Version]
- Box, N.F.; Wyeth, J.R.; O’Gorman, L.E.; Martin, N.G.; Sturm, R.A. Characterization of melanocyte stimulating hormone receptor variant alleles in twins with red hair. Hum. Mol. Genet. 1997, 6, 1891–1897. [Google Scholar] [CrossRef] [Green Version]
- Robles-Espinoza, C.D.; Roberts, N.D.; Chen, S.; Leacy, F.P.; Alexandrov, L.B.; Pornputtapong, N.; Halaban, R.; Krauthammer, M.; Cui, R.; Bishop, D.T.; et al. Germline MC1R status influences somatic mutation burden in melanoma. Nat. Commun. 2016, 7, 12064. [Google Scholar] [CrossRef] [Green Version]
- Palmer, J.S.; Duffy, D.; Box, N.; Aitken, J.; O’Gorman, L.E.; Green, A.C.; Hayward, N.K.; Martin, N.; Sturm, R.A. Melanocortin-1 Receptor Polymorphisms and Risk of Melanoma: Is the Association Explained Solely by Pigmentation Phenotype? Am. J. Hum. Genet. 2000, 66, 176–186. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, C.; ter Huurne, J.; Berkhout, M.; Gruis, N.; Bastiaens, M.; Bergman, W.; Willemze, R.; Bavinck, J.N.B. Melanocortin 1 Receptor (MC1R) Gene Variants are Associated with an Increased Risk for Cutaneous Melanoma Which is Largely Independent of Skin Type and Hair Color. J. Investig. Dermatol. 2001, 117, 294–300. [Google Scholar] [CrossRef] [Green Version]
- Matichard, E.; Verpillat, P.; Meziani, R.; Gérard, B.; Descamps, V.; Legroux, E.; Burnouf, M.; Bertrand, G.; Bouscarat, F.; Archimbaud, A.; et al. Melanocortin 1 receptor (MC1R) gene variants may increase the risk of melanoma in France independently of clinical risk factors and UV exposure. J. Med. Genet. 2004, 41, e13. [Google Scholar] [CrossRef]
- Demenais, F.; Mohamdi, H.; Chaudru, V.; Goldstein, A.M.; Bishop, J.A.N.; Bishop, D.T.; Kanetsky, P.A.; Hayward, N.K.; Gillanders, E.; Elder, D.E.; et al. Association of MC1R Variants and Host Phenotypes with Melanoma Risk in CDKN2A Mutation Carriers: A GenoMEL Study. J. Natl. Cancer Inst. 2010, 102, 1568–1583. [Google Scholar] [CrossRef] [Green Version]
- Bertolotto, C.; Lesueur, F.; Giuliano, S.; Strub, T.; De Lichy, M.; Bille, K.; Dessen, P.; d’Hayer, B.; Mohamdi, H.; Remenieras, A.; et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011, 480, 94–98. [Google Scholar] [CrossRef]
- Yokoyama, S.; Woods, S.L.; Boyle, G.M.; Aoude, L.G.; MacGregor, S.; Zismann, V.; Gartside, M.; Cust, A.E.; Haq, R.; Harland, M.; et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011, 480, 99–103. [Google Scholar] [CrossRef] [Green Version]
- Ghiorzo, P.; Pastorino, L.; Queirolo, P.; Bruno, W.; Tibiletti, M.G.; Nasti, S.; Andreotti, V.; Paillerets, B.B.-D.; Scarrà, G.B.; Genoa Pancreatic Cancer Study Group. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: Associations with histological subtypes and family cancer history. Pigment Cell Melanoma Res. 2012, 26, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Njauw, C.-N.J.; Kim, I.; Piris, A.; Gabree, M.; Taylor, M.; Lane, A.M.; DeAngelis, M.M.; Gragoudas, E.; Duncan, L.M.; Tsao, H. Germline BAP1 Inactivation Is Preferentially Associated with Metastatic Ocular Melanoma and Cutaneous-Ocular Melanoma Families. PLoS ONE 2012, 7, e35295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoude, L.G.; Vajdic, C.; Kricker, A.; Armstrong, B.; Hayward, N.K. Prevalence of germline BAP1 mutation in a population-based sample of uveal melanoma cases. Pigment Cell Melanoma Res. 2012, 26, 278–279. [Google Scholar] [CrossRef] [PubMed]
- Psaty, E.L.; Scope, A.; Halpern, A.C.; Marghoob, A.A. Defining the patient at high risk for melanoma. Int. J. Dermatol. 2010, 49, 362–376. [Google Scholar] [CrossRef] [PubMed]
- Goggins, W.B.; Tsao, H. A population-based analysis of risk factors for a second primary cutaneous melanoma among melanoma survivors. Cancer 2003, 97, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary melanoma: Update on syndromes and management Genetics of familial atypical multipleelanoma syndrome. J. Am. Acad. Dermatol. 2016, 74, 411–420. [Google Scholar] [CrossRef] [Green Version]
- Vranic, S.; Serman, N.; Glibo, M.; Serman, L.; Mokos, Z.B. Genetic risk factors in melanoma etiopathogenesis and the role of genetic counseling: A concise review. Bosn. J. Basic Med Sci. 2022, 22, 673–682. [Google Scholar] [CrossRef]
- Marghoob, N.G.; Liopyris, K.; Jaimes, N. Dermoscopy: A Review of the Structures That Facilitate Melanoma Detection. J. Am. Osteopath. Assoc. 2019, 119, 380–390. [Google Scholar] [CrossRef]
- Associazione Italiana di Oncologia Medica, Linee Guida Melanoma. 2020. Available online: https://www.aiom.it/wp-content/uploads/2020/10/2020_LG_AIOM_Melanoma.pdf (accessed on 5 January 2021).
- Puig, S.; Malvehy, J.; Badenas, C.; Ruiz, A.; Jimenez, D.; Cuellar, F.; Azon, A.; Gonzàlez, U.; Castel, T.; Campoy, A.; et al. Role of the CDKN2A Locus in Patients with Multiple Primary Melanomas. J. Clin. Oncol. 2005, 23, 3043–3051. [Google Scholar] [CrossRef]
- Taylor, N.; Handorf, E.A.; Mitra, N.; Avril, M.-F.; Azizi, E.; Bergman, W.; Scarrà, G.B.; Bishop, D.T.; Paillerets, B.B.-D.; Calista, D.; et al. Phenotypic and Histopathological Tumor Characteristics According to CDKN2A Mutation Status among Affected Members of Melanoma Families. J. Investig. Dermatol. 2016, 136, 1066–1069. [Google Scholar] [CrossRef] [Green Version]
- Van der Rhee, J.I.; Krijnen, P.; Gruis, N.A.; de Snoo, F.A.; Vasen, H.F.; Putter, H.; Kukutsch, N.A.; Bergman, W. Clinical and histologic characteristics of malignant melanoma in families with a germline mutation in CDKN2A. J. Am. Acad. Dermatol. 2011, 65, 281–288. [Google Scholar] [CrossRef]
- Florell, S.R.; Meyer, L.J.; Boucher, K.M.; Porter-Gill, P.A.; Hart, M.; Erickson, J.; Cannon-Albright, L.A.; Pershing, L.K.; Harris, R.M.; Samlowski, W.E.; et al. Longitudinal Assessment of the Nevus Phenotype in a Melanoma Kindred. J. Investig. Dermatol. 2004, 123, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Quint, K.D.; van der Rhee, J.I.; Gruis, N.A.; Ter Huurne, J.A.; Wolterbeek, R.; van der Stoep, N.; Bergman, W.; Kukutsch, N.A. Melanocortin 1 receptor (MC1R) variants in high melanoma risk patients are associated with specific dermoscopic ABCD features. Acta Derm. Venereol. 2012, 92, 587–592. [Google Scholar] [CrossRef] [Green Version]
- Sturm, R.A.; Fox, C.; McClenahan, P.; Jagirdar, K.; Ibarrola-Villava, M.; Banan, P.; Abbott, N.C.; Ribas, G.; Gabrielli, B.; Duffy, D.L.; et al. Phenotypic Characterization of Nevus and Tumor Patterns in MITF E318K Mutation Carrier Melanoma Patients. J. Investig. Dermatol. 2014, 134, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Ciccarese, G.; Dalmasso, B.; Bruno, W.; Queirolo, P.; Pastorino, L.; Andreotti, V.; Spagnolo, F.; Tanda, E.; Ponti, G.; Massone, C.; et al. Clinical, pathological and dermoscopic phenotype of MITF p.E318K carrier cutaneous melanoma patients. J. Transl. Med. 2020, 18, 78. [Google Scholar] [CrossRef]
- Carbone, M.; Yang, H.; Pass, H.; Krausz, T.; Testa, J.R.; Gaudino, G. BAP1 and cancer. Nat. Rev. Cancer 2013, 13, 153–159. [Google Scholar] [CrossRef]
- Cavallo, F.; Roccuzzo, G.; Merli, M.; Avallone, G.; Zocchi, L.; Ogliara, P.; Pasini, B.; Quaglino, P.; Ribero, S. BRCA1-associated protein 1 c.368delG mutation leads to the development of multiple BAPomas and cutaneous melanomas: A novel pathogenic variant in BRCA1-associated protein tumor predisposition syndrome. Melanoma Res. 2022, 32, 390–392. [Google Scholar] [CrossRef]
- Haugh, A.M.; Njauw, C.N.; Bubley, J.A.; Verzì, A.E.; Zhang, B.; Kudalkar, E.; VandenBoom, T.; Walton, K.; Swick, B.L.; Kumar, R.; et al. Genotypic and Phenotypic Features of BAP1 Cancer Syndrome: A Report of 8 New Families and Review of Cases in the Literature. JAMA Dermatol. 2017, 153, 999–1006. [Google Scholar] [CrossRef]
- Pellegrini, C.; Maturo, M.G.M.; Martorelli, C.C.; Suppa, M.; Antonini, A.A.; Kostaki, D.D.; Verna, L.; Landi, M.T.M.; Peris, K.; Fargnoli, M.M.C.M. Characterization of melanoma susceptibility genes in high-risk patients from Central Italy. Melanoma Res. 2017, 27, 258–267. [Google Scholar] [CrossRef]
- Bassoli, S.; Maurichi, A.; Rodolfo, M.; Casari, A.; Frigerio, S.; Pupelli, G.; Farnetani, F.; Pelosi, G.; Santinami, M.; Pellacani, G. CDKN2A and MC1R variants influence dermoscopic and confocal features of benign melanocytic lesions in multiple melanoma patients. Exp. Dermatol. 2013, 22, 411–416. [Google Scholar] [CrossRef]
- Vallone, M.G.; Tell-Marti, G.; Potrony, M.; Rebollo-Morell, A.; Badenas, C.; Puig-Butille, J.A.; Gimenez-Xavier, P.; Carrera, C.; Malvehy, J.; Puig, S. Melanocortin 1 receptor (MC1R) polymorphisms’ influence on size and dermoscopic features of nevi. Pigment Cell Melanoma Res. 2017, 31, 39–50. [Google Scholar] [CrossRef]
- Robertson, G.P.; Furnari, F.B.; Miele, M.E.; Glendening, M.J.; Welch, D.R.; Fountain, J.W.; Lugo, T.G.; Huang, H.-J.S.; Cavenee, W.K. In vitro loss of heterozygosity targets the PTEN/MMAC1 gene in melanoma. Proc. Natl. Acad. Sci. USA 1998, 95, 9418–9423. [Google Scholar] [CrossRef]
- Masaki, T.; Wang, Y.; DiGiovanna, J.J.; Khan, S.G.; Raffeld, M.; Beltaifa, S.; Hornyak, T.J.; Darling, T.N.; Lee, C.-C.R.; Kraemer, K.H. High frequency of PTEN mutations in nevi and melanomas from xeroderma pigmentosum patients. Pigment Cell Melanoma Res. 2014, 27, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.; Maiques, O.; Barcelo, C.; Romero, M.; Santacana, M.; Gómez, I.; Cuevas, D.; Velasco, A.; Vea, A.; Macia, A.; et al. Differential Immunoexpression of BRAF/V600E, Senescence Markers, PTEN, and T-type Calcium Channels in Acquired Naevi According to their Histopathological and Dermoscopic Classification. Acta Dermatol. Venereol. 2021, 101, adv00597. [Google Scholar] [CrossRef] [PubMed]
- Ribero, S.; Carrera, C.; Tell-Marti, G.; Pastorino, C.; Badenas, C.; Garcia, A.; Malvehy, J.; Puig, S. Amelanotic melanoma in oculocutaneous albinism: A genetic, dermoscopic and reflectance confocal microscopy study. Br. J. Dermatol. 2017, 177, e333–e335. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, S.; Funasaka, Y.; Otsuka, Y.; Oyama, S.; Ito, M.; Osada, S.; Ueno, T.; Okamura, K.; Hozumi, Y.; Suzuki, T.; et al. Melanotic Malignant Melanoma in Oculocutaneous Albinism Type 4. Acta Dermatol. Venereol. 2017, 97, 287–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conde-Perez, A.; Larue, L. PTEN and melanomagenesis. Futur. Oncol. 2012, 8, 1109–1120. [Google Scholar] [CrossRef]
- Nielsen, K.; Harbst, K.; Måsbäck, A.; Jönsson, G.; Borg, Å.; Olsson, H.; Ingvar, C. Swedish CDKN2A mutation carriers do not present the atypical mole syndrome phenotype. Melanoma Res. 2010, 20, 266–272. [Google Scholar] [CrossRef]
- Rayner, J.E.; Duffy, D.L.; Smit, D.J.; Jagirdar, K.; Lee, K.J.; De’Ambrosis, B.; Smithers, B.M.; McMeniman, E.K.; McInerney-Leo, A.M.; Schaider, H.; et al. Germline and somatic albinism variants in amelanotic/hypomelanotic melanoma: Increased carriage of TYR and OCA2 variants. PLoS ONE 2020, 15, e0238529. [Google Scholar] [CrossRef]
- Bruno, W.; Pastorino, L.; Ghiorzo, P.; Andreotti, V.; Martinuzzi, C.; Menin, C.; Elefanti, L.; Stagni, C.; Vecchiato, A.; Rodolfo, M.; et al. Multiple primary melanomas (MPMs) and criteria for genetic assessment: MultiMEL, a multicenter study of the Italian Melanoma Intergroup. J. Am. Acad. Dermatol. 2016, 74, 325–332. [Google Scholar] [CrossRef]
- Cuéllar, F.; Puig, S.; Kolm, I.; Puig-Butille, J.; Zaballos, P.; Martí-Laborda, R.; Badenas, C.; Malvehy, J. Dermoscopic features of melanomas associated with MC1R variants in Spanish CDKN2A mutation carriers. Br. J. Dermatol. 2009, 160, 448–453. [Google Scholar] [CrossRef]
- Del Chiaro, M.; Verbeke, C.S.; Kartalis, N.; Mucelli, R.P.; Gustafsson, P.; Hansson, J.; Haas, S.L.; Segersvärd, R.; Andren-Sandberg, Å.; Löhr, J.M. Short-term results of a magnetic resonance imaging-based Swedish screening program for individuals at risk for pancreatic cancer. JAMA Surg. 2015, 150, 512–518. [Google Scholar] [CrossRef] [Green Version]
- Ipenburg, N.A.; Gruis, N.A.; Bergman, W.; Van Kester, M.S. The absence of multiple atypical nevi in germline CDKN2A mutations: Comment on “Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome”. J. Am. Acad. Dermatol. 2016, 75, e157. [Google Scholar] [CrossRef]
- Tucker, M.A.; Fraser, M.C.; Goldstein, A.M.; Struewing, J.P.; King, M.A.; Crawford, J.T.; Chiazze, E.A.; Zametkin, D.P.; Fontaine, L.S.; Clark, W.H., Jr. A natural history of melanomas and dysplastic nevi: An atlas of lesions in melanoma-prone families. Cancer 2002, 943, 192–209. [Google Scholar] [CrossRef]
- Harland, M.; Holland, E.A.; Ghiorzo, P.; Mantelli, M.; Bianchi-Scarrà, G.; Goldstein, A.M.; Tucker, M.A.; Ponder, B.A.; Mann, G.J.; Bishop, D.T.; et al. Mutation screening of the CDKN2A promoter in melanoma families. Genes Chromosom. Cancer 2000, 28, 45–57. [Google Scholar] [CrossRef]
- Bassoli, S.; Pellegrini, C.; Longo, C.; Di Nardo, L.; Farnetani, F.; Pellacani, G.; Fargnoli, M. 592 Clinical, dermoscopic and confocal features of nevi and melanomas in a multiple primary melanoma patient with the MITF p.E318K homozygous mutation. J. Investig. Dermatol. 2017, 137, S293. [Google Scholar] [CrossRef] [Green Version]
- Berwick, M.; MacArthur, J.; Orlow, I.; Kanetsky, P.; Begg, C.B.; Luo, L.; Reiner, A.; Sharma, A.; Armstrong, B.K.; Kricker, A.; et al. MITF E318K’s effect on melanoma risk independent of, but modified by, other risk factors. Pigment Cell Melanoma Res. 2015, 27, 485–488. [Google Scholar] [CrossRef] [Green Version]
- Castro-Vega, L.J.; Kiando, S.R.; Burnichon, N.; Buffet, A.; Amar, L.; Simian, C.; Berdelou, A.; Galan, P.; Schlumberger, M.; Bouatia-Naji, N.; et al. The MITF, p.E318K variant, as a risk factor for pheochromocytoma and paraganglioma. J. Clin. Endocrinol. Metab. 2016, 101, 4764–4768. [Google Scholar] [CrossRef] [Green Version]
- Toussi, A.; Mans, N.; Welborn, J.; Kiuru, M. Germline mutations predisposing to melanoma. J. Cutan. Pathol. 2020, 47, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Höiom, V.; Tuominen, R.; Käller, M.; Lindén, D.; Ahmadian, A.; Månsson-Brahme, E.; Egyhazi, S.; Sjöberg, K.; Lundeberg, J.; Hansson, J. MC1R variation and melanoma risk in the Swedish population in relation to clinical and pathological parameters. Pigment Cell Melanoma Res. 2009, 22, 196–204. [Google Scholar] [CrossRef]
- Puig-Butillé, J.; Carrera, C.; Kumar, R.; Garcia-Casado, Z.; Badenas, C.; Aguilera, P.; Malvehy, J.; Nagore, E.; Puig, S. Distribution of MC1R variants among melanoma subtypes: P.R163Q is associated with lentigo maligna melanoma in a Mediterranean population. Br. J. Dermatol. 2013, 169, 804–811. [Google Scholar] [CrossRef]
- Raimondi, S.; Sera, F.; Gandini, S.; Iodice, S.; Caini, S.; Maisonneuve, P.; Fargnoli, M.C. MC1R variants, melanoma and red hair color phenotype: A meta-analysis. Int. J. Cancer 2008, 122, 2753–2760. [Google Scholar] [CrossRef]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrera, C.; Palou, J.; Malvehy, J.; Segura, S.; Aguilera, P.; Salerni, G.; Lovatto, L.; Puig-Butille, J.; Alos, L.; Puig, S. Early stages of melanoma on the limbs of high-risk patients: Clinical, dermoscopic, reflectance confocal microscopy and histopathological characterization for improved recognition. Acta Derm. Venereol. 2011, 91, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, A.N.; Fraga-Braghiroli, N.; Ramos, J.G.R.; Scope, A.; Fernandes, J.D. Dermoscopy of naevi in patients with oculocutaneous albinism. Clin. Exp. Dermatol. 2019, 44, e196–e199. [Google Scholar] [CrossRef] [PubMed]




| Mutation | Frequency [25] | Function [25] | Clinical Features [25,50,51,52,53,54,55,56] | Dermoscopic Features [25,50,51,52,53,54,55,56] |
|---|---|---|---|---|
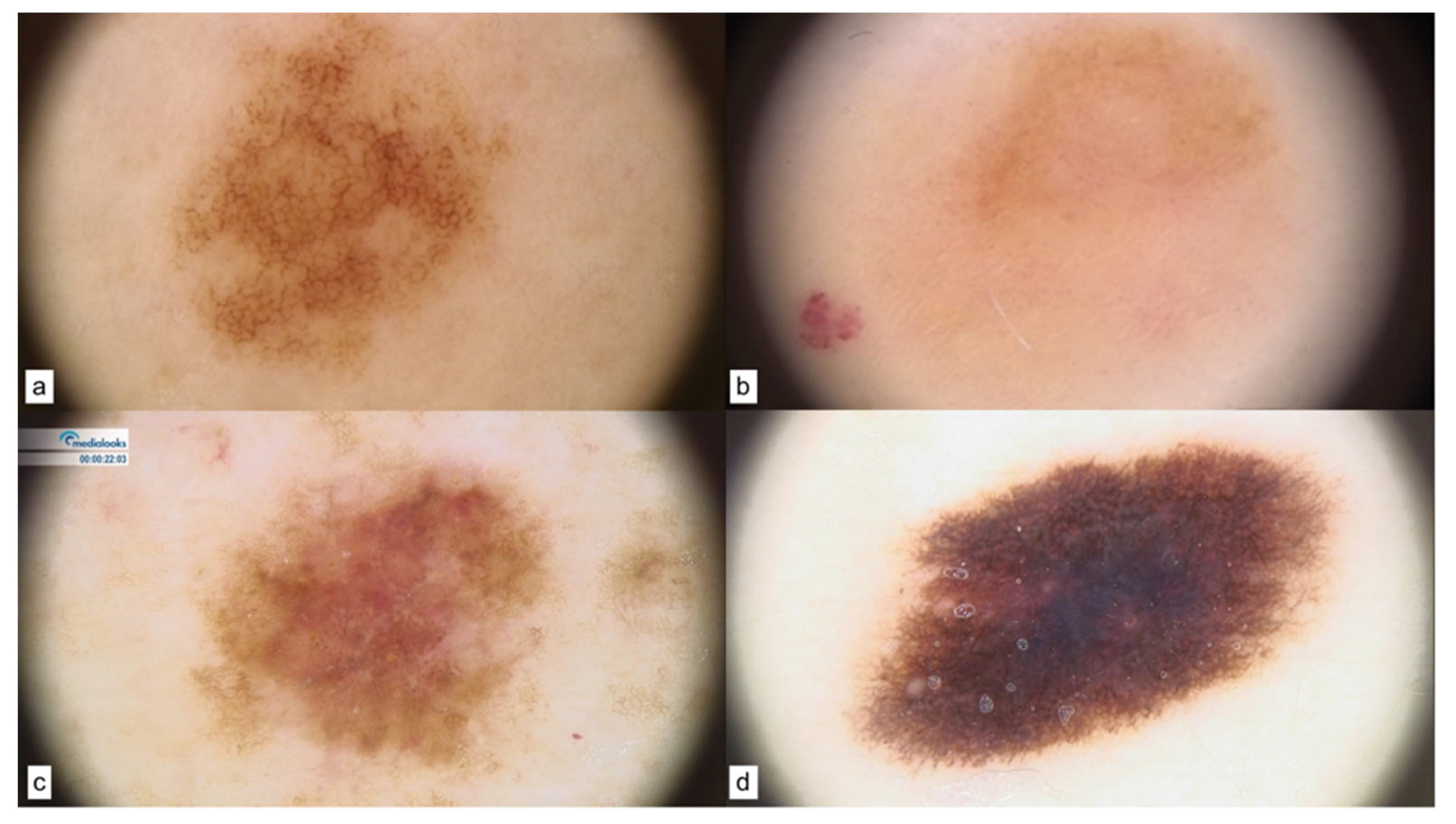
| CDKN2A | 20–40% of FM | Tumor suppressor | -Minor age at melanoma diagnosis [25] -Multiple melanomas [51] -Tendency to SSM in absence of sunburns [52] -Low proportion of acral-nodular melanoma [53] -High TNN, atypical and increasing size nevi [54] | -Unstructured areas (MC1R variants) [55] -Streaks and pigmented network [55] |
| MITF | 1–5% of FM | Regulates melanocyte development | -Minor age at melanoma diagnosis [25] -High TNN [56] -Non-blue eye color [41] -Amelanotic and nodular melanomas (back, leg, arm, abdomen) [42,57] | -Reticular pattern in nevi [56] -Three patterns in dysplastic nevi and melanomas [57]: I (non-specific) II (globular-homogeneous) III (reticular-homogeneous) |
| BAP1 | <1% of FM | Tumor suppressor | -Uveal melanoma at younger age [58] -Multiple BAP1-inactivated melanocytic tumors (BIMTs): nodular features [25,59] | Five dermoscopic patterns [25,60]: I (structureless pink-to-tan with irregular dots/globules located eccentrically) II (structureless pink-to-tan with peripheral vessels) III (structureless pink-to-tan) IV (network with raised, structureless, pink-to-tan areas) V (globular) |
| MC1R | 70–90% of FM | Melanin production | -Red hair color phenotype [25,37] -Melanomas on trunk-arms [61] -Hypopigmented nevi [62] -Larger nevi and melanomas [63] | -Hypopigmentation [62] -Few dermoscopic structures [62] -Vascular pattern [62] |
| PTEN | NA | Tumor suppressor | -Loss of heterozygosity observed in approximately 30% of human melanomas [64] -Higher mutation frequency in pigmented nevi and melanomas of Xeroderma Pigmentosum patients [65] | -Multicomponent pattern associated with lower PTEN expression [66] |
| TYR (OCA1) | NA | Tyrosinase | -NA | -Hypopigmentation, vascular pattern [67] |
| OCA2 | NA | Melanocyte-specific transporter protein | -NA | -Hypopigmentation, amelanotic melanoma [67] |
| SLC45A2 (OCA 4) | NA | Membrane-associated transporter protein | -NA | -Amelanotic melanoma [67] -Melanotic melanoma in case of increased expression of MITF [68] |
| Nucleotide | Protein Change | Mutation | |
|---|---|---|---|
| CDKN2A |
|
|
|
| MITF |
|
|
|
| MC1R |
|
|
|
| BAP1 | c.368delG | p.Ser123Thrfs*64 | nonsense |
| TYR |
|
|
|
| SLC45A |
|
|
|
| OCA2 | c.1327G>A | p.Val443Ile | missense |
| PTEN | c.801G>A | p.Lys267Asn | missense |
| Mutation | N° pts | Sex | Age, Mean (Range) | Previous Melanomas, Mean (Range) | TNN, Mean (Range) | Body Site | Melanoma Type | Nevi Dermoscopy | Melanoma Dermoscopy |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2A | 12 | 7 F 5 M | 43 (36–52) | 6 (2–8) | 21 (154–270) | Dorsum and upper limbs | MIS | -Pigmented reticular pattern -Hypopigmented | -Unstructured areas -Blotches -Atypical network -Pinkish areas -Atypical vascular structures |
| MITF | 3 | 3 F | 71 (62–80) | 7 (5–9) | 260 (130–390) | Upper limbs | 2 MIS SSM 0.7 mm | -Pigmented reticular pattern | Non-specific pattern |
| BAP1 | 1 | F | 34 | 2 melanomas 1 BAPomas 1 AST | 75 | Dorsum | SSM 0.4 mm | -Hypomelanotic and amelanotic pattern | -Structureless pink-to-tan -Atypical vascular structures |
| MC1R | 3 | 2F 1M | 57 (40–72) | 1 | 70 (50–90) | Dorsum | SSM 0.5 mm | -Pigmented reticular pattern | -Atypical, pigmented network |
| PTEN | 1 | M | 56 | 1 | 150 | Dorsum | SSM 0.8 mm | -Pigmented reticular pattern -Hypopigmented | -Atypical, pigmented network |
| TYR (OCA1) | 2 | 2 F | 45 (43–47) | 2 (1–3) | 159 (48–270) | Upper limbs, Trunk | SSM 0.5 mm MIS | -Reticular pattern -Hypopigmented | -Atypical, pigmented network |
| OCA2 | 1 | M | 58 | 3 | 300 | Trunk | MIS | Pigmented reticular pattern | -Atypical pigmented network -Pinkish areas |
| SLC45A2 (OCA 4) | 2 | 1 F 1 M | 25 (26–29) | 1 (1–1) | 221 (172–270) | Trunk | SSM 0.5 mm, SSM 2.3 mm | Globular and reticular pattern | -Atypical pigmented network -Streaks |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roccuzzo, G.; Giordano, S.; Granato, T.; Cavallo, F.; Mastorino, L.; Avallone, G.; Pasini, B.; Quaglino, P.; Ribero, S. Phenotypic and Dermoscopic Patterns of Familial Melanocytic Lesions: A Pilot Study in a Third-Level Center. Cancers 2023, 15, 3772. https://doi.org/10.3390/cancers15153772
Roccuzzo G, Giordano S, Granato T, Cavallo F, Mastorino L, Avallone G, Pasini B, Quaglino P, Ribero S. Phenotypic and Dermoscopic Patterns of Familial Melanocytic Lesions: A Pilot Study in a Third-Level Center. Cancers. 2023; 15(15):3772. https://doi.org/10.3390/cancers15153772
Chicago/Turabian StyleRoccuzzo, Gabriele, Silvia Giordano, Thomas Granato, Francesco Cavallo, Luca Mastorino, Gianluca Avallone, Barbara Pasini, Pietro Quaglino, and Simone Ribero. 2023. "Phenotypic and Dermoscopic Patterns of Familial Melanocytic Lesions: A Pilot Study in a Third-Level Center" Cancers 15, no. 15: 3772. https://doi.org/10.3390/cancers15153772
APA StyleRoccuzzo, G., Giordano, S., Granato, T., Cavallo, F., Mastorino, L., Avallone, G., Pasini, B., Quaglino, P., & Ribero, S. (2023). Phenotypic and Dermoscopic Patterns of Familial Melanocytic Lesions: A Pilot Study in a Third-Level Center. Cancers, 15(15), 3772. https://doi.org/10.3390/cancers15153772









