Expression of TILs and Patterns of Gene Expression from Paired Samples of Malignant Pleural Mesothelioma (MPM) Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Study Outcomes
2.3. Statistical Analysis
2.4. Tumor Tissue
2.5. DNA Sequencing of FFPE Tumor Samples
2.6. RNA Library Preparation from FFPE Tumor Samples
3. Results
3.1. Patient Population
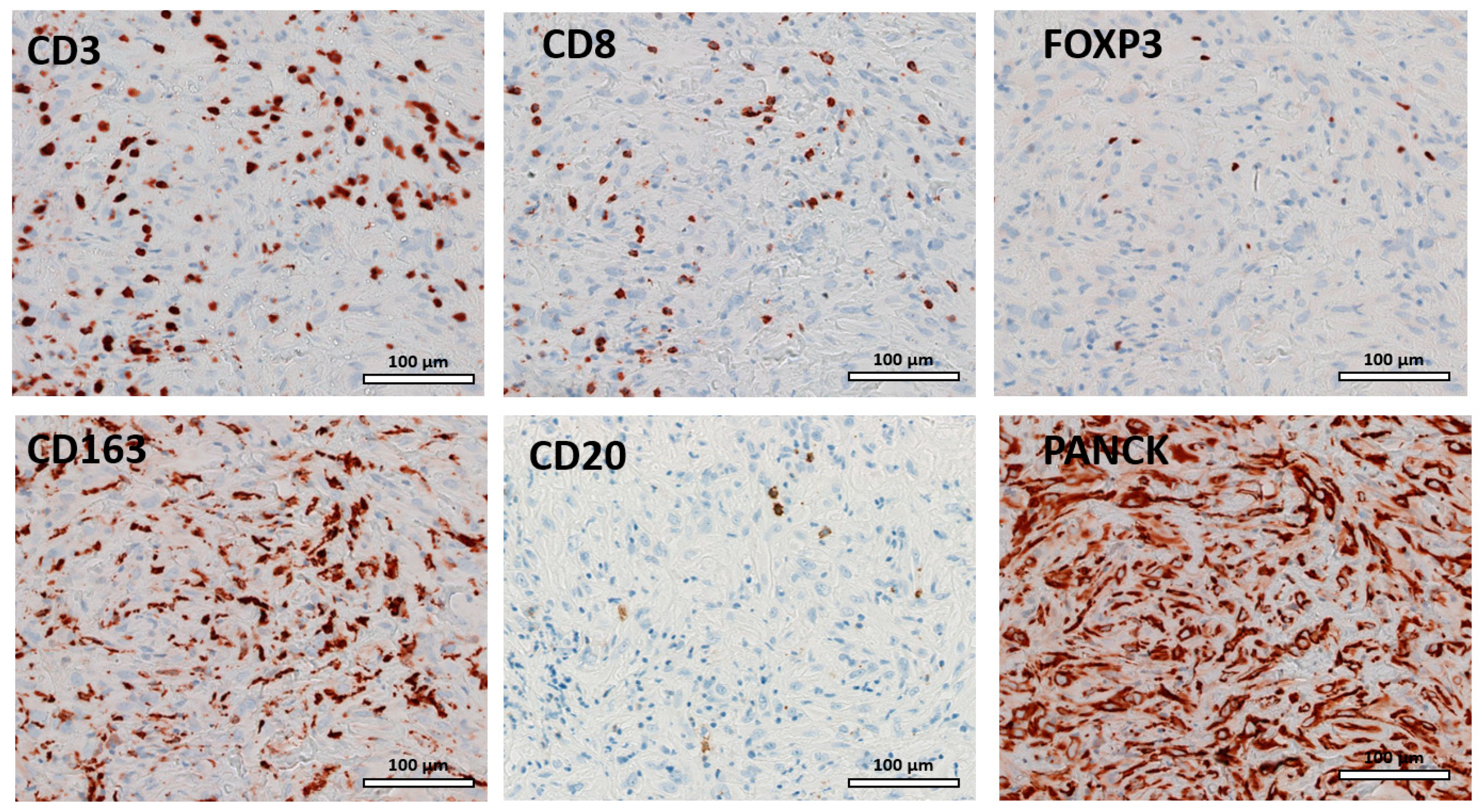
3.2. TIL
3.3. PD-L1
3.4. Survival
3.5. Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janes, S.M.; Alrifai, D.; Fennell, D.A. Perspectives on the Treatment of Malignant Pleural Mesothelioma. N. Engl. J. Med. 2021, 385, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- NCCN Guidelines, Version 1.2023. Available online: http://NCCN.org/guidelines (accessed on 14 March 2023).
- Vogelzang, N.J.; Rusthoven, J.J.; Symanowski, J.; Denham, C.; Kaukel, E.; Ruffie, P.; Gatzemeier, U.; Boyer, M.; Emri, S.; Manegold, C.; et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003, 21, 2636–2644. [Google Scholar] [CrossRef] [PubMed]
- Zalcman, G.; Mazieres, J.; Margery, J.; Greillier, L.; Audigier-Valette, C.; Moro-Sibilot, D.; Molinier, O.; Corre, R.; Monnet, I.; Gounant, V.; et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): A randomised, controlled, open-label, phase 3 trial. Lancet 2016, 387, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; DuBois, R.N. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis 2015, 36, 1085–1093. [Google Scholar] [CrossRef]
- Carbone, M.; Yang, H. Molecular pathways: Targeting mechanisms of asbestos and erionite carcinogenesis in mesothelioma. Clin. Cancer Res. 2012, 18, 598–604. [Google Scholar] [CrossRef]
- Baas, P.; Scherpereel, A.; Nowak, A.K.; Fujimoto, N.; Peters, S.; Tsao, A.S.; Mansfield, A.S.; Popat, S.; Jahan, T.; Antonia, S.; et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): A multicentre, randomised, open-label, phase 3 trial. Lancet 2021, 397, 375–386. [Google Scholar] [CrossRef]
- Peters, S.; Scherpereel, A.; Cornelissen, R.; Oulkhouir, Y.; Greillier, L.; Kaplan, M.A.; Talbot, T.; Monnet, I.; Hiret, S.; Baas, P.; et al. First-line nivolumab plus ipilimumab versus chemotherapy in patients with unresectable malignant pleural mesothelioma: 3-year outcomes from CheckMate 743. Ann. Oncol. 2022, 33, 488–499. [Google Scholar] [CrossRef]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef]
- de Reyniès, A.; Jaurand, M.C.; Renier, A.; Couchy, G.; Hysi, I.; Elarouci, N.; Galateau-Sallé, F.; Copin, M.C.; Hofman, P.; Cazes, A.; et al. Molecular classification of malignant pleural mesothelioma: Identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin. Cancer Res. 2014, 20, 1323–1334. [Google Scholar] [CrossRef]
- Zhang, M.; Luo, J.L.; Sun, Q.; Harber, J.; Dawson, A.G.; Nakas, A.; Busacca, S.; Sharkey, A.J.; Waller, D.; Sheaff, M.T.; et al. Clonal architecture in mesothelioma is prognostic and shapes the tumour microenvironment. Nat. Commun. 2021, 12, 1751. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Aerts, J.G.; Popat, S.; Fennell, D.A. Novel insights into mesothelioma biology and implications for therapy. Nat. Rev. Cancer 2017, 17, 475–488. [Google Scholar] [CrossRef]
- Coussens, L.M.; Zitvogel, L.; Palucka, A.K. Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science 2013, 339, 286–291. [Google Scholar] [CrossRef]
- McGuigan, A.J.; Coleman, H.G.; McCain, R.S.; Kelly, P.J.; Johnston, D.I.; Taylor, M.A.; Turkington, R.C. Immune cell infiltrates as prognostic biomarkers in pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. J. Pathol. Clin. Res. 2021, 7, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Idos, G.E.; Kwok, J.; Bonthala, N.; Kysh, L.; Gruber, S.B.; Qu, C. The Prognostic Implications of Tumor Infiltrating Lymphocytes in Colorectal Cancer: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 3360. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Wang, Z.; Qu, X.; Zhang, Z. Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: A systematic review and meta-analysis. BMC Cancer 2020, 20, 179. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.Q.; Yu, Y.F.; Ou, Q.Y.; Li, X.Y.; Zhong, R.Z.; Xie, C.M.; Hu, Q.G. Prognostic and predictive value of tumor-infiltrating lymphocytes for clinical therapeutic research in patients with non-small cell lung cancer. Oncotarget 2016, 7, 13765–13781. [Google Scholar] [CrossRef]
- Gooden, M.J.; de Bock, G.H.; Leffers, N.; Daemen, T.; Nijman, H.W. The prognostic influence of tumour-infiltrating lymphocytes in cancer: A systematic review with meta-analysis. Br. J. Cancer 2011, 105, 93–103. [Google Scholar] [CrossRef]
- Anraku, M.; Cunningham, K.S.; Yun, Z.; Tsao, M.S.; Zhang, L.; Keshavjee, S.; Johnston, M.R.; de Perrot, M. Impact of tumor-infiltrating T cells on survival in patients with malignant pleural mesothelioma. J. Thorac. Cardiovasc. Surg. 2008, 135, 823–829. [Google Scholar] [CrossRef]
- Yamada, N.; Oizumi, S.; Kikuchi, E.; Shinagawa, N.; Konishi-Sakakibara, J.; Ishimine, A.; Aoe, K.; Gemba, K.; Kishimoto, T.; Torigoe, T.; et al. CD8+ tumor-infiltrating lymphocytes predict favorable prognosis in malignant pleural mesothelioma after resection. Cancer Immunol. Immunother. 2010, 59, 1543–1549. [Google Scholar] [CrossRef]
- Losi, L.; Bertolini, F.; Guaitoli, G.; Fabbiani, L.; Banchelli, F.; Ambrosini-Spaltro, A.; Botticelli, L.; Scurani, L.; Baldessari, C.; Barbieri, F.; et al. Role of evaluating tumor-infiltrating lymphocytes, programmed death-1 ligand 1 and mismatch repair proteins expression in malignant mesothelioma. Int. J. Oncol. 2019, 55, 1157–1164. [Google Scholar] [CrossRef]
- Ujiie, H.; Kadota, K.; Nitadori, J.I.; Aerts, J.G.; Woo, K.M.; Sima, C.S.; Travis, W.D.; Jones, D.R.; Krug, L.M.; Adusumilli, P.S. The tumoral and stromal immune microenvironment in malignant pleural mesothelioma: A comprehensive analysis reveals prognostic immune markers. Oncoimmunology 2015, 4, e1009285. [Google Scholar] [CrossRef] [PubMed]
- Fusco, N.; Vaira, V.; Righi, I.; Sajjadi, E.; Venetis, K.; Lopez, G.; Cattaneo, M.; Castellani, M.; Rosso, L.; Nosotti, M.; et al. Characterization of the immune microenvironment in malignant pleural mesothelioma reveals prognostic subgroups of patients. Lung Cancer 2020, 150, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.S.; Wang, L.; Felizola, S.J.; Ueno, T.; Toi, M.; Loo, W.; Chow, L.W.; Suzuki, T.; Sasano, H. Changes of tumor infiltrating lymphocyte subtypes before and after neoadjuvant endocrine therapy in estrogen receptor-positive breast cancer patients-an immunohistochemical study of Cd8+ and Foxp3+ using double immunostaining with correlation to the pathobiological response of the patients. Int. J. Biol. Markers 2012, 27, e295–e304. [Google Scholar]
- Griguolo, G.; Serna, G.; Pascual, T.; Fasani, R.; Guardia, X.; Chic, N.; Paré, L.; Pernas, S.; Muñoz, M.; Oliveira, M.; et al. Immune microenvironment characterisation and dynamics during anti-HER2-based neoadjuvant treatment in HER2-positive breast cancer. NPJ Precis. Oncol. 2021, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Grabosh, S.; Zeng, F.; Zhang, L.; Strange, M.; Brozick, J.; Edwards, R.P.; Vlad, A. PD-L1 biology in response to chemotherapy in vitro and in vivo in ovarian cancer. J. Immuno. Therapy Cancer 2015, 3 (Suppl. S2), P302. [Google Scholar] [CrossRef]
- Katsuya, Y.; Horinouchi, H.; Asao, T.; Kitahara, S.; Goto, Y.; Kanda, S.; Fujiwara, Y.; Nokihara, H.; Yamamoto, N.; Watanabe, S.; et al. Expression of programmed death 1 (PD-1) and its ligand (PD-L1) in thymic epithelial tumors: Impact on treatment efficacy and alteration in expression after chemotherapy. Lung Cancer 2016, 99, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Gassner, F.J.; Zaborsky, N.; Neureiter, D.; Huemer, M.; Melchardt, T.; Egle, A.; Rebhandl, S.; Catakovic, K.; Hartmann, T.N.; Greil, R.; et al. Chemotherapy-induced augmentation of T cells expressing inhibitory receptors is reversed by treatment with lenalidomide in chronic lymphocytic leukemia. Haematologica 2014, 99, 67–69. [Google Scholar] [CrossRef]
- Peng, J.; Hamanishi, J.; Matsumura, N.; Abiko, K.; Murat, K.; Baba, T.; Yamaguchi, K.; Horikawa, N.; Hosoe, Y.; Murphy, S.K.; et al. Chemotherapy Induces Programmed Cell Death-Ligand 1 Overexpression via the Nuclear Factor-kappaB to Foster an Immunosuppressive Tumor Microenvironment in Ovarian Cancer. Cancer Res. 2015, 75, 5034–5045. [Google Scholar] [CrossRef]
- Pasello, G.; Zago, G.; Lunardi, F.; Urso, L.; Kern, I.; Vlacic, G.; Grosso, F.; Mencoboni, M.; Ceresoli, G.L.; Schiavon, M.; et al. Malignant pleural mesothelioma immune microenvironment and checkpoint expression: Correlation with clinical-pathological features and intratumor heterogeneity over time. Ann. Oncol. 2018, 29, 1258–1265. [Google Scholar] [CrossRef]
- Mark, M.; Rusakiewicz, S.; Früh, M.; Hayoz, S.; Grosso, F.; Pless, M.; Zucali, P.; Ceresoli, G.L.; Maconi, A.; Schneider, M.; et al. Long-term benefit of lurbinectedin as palliative chemotherapy in progressive malignant pleural mesothelioma (MPM): Final efficacy and translational data of the SAKK 17/16 study. ESMO Open 2022, 7, 100446. [Google Scholar] [CrossRef] [PubMed]
- Valpione, S.; Galvani, E.; Tweedy, J.; Mundra, P.A.; Banyard, A.; Middlehurst, P.; Barry, J.; Mills, S.; Salih, Z.; Weightman, J.; et al. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat. Cancer. 2020, 1, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Fairfax, B.P.; Taylor, C.A.; Watson, R.A.; Nassiri, I.; Danielli, S.; Fang, H.; Mahé, E.A.; Cooper, R.; Woodcock, V.; Traill, Z.; et al. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 2020, 26, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, C.; Cai, X.; Xie, Z.; Zhou, L.; Cheng, B.; Zhong, R.; Xiong, S.; Li, J.; Chen, Z.; et al. The association between CD8+ tumor-infiltrating lymphocytes and the clinical outcome of cancer immunotherapy: A systematic review and meta-analysis. EClinicalMedicine 2021, 41, 101134. [Google Scholar] [CrossRef] [PubMed]
- Quispel-Janssen, J.; van der Noort, V.; de Vries, J.F.; Zimmerman, M.; Lalezari, F.; Thunnissen, E.; Monkhorst, K.; Schouten, R.; Schunselaar, L.; Disselhorst, M.; et al. Programmed Death 1 Blockade with Nivolumab in Patients with Recurrent Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2018, 13, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Serna, G.; Simonetti, S.; Fasani, R.; Pagliuca, F.; Guardia, X.; Gallego, P.; Nuciforo, P. Sequential immunohistochemistry and virtual image reconstruction using a single slide for quantitative KI67 measurement in breast cancer. Breast 2020, 53, 102–110. [Google Scholar] [CrossRef]
- Camacho, J.; Rábano, A.; Marazuela, P.; Bonaterra-Pastra, A.; Serna, G.; Moliné, T.; Ramón YCajal, S.; Martínez-Sáez, E.; Hernández-Guillamon, M. Association of CD2AP neuronal deposits with Braak neurofibrillary stage in Alzheimer’s disease. Brain Pathol. 2022, 32, e13016. [Google Scholar] [CrossRef]
- Chee, S.J.; Lopez, M.; Mellows, T.; Gankande, S.; Moutasim, K.A.; Harris, S.; Clarke, J.; Vijayanand, P.; Thomas, G.J.; Ottensmeier, C.H. Evaluating the effect of immune cells on the outcome of patients with mesothelioma. Br. J. Cancer 2017, 117, 1341–1348. [Google Scholar] [CrossRef]
- Xu, X.; Cheng, L.; Fan, Y.; Mao, W. Tumor Microenvironment-Associated Immune-Related Genes for the Prognosis of Malignant Pleural Mesothelioma. Front. Oncol. 2020, 10, 544789. [Google Scholar] [CrossRef]
- Marcq, E.; Siozopoulou, V.; De Waele, J.; van Audenaerde, J.; Zwaenepoel, K.; Santermans, E.; Hens, N.; Pauwels, P.; van Meerbeeck, J.P.; Smits, E.L. Prognostic and predictive aspects of the tumor immune microenvironment and immune checkpoints in malignant pleural mesothelioma. Oncoimmunology 2016, 6, e1261241. [Google Scholar] [CrossRef]
- Dammeijer, F.; De Gooijer, C.J.; van Gulijk, M.; Lukkes, M.; Klaase, L.; Lievense, L.A.; Waasdorp, C.; Jebbink, M.; Bootsma, G.P.; Stigt, J.A.; et al. Immune monitoring in mesothelioma patients identifies novel immune-modulatory functions of gemcitabine associating with clinical response. EBioMedicine 2021, 64, 103160. [Google Scholar] [CrossRef] [PubMed]
- Raghav, K.; Liu, S.; Overman, M.J.; Willett, A.F.; Knafl, M.; Fu, S.C.; Malpica, A.; Prasad, S.; Royal, R.E.; Scally, C.P.; et al. Efficacy, Safety, and Biomarker Analysis of Combined PD-L1 (Atezolizumab) and VEGF (Bevacizumab) Blockade in Advanced Malignant Peritoneal Mesothelioma. Cancer Discov. 2021, 11, 2738–2747. [Google Scholar] [CrossRef]
- Sottile, R.; Tannazi, M.; Johansson, M.H.; Cristiani, C.M.; Calabró, L.; Ventura, V.; Cutaia, O.; Chiarucci, C.; Covre, A.; Garofalo, C.; et al. NK- and T-cell subsets in malignant mesothelioma patients: Baseline pattern and changes in the context of anti-CTLA-4 therapy. Int. J. Cancer 2019, 145, 2238–2248. [Google Scholar] [CrossRef]
- Mankor, J.M.; Disselhorst, M.J.; Poncin, M.; Baas, P.; Aerts, J.G.J.V.; Vroman, H. Efficacy of nivolumab and ipilimumab in patients with malignant pleural mesothelioma is related to a subtype of effector memory cytotoxic T cells: Translational evidence from two clinical trials. EBioMedicine 2020, 62, 103040. [Google Scholar] [CrossRef]
- Ranki, T.; Pesonen, S.; Hemminki, A.; Partanen, K.; Kairemo, K.; Alanko, T.; Lundin, J.; Linder, N.; Turkki, R.; Ristimäki, A.; et al. Phase I study with ONCOS-102 for the treatment of solid tumors-an evaluation of clinical response and exploratory analyses of immune markers. J. Immunother. Cancer 2016, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Patil, N.S.; Righi, L.; Koeppen, H.; Zou, W.; Izzo, S.; Grosso, F.; Libener, R.; Loiacono, M.; Monica, V.; Buttigliero, C.; et al. Molecular and Histopathological Characterization of the Tumor Immune Microenvironment in Advanced Stage of Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2018, 13, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Jang, H.J.; Choi, J.M.; Zhang, J.; de Rosen, V.L.; Wheeler, T.M.; Lee, J.S.; Tu, T.; Jindra, P.T.; Kerman, R.H.; et al. Comprehensive immunoproteogenomic analyses of malignant pleural mesothelioma. JCI Insight 2018, 3, e98575. [Google Scholar] [CrossRef]
- Blum, Y.; Meiller, C.; Quetel, L.; Elarouci, N.; Ayadi, M.; Tashtanbaeva, D.; Armenoult, L.; Montagne, F.; Tranchant, R.; Renier, A.; et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat. Commun. 2019, 10, 1333. [Google Scholar] [CrossRef]
- Quetel, L.; Meiller, C.; Assié, J.B.; Blum, Y.; Imbeaud, S.; Montagne, F.; Tranchant, R.; de Wolf, J.; Caruso, S.; Copin, M.C.; et al. Genetic alterations of malignant pleural mesothelioma: Association with tumor heterogeneity and overall survival. Mol. Oncol. 2020, 14, 1207–1223. [Google Scholar] [CrossRef]
- Meiller, C.; Montagne, F.; Hirsch, T.Z.; Caruso, S.; de Wolf, J.; Bayard, Q.; Assié, J.B.; Meunier, L.; Blum, Y.; Quetel, L.; et al. Multi-site tumor sampling highlights molecular intra-tumor heterogeneity in malignant pleural mesothelioma. Genome. Med. 2021, 13, 113. [Google Scholar] [CrossRef]





| Antibody | Company | Reference | Antigen Retrieval | Primary Ab | Detection Kit | Detection Kit Incubation Time | Chromogen | Counterstaining |
|---|---|---|---|---|---|---|---|---|
| PD-L1 | ROCHE | 741–4905 | Ultra CC1 64 min 100 °C | 16 min 36 °C RTU | OV OptiView DAB | OV HQ Universal linker 8 min OV HRP Multimer 8 min | OV DAB + OV H202 | Hematoxylin II 8 min + bluing reagent 4 min |
| CD20 | ROCHE | 760–2531 | Ultra CC1 36 min 95 °C | 32 min RT RTU | UV UltraView DAB | UV HRP UNIV MULT 8 min | UV DAB + UV DAB H2O2 8 min | Hematoxylin II 4 min + bluing reagent 4 min |
| Antibody | Company | Reference | Antigen Retrieval | Primary Antibody | Secondary Antibody | Chromogen |
|---|---|---|---|---|---|---|
| FOXP3 | ABCAM | AB99963 | CC1 92′ 95° | 1 h | HRP RB 20′ | AECPLUS. 150UL |
| CD8/144B | DAKO | M7103 | CC2 8′ 100° CC1 40′ 95° | 32′ 37° 1/100 | HRP MS 8′ | AECPLUS. 200UL |
| CD3(2GV6) | ROCHE | 790–4341 | CC2 8′ 100° CC1 40′ 95° | 40′ 36° | HRP RB 8′ | AECPLUS. 200UL |
| CD163 | ROCHE | 760–4437 | CC2 8′ 100° CC1 64′ 95° | 48′ 37° | HRP MS 8′ | AECPLUS. 200UL |
| KI67 | ROCHE | 790–4286 | CC2 8′ CC1 64′ 95° | 52′ 37° | HRP RB 8′ | AECPLUS. 200UL |
| PanCK (AE1/AE3) | PALEX | PDM072 | CC2 8′ 100° CC1 40′ 95° | 24′ 36° | HRP MS 8′ | AECPLUS. 200UL |
| Gender | Age | Histology | Clinical Stage | Asbestos Exposure | Time between Biopsies (Months) | Treatment between Biopsies | OS (Months) | |
|---|---|---|---|---|---|---|---|---|
| Patient 1 | F | 53 | Epithelioid | II | Yes | 7 | None | 25 |
| Patient 2 | F | 54 | Epithelioid | II | Yes | 10 | Cisplatin–pemetrexed Anetumab–ravtansine | 21 |
| Patient 3 | M | 71 | Epithelioid | II | Yes | 10 | Cisplatin–pemetrexed Oncolytic virus | 39 |
| Patient 4 | M | 62 | Epithelioid | III | Yes | 10 | Cisplatin–pemetrexed | 23 |
| Patient 5 | M | 60 | Epithelioid | III | Yes | 14 | Cisplatin–pemetrexed Oncolytic virus | 31 |
| Patient 6 | F | 83 | Epithelioid | III | Yes | 8 | None | 17 |
| Patient 7 | M | 84 | Biphasic | III | Yes | 1 | None | 19 |
| Patient 8 | M | 62 | Epithelioid | II | No | 3 | Cisplatin–pemetrexed | 43 |
| Patient 9 | M | 69 | Epithelioid | III | Yes | 33 | Cisplatin–pemetrexed–bevacizumab, pembrolizumab | 34 |
| Patient 10 | M | 59 | Epithelioid | III | Yes | 16 | Cisplatin–pemetrexed, vinorelbine | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cedres, S.; Serna, G.; Gonzalez-Medina, A.; Valdivia, A.; Assaf-Pastrana, J.D.; Iranzo, P.; Callejo, A.; Pardo, N.; Navarro, A.; Martinez-Marti, A.; et al. Expression of TILs and Patterns of Gene Expression from Paired Samples of Malignant Pleural Mesothelioma (MPM) Patients. Cancers 2023, 15, 3611. https://doi.org/10.3390/cancers15143611
Cedres S, Serna G, Gonzalez-Medina A, Valdivia A, Assaf-Pastrana JD, Iranzo P, Callejo A, Pardo N, Navarro A, Martinez-Marti A, et al. Expression of TILs and Patterns of Gene Expression from Paired Samples of Malignant Pleural Mesothelioma (MPM) Patients. Cancers. 2023; 15(14):3611. https://doi.org/10.3390/cancers15143611
Chicago/Turabian StyleCedres, Susana, Garazi Serna, Alberto Gonzalez-Medina, Augusto Valdivia, Juan David Assaf-Pastrana, Patricia Iranzo, Ana Callejo, Nuria Pardo, Alejandro Navarro, Alex Martinez-Marti, and et al. 2023. "Expression of TILs and Patterns of Gene Expression from Paired Samples of Malignant Pleural Mesothelioma (MPM) Patients" Cancers 15, no. 14: 3611. https://doi.org/10.3390/cancers15143611
APA StyleCedres, S., Serna, G., Gonzalez-Medina, A., Valdivia, A., Assaf-Pastrana, J. D., Iranzo, P., Callejo, A., Pardo, N., Navarro, A., Martinez-Marti, A., Priano, I., Fasani, R., Guardia, X., Gonzalo, J., Carbonell, C., Frigola, J., Amat, R., Navarro, V., Dienstmann, R., ... Felip, E. (2023). Expression of TILs and Patterns of Gene Expression from Paired Samples of Malignant Pleural Mesothelioma (MPM) Patients. Cancers, 15(14), 3611. https://doi.org/10.3390/cancers15143611











