p53-Dependent Cytoprotective Mechanisms behind Resistance to Chemo-Radiotherapeutic Agents Used in Cancer Treatment
Abstract
Simple Summary
Abstract
1. Introduction
2. p53
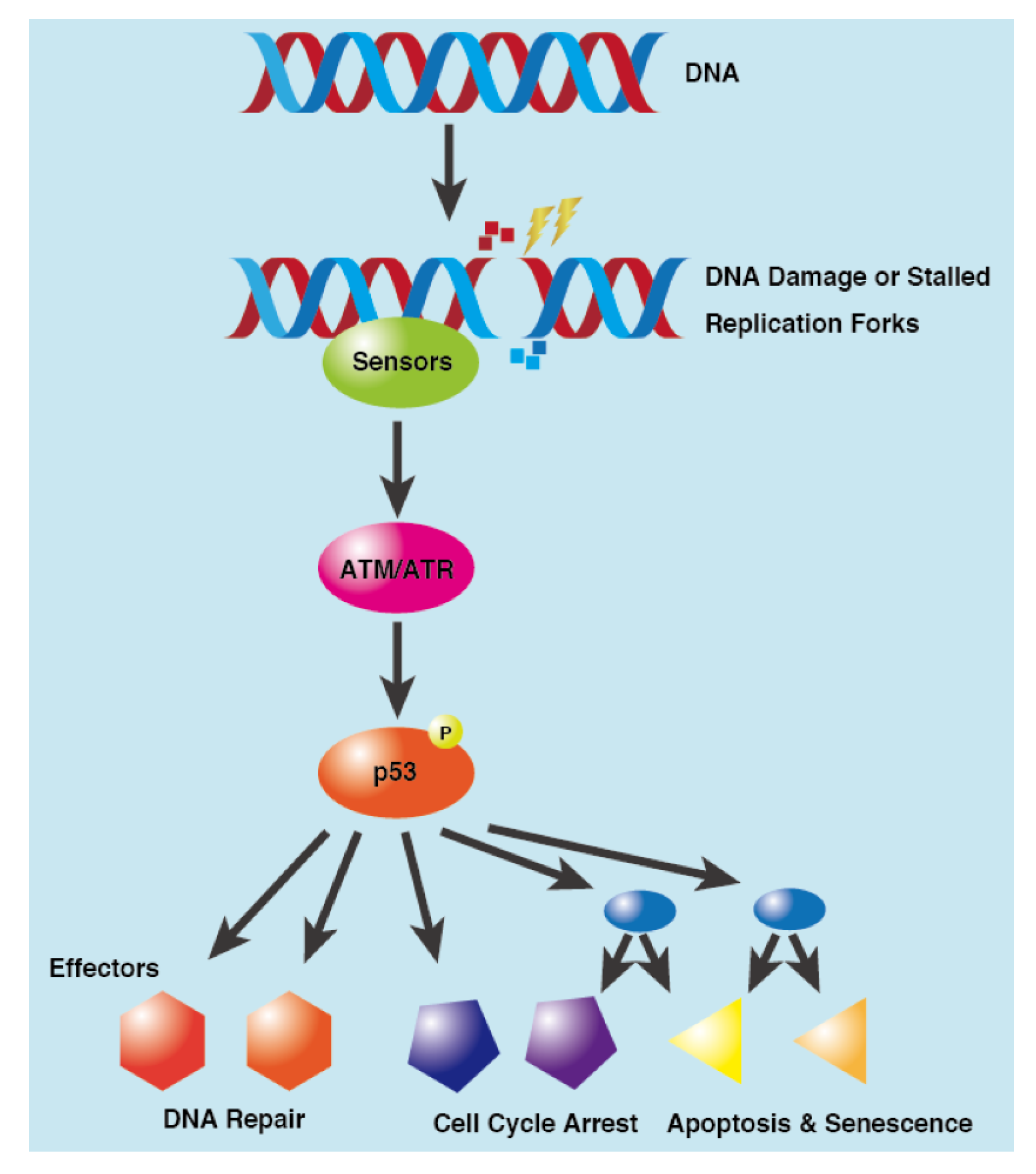
3. DNA Damage Response
4. Resistance to Chemoradiotherapy in Cancer
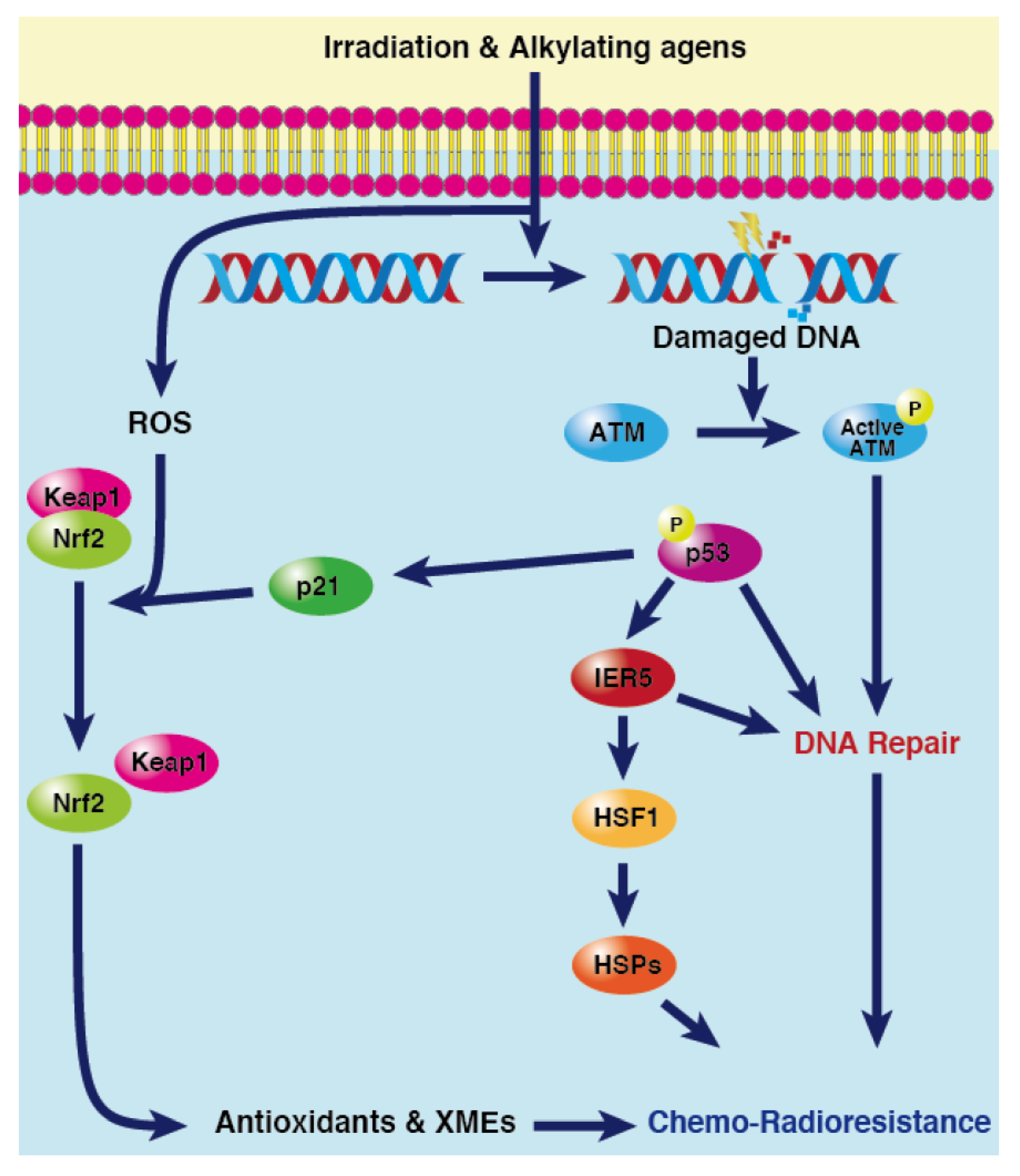
4.1. Role of p53-mediated DNA Repair in MDR
4.2. Role of p53/IER5/HSF1 Pathway in MDR
- IER5 is a Target Gene of p53
- IER5 Protects Normal and Cancer Cells from Stress via p53/IER5/HSF1 Axis
4.3. Role of p53-p21 Pathway in MDR
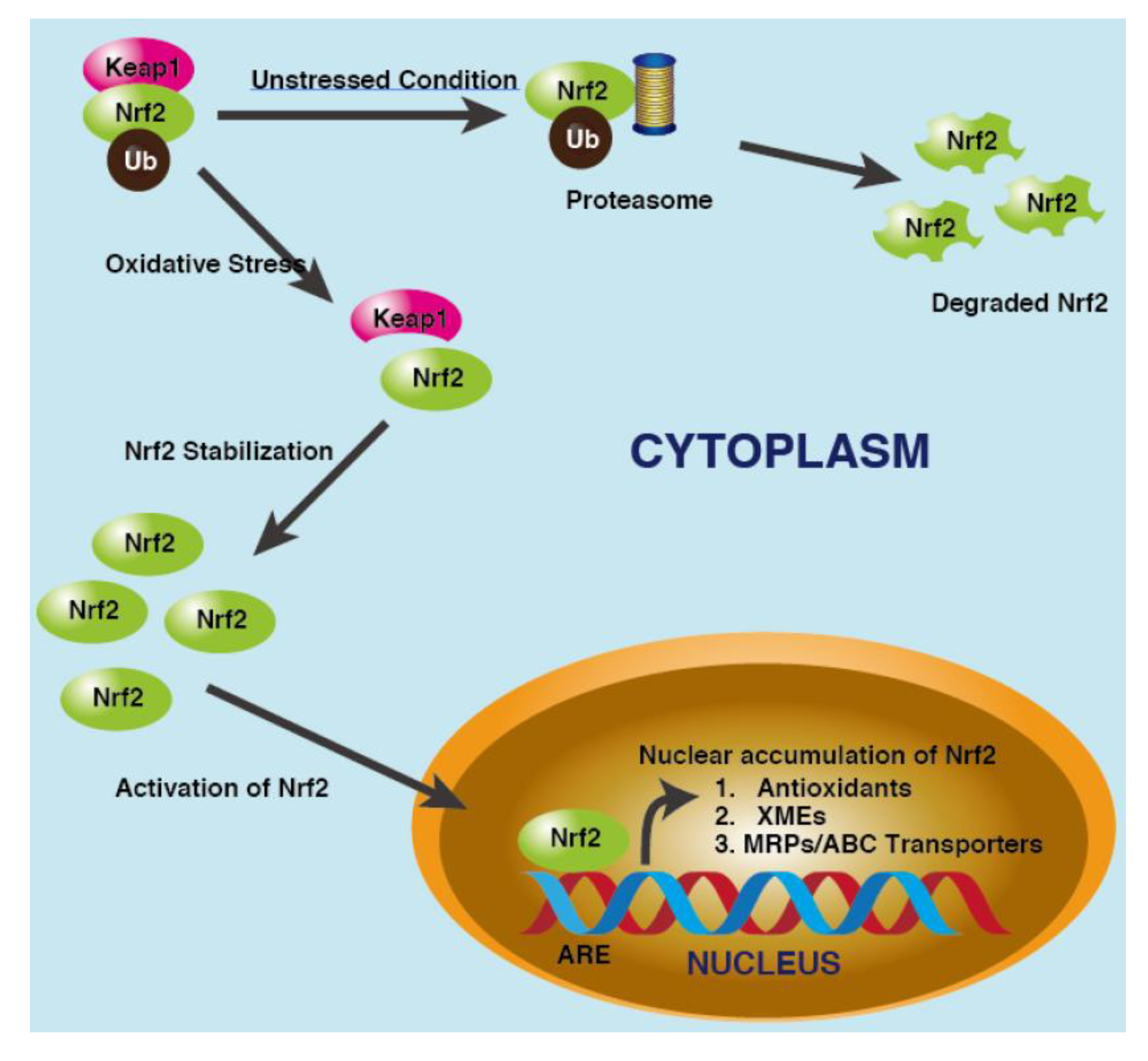
- NRF2
- p53/p21/NRF2 Pathway
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Silva, J.L.; Cino, E.A.; Soares, I.N.; Ferreira, V.F.; De Oliveira, G.A.P. Targeting the Prion-like Aggregation of Mutant p53 to Combat Cancer. Acc. Chem. Res. 2018, 51, 181–190. [Google Scholar] [CrossRef]
- Capodanno, Y.; Chen, Y.; Schrader, J.; Tomosugi, M.; Sumi, S.; Yokoyama, A.; Hiraoka, N.; Ohki, R. Cross-talk among MEN1, p53 and Notch regulates the proliferation of pancreatic neuroendocrine tumour cells by modulating INSM1 expression and subcellular localization. Neoplasia 2021, 23, 979–992. [Google Scholar] [CrossRef]
- Levine, A.J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 2020, 20, 471–480. [Google Scholar] [CrossRef]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Cheok, C.F.; Lane, D.P. Exploiting the p53 Pathway for Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026310. [Google Scholar] [CrossRef]
- Venot, C.; Maratrat, M.; Sierra, V.; Conseiller, E.; Debussche, L. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 1999, 18, 2405–2410. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lu, X. Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2002, 2, 594–604. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, S.; Jiang, J.; Chen, X. Definition of the p53 functional domains necessary for inducing apoptosis. J. Biol. Chem. 2000, 275, 39927–39934. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumourigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef]
- Prives, C. Signaling to p53: Breaking the MDM2-p53 circuit. Cell 1998, 95, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Honda, R.; Tanaka, H.; Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997, 420, 25–27. [Google Scholar] [CrossRef]
- Yin, Y.; Stephen, C.W.; Luciani, M.G.; Fahraeus, R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 2002, 4, 462–467. [Google Scholar] [CrossRef]
- Ohki, R.; Kawase, T.; Ohta, T.; Ichikawa, H.; Taya, Y. Dissecting functional roles of p53 N-terminal transactivation domains by microarray expression analysis. Cancer Sci. 2007, 98, 189–200. [Google Scholar] [CrossRef]
- Suzuki, S.; Tsutsumi, S.; Chen, Y.; Ozeki, C.; Okabe, A.; Kawase, T.; Aburatani, H.; Ohki, R. Identification and characterization of the binding sequences and target genes of p53 lacking the 1st transactivation domain. Cancer Sci. 2020, 111, 451–466. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Saito, S.; Higashimoto, Y.; Roy, S.; Anderson, C.W.; Appella, E. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 2000, 275, 9278–9283. [Google Scholar] [CrossRef]
- Shieh, S.Y.; Taya, Y.; Prives, C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999, 18, 1815–1823. [Google Scholar] [CrossRef]
- Unger, T.; Juven-Gershon, T.; Moallem, E.; Berger, M.; Vogt Sionov, R.; Lozano, G.; Oren, M.; Haupt, Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999, 18, 1805–1814. [Google Scholar] [CrossRef]
- Dumaz, N.; Milne, D.M.; Meek, D.W. Protein kinase CK1 is a p53-threonine 18 kinase which requires prior phosphorylation of serine 15. FEBS Lett. 1999, 463, 312–316. [Google Scholar] [CrossRef]
- Banin, S.; Moyal, L.; Shieh, S.; Taya, Y.; Anderson, C.W.; Chessa, L.; Smorodinsky, N.I.; Prives, C.; Reiss, Y.; Shiloh, Y.; et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998, 281, 1674–1677. [Google Scholar] [CrossRef]
- Shieh, S.Y.; Ahn, J.; Tamai, K.; Taya, Y.; Prives, C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000, 14, 289–300. [Google Scholar] [CrossRef]
- Higashimoto, Y.; Saito, S.; Tong, X.H.; Hong, A.; Sakaguchi, K.; Appella, E.; Anderson, C.W. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J. Biol. Chem. 2000, 275, 23199–23203. [Google Scholar] [CrossRef] [PubMed]
- Soubeyrand, S.; Schild-Poulter, C.; Haché, R.J. Structured DNA promotes phosphorylation of p53 by DNA-dependent protein kinase at serine 9 and threonine 18. Eur. J. Biochem. 2004, 271, 3776–3784. [Google Scholar] [CrossRef] [PubMed]
- Chehab, N.H.; Malikzay, A.; Stavridi, E.S.; Halazonetis, T.D. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA 1999, 96, 13777–13782. [Google Scholar] [CrossRef]
- Matlashewski, G.J.; Tuck, S.; Pim, D.; Lamb, P.; Schneider, J.; Crawford, L.V. Primary structure polymorphism at amino acid residue 72 of human p53. Mol. Cell Biol. 1987, 7, 961–963. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Wu, M.T.; Miller, D.; Wain, J.C.; Kelsey, K.T.; Wiencke, J.K.; Christiani, D.C. The p53 codon 72 polymorphism and lung cancer risk. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1037–1042. [Google Scholar]
- Tommiska, J.; Eerola, H.; Heinonen, M.; Salonen, L.; Kaare, M.; Tallila, J.; Ristimaki, A.; von Smitten, K.; Aittomaki, K.; Heikkila, P.; et al. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin. Cancer Res. 2005, 11, 5098–5103. [Google Scholar] [CrossRef]
- Ozeki, C.; Sawai, Y.; Shibata, T.; Kohno, T.; Okamoto, K.; Yokota, J.; Tashiro, F.; Tanuma, S.; Sakai, R.; Kawase, T.; et al. Cancer susceptibility polymorphism of p53 at codon 72 affects phosphorylation and degradation of p53 protein. J. Biol. Chem. 2011, 286, 18251–18260. [Google Scholar] [CrossRef]
- el-Deiry, W.S.; Kern, S.E.; Pietenpol, J.A.; Kinzler, K.W.; Vogelstein, B. Definition of a consensus binding site for p53. Nat. Genet. 1992, 1, 45–49. [Google Scholar] [CrossRef]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef] [PubMed]
- Oda, E.; Ohki, R.; Murasawa, H.; Nemoto, J.; Shibue, T.; Yamashita, T.; Tokino, T.; Taniguchi, T.; Tanaka, N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000, 288, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Ohki, R.; Nemoto, J.; Murasawa, H.; Oda, E.; Inazawa, J.; Tanaka, N.; Taniguchi, T. Reprimo, a new candidate mediator of the p53-mediated cell cycle arrest at the G2 phase. J. Biol. Chem. 2000, 275, 22627–22630. [Google Scholar] [CrossRef] [PubMed]
- Kawase, T.; Ichikawa, H.; Ohta, T.; Nozaki, N.; Tashiro, F.; Ohki, R.; Taya, Y. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene 2008, 27, 3797–3810. [Google Scholar] [CrossRef] [PubMed]
- Ezawa, I.; Sawai, Y.; Kawase, T.; Okabe, A.; Tsutsumi, S.; Ichikawa, H.; Kobayashi, Y.; Tashiro, F.; Namiki, H.; Kondo, T.; et al. Novel p53 target gene FUCA1 encodes a fucosidase and regulates growth and survival of cancer cells. Cancer Sci. 2016, 107, 734–745. [Google Scholar] [CrossRef]
- Asano, Y.; Kawase, T.; Okabe, A.; Tsutsumi, S.; Ichikawa, H.; Tatebe, S.; Kitabayashi, I.; Tashiro, F.; Namiki, H.; Kondo, T.; et al. IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumourigenesis. Sci. Rep. 2016, 6, 19174. [Google Scholar] [CrossRef]
- Chen, Y.; Takikawa, M.; Tsutsumi, S.; Yamaguchi, Y.; Okabe, A.; Shimada, M.; Kawase, T.; Sada, A.; Ezawa, I.; Takano, Y.; et al. PHLDA1, another PHLDA family protein that inhibits Akt. Cancer Sci. 2018, 109, 3532–3542. [Google Scholar] [CrossRef]
- Kawase, T.; Ohki, R.; Shibata, T.; Tsutsumi, S.; Kamimura, N.; Inazawa, J.; Ohta, T.; Ichikawa, H.; Aburatani, H.; Tashiro, F.; et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 2009, 136, 535–550. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Novak, B.; Sible, J.C.; Tyson, J.J. Checkpoints in the Cell Cycle. In Encyclopedia of Systems Biology; Wiley: Hoboken, NJ, USA, 2003; pp. 254–259. [Google Scholar]
- Finn, K.; Lowndes, N.F.; Grenon, M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell Mol. Life Sci. 2012, 69, 1447–1473. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef]
- Shiloh, Y.; Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197–210. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Chen, J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle 2010, 9, 472–478. [Google Scholar] [CrossRef]
- Hofmann, T.G.; Glas, C.; Bitomsky, N. HIPK2: A tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. Bioessays 2013, 35, 55–64. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumour Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef] [PubMed]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef] [PubMed]
- Haider, T.; Pandey, V.; Banjare, N.; Gupta, P.N.; Soni, V. Drug resistance in cancer: Mechanisms and tackling strategies. Pharmacol. Rep. 2020, 72, 1125–1151. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.L.; Lan, L.; Zou, L. DNA repair defects in cancer and therapeutic opportunities. Genes Dev. 2022, 36, 278–293. [Google Scholar] [CrossRef]
- Janic, A.; Valente, L.J.; Wakefield, M.J.; Di Stefano, L.; Milla, L.; Wilcox, S.; Yang, H.; Tai, L.; Vandenberg, C.J.; Kueh, A.J.; et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 2018, 24, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.B.; Schumacher, B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef]
- Roy, S.; Tomaszowski, K.H.; Luzwick, J.W.; Park, S.; Li, J.; Murphy, M.; Schlacher, K. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. Elife 2018, 7, e31723. [Google Scholar] [CrossRef]
- Wang, Y.H.; Ho, T.L.F.; Hariharan, A.; Goh, H.C.; Wong, Y.L.; Verkaik, N.S.; Lee, M.Y.; Tam, W.L.; van Gent, D.C.; Venkitaraman, A.R.; et al. Rapid recruitment of p53 to DNA damage sites directs DNA repair choice and integrity. Proc. Natl. Acad. Sci. USA 2022, 119, e2113233119. [Google Scholar] [CrossRef]
- Lodovichi, S.; Cervelli, T.; Pellicioli, A.; Galli, A. Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach. Int. J. Mol. Sci. 2020, 21, 6684. [Google Scholar] [CrossRef]
- Oliver, T.G.; Mercer, K.L.; Sayles, L.C.; Burke, J.R.; Mendus, D.; Lovejoy, K.S.; Cheng, M.H.; Subramanian, A.; Mu, D.; Powers, S.; et al. Chronic cisplatin treatment promotes enhanced damage repair and tumour progression in a mouse model of lung cancer. Genes Dev. 2010, 24, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.E.; Milum, K.; Han, C.; Huang, Y.W.; Wani, G.; Thomale, J.; Wani, A.A. Differential contributory roles of nucleotide excision and homologous recombination repair for enhancing cisplatin sensitivity in human ovarian cancer cells. Mol. Cancer 2011, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Stefanski, C.D.; Keffler, K.; McClintock, S.; Milac, L.; Prosperi, J.R. APC loss affects DNA damage repair causing doxorubicin resistance in breast cancer cells. Neoplasia 2019, 21, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef]
- Xiong, Q.; Jiang, X.; Liu, X.; Zhou, P.; Ding, K. Prediction of IER5 structure and function using a bioinformatics approach. Mol. Med. Rep. 2019, 19, 4631–4636. [Google Scholar] [CrossRef]
- Williams, M.; Lyu, M.S.; Yang, Y.L.; Lin, E.P.; Dunbrack, R.; Birren, B.; Cunningham, J.; Hunter, K. Ier5, a novel member of the slow-kinetics immediate-early genes. Genomics 1999, 55, 327–334. [Google Scholar] [CrossRef]
- Yamano, S.; Kimura, M.; Chen, Y.; Imamoto, N.; Ohki, R. Nuclear import of IER5 is mediated by a classical bipartite nuclear localization signal and is required for HSF1 full activation. Exp. Cell Res. 2020, 386, 111686. [Google Scholar] [CrossRef]
- Kawase, T.; Chen, Y.; Ohki, R. IER5 Is a p53-Regulated Activator of HSF1 That Contributes to Promotion of Cancer. In Heat Shock Proteins; Springer: Berlin/Heidelberg, Germany, 2019; Volume 17, p. 20. [Google Scholar]
- Zheng, J.J.; He, Y.; Liu, Y.; Li, F.S.; Cui, Z.; Du, X.M.; Wang, C.P.; Wu, Y.M. Novel role of PAF1 in attenuating radiosensitivity in cervical cancer by inhibiting IER5 transcription. Radiat. Oncol. 2020, 15, 131. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, D.; Zeng, F.; Zhang, Y.; Zhu, G.; Ma, Y.; Song, B.; Lui, S.; Wu, M. High IER5 Gene Expression Is Associated with Poor Prognosis in Glioma Patients. Front. Cell Dev. Biol. 2021, 9, 679684. [Google Scholar] [CrossRef]
- Cirelli, C.; Tononi, G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000, 885, 303–321. [Google Scholar] [CrossRef]
- Li, M.J.; Wang, W.W.; Chen, S.W.; Shen, Q.; Min, R. Radiation dose effect of DNA repair-related gene expression in mouse white blood cells. Med. Sci. Monit. 2011, 17, BR290–BR297. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Hon, C.C.; Sit, W.H.; Chow, K.Y.; Hui, R.K.; Law, I.K.; Ng, V.W.; Yang, X.T.; Leung, F.C.; Wan, J.M. Molecular characterization of Coriolus versicolor PSP-induced apoptosis in human promyelotic leukemic HL-60 cells using cDNA microarray. Int. J. Oncol. 2005, 27, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Kushima, K.; Aoki, Y.; Bialer, M.; Fujiwara, M. Identification of early-responsive genes correlated to valproic acid-induced neural tube defects in mice. Birth Defects Res. A Clin. Mol. Teratol. 2005, 73, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Kis, E.; Szatmari, T.; Keszei, M.; Farkas, R.; Esik, O.; Lumniczky, K.; Falus, A.; Safrany, G. Microarray analysis of radiation response genes in primary human fibroblasts. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 1506–1514. [Google Scholar] [CrossRef]
- Ding, K.K.; Shang, Z.F.; Hao, C.; Xu, Q.Z.; Shen, J.J.; Yang, C.J.; Xie, Y.H.; Qiao, C.; Wang, Y.; Xu, L.L.; et al. Induced expression of the IER5 gene by gamma-ray irradiation and its involvement in cell cycle checkpoint control and survival. Radiat. Environ. Biophys. 2009, 48, 205–213. [Google Scholar] [CrossRef]
- Tavakoli, H.; Manoochehri, M.; Modarres Mosalla, S.M.; Ghafori, M.; Karimi, A.A. Dose-dependent and gender-related radiation-induced transcription alterations of Gadd45a and Ier5 inhuman lymphocytes exposed to gamma ray emitted by (60)Co. Radiat. Prot. Dosim. 2013, 154, 37–44. [Google Scholar] [CrossRef]
- Skorokhod, A.; Bachmann, J.; Giese, N.A.; Martignoni, M.E.; Krakowski-Roosen, H. Real-imaging cDNA-AFLP transcript profiling of pancreatic cancer patients: Egr-1 as a potential key regulator of muscle cachexia. BMC Cancer 2012, 12, 265. [Google Scholar] [CrossRef]
- Wouters, J.; Stas, M.; Govaere, O.; Van den Eynde, K.; Vankelecom, H.; van den Oord, J.J. Gene expression changes in melanoma metastases in response to high-dose chemotherapy during isolated limb perfusion. Pigment Cell Melanoma Res. 2012, 25, 454–465. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Kawabata, S.; Sakurai, H. HSF1 transcriptional activity is modulated by IER5 and PP2A/B55. FEBS Lett. 2015, 589, 1150–1155. [Google Scholar] [CrossRef]
- Toma-Jonik, A.; Vydra, N.; Janus, P.; Widlak, W. Interplay between HSF1 and p53 signaling pathways in cancer initiation and progression: Non-oncogene and oncogene addiction. Cell Oncol. 2019, 42, 579–589. [Google Scholar] [CrossRef]
- Yu, X.P.; Wu, Y.M.; Liu, Y.; Tian, M.; Wang, J.D.; Ding, K.K.; Ma, T.; Zhou, P.K. IER5 is involved in DNA Double-Strand Breaks Repair in Association with PAPR1 in Hela Cells. Int. J. Med. Sci. 2017, 14, 1292–1300. [Google Scholar] [CrossRef]
- Nakamura, S.; Nagata, Y.; Tan, L.; Takemura, T.; Shibata, K.; Fujie, M.; Fujisawa, S.; Tanaka, Y.; Toda, M.; Makita, R.; et al. Transcriptional repression of Cdc25B by IER5 inhibits the proliferation of leukemic progenitor cells through NF-YB and p300 in acute myeloid leukemia. PLoS ONE 2011, 6, e28011. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhao, X.; Xiong, Q.; Jiang, X.; Liu, X.; Ding, K.; Zhou, P. Cdc25B is transcriptionally inhibited by IER5 through the NF-YB transcription factor in irradiation-treated HeLa cells. Toxicol. Res. 2021, 10, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Takeuchi, H.; Sakurai, H. PP2A-B55 and its adapter proteins IER2 and IER5 regulate the activity of RB family proteins and the expression of cell cycle-related genes. FEBS J. 2023, 290, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef]
- Koster, R.; di Pietro, A.; Timmer-Bosscha, H.; Gibcus, J.H.; van den Berg, A.; Suurmeijer, A.J.; Bischoff, R.; Gietema, J.A.; de Jong, S. Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J. Clin. Investig. 2010, 120, 3594–3605. [Google Scholar] [CrossRef]
- Xia, X.; Ma, Q.; Li, X.; Ji, T.; Chen, P.; Xu, H.; Li, K.; Fang, Y.; Weng, D.; Weng, Y.; et al. Cytoplasmic p21 is a potential predictor for cisplatin sensitivity in ovarian cancer. BMC Cancer 2011, 11, 399. [Google Scholar] [CrossRef]
- Johnson, G.G.; Sherrington, P.D.; Carter, A.; Lin, K.; Liloglou, T.; Field, J.K.; Pettitt, A.R. A novel type of p53 pathway dysfunction in chronic lymphocytic leukemia resulting from two interacting single nucleotide polymorphisms within the p21 gene. Cancer Res. 2009, 69, 5210–5217. [Google Scholar] [CrossRef]
- Roman-Gomez, J.; Castillejo, J.A.; Jimenez, A.; Gonzalez, M.G.; Moreno, F.; Rodriguez Mdel, C.; Barrios, M.; Maldonado, J.; Torres, A. 5′ CpG island hypermethylation is associated with transcriptional silencing of the p21(CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia. Blood 2002, 99, 2291–2296. [Google Scholar] [CrossRef]
- Anuranjani; Bala, M. Concerted action of Nrf2-ARE pathway, MRN complex, HMGB1 and inflammatory cytokines—Implication in modification of radiation damage. Redox Biol. 2014, 2, 832–846. [Google Scholar] [CrossRef]
- Ahmadinejad, F.; Geir Moller, S.; Hashemzadeh-Chaleshtori, M.; Bidkhori, G.; Jami, M.S. Molecular Mechanisms behind Free Radical Scavengers Function against Oxidative Stress. Antioxidants 2017, 6, 51. [Google Scholar] [CrossRef]
- No, J.H.; Kim, Y.B.; Song, Y.S. Targeting nrf2 signaling to combat chemoresistance. J. Cancer Prev. 2014, 19, 111–117. [Google Scholar] [CrossRef]
- Nguyen, T.; Huang, H.C.; Pickett, C.B. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 2000, 275, 15466–15473. [Google Scholar] [CrossRef] [PubMed]
- Jyrkkanen, H.K.; Kuosmanen, S.; Heinaniemi, M.; Laitinen, H.; Kansanen, E.; Mella-Aho, E.; Leinonen, H.; Yla-Herttuala, S.; Levonen, A.L. Novel insights into the regulation of antioxidant-response-element-mediated gene expression by electrophiles: Induction of the transcriptional repressor BACH1 by Nrf2. Biochem. J. 2011, 440, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, J.W.; Niture, S.K.; Jaiswal, A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009, 47, 1304–1309. [Google Scholar] [CrossRef]
- Canning, P.; Sorrell, F.J.; Bullock, A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015, 88, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Jeddi, F.; Soozangar, N.; Sadeghi, M.R.; Somi, M.H.; Samadi, N. Contradictory roles of Nrf2/Keap1 signaling pathway in cancer prevention/promotion and chemoresistance. DNA Repair 2017, 54, 13–21. [Google Scholar] [CrossRef]
- Chen, W.; Sun, Z.; Wang, X.J.; Jiang, T.; Huang, Z.; Fang, D.; Zhang, D.D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009, 34, 663–673. [Google Scholar] [CrossRef]
- Villeneuve, N.F.; Sun, Z.; Chen, W.; Zhang, D.D. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle 2009, 8, 3255–3256. [Google Scholar] [CrossRef]
- Dodson, M.; de la Vega, M.R.; Cholanians, A.B.; Schmidlin, C.J.; Chapman, E.; Zhang, D.D. Modulating NRF2 in Disease: Timing Is Everything. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 555–575. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, K.; Thiyagarajan, P.; Rathna Nandhini, J.; Mishra, R.; Nagini, S. Chemopreventive effects of diverse dietary phytochemicals against DMBA-induced hamster buccal pouch carcinogenesis via the induction of Nrf2-mediated cytoprotective antioxidant, detoxification, and DNA repair enzymes. Biochimie 2013, 95, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Lu, Z.N.; Guo, X.L. Regulation and role of nuclear factor-E2-related factor 2 (Nrf2) in multidrug resistance of hepatocellular carcinoma. Chem. Biol. Interact. 2018, 280, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Steed, A.; Co, M.; Chen, X. Cancer stem cells, epithelial-mesenchymal transition, ATP and their roles in drug resistance in cancer. Cancer Drug Resist. 2021, 4, 684–709. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zheng, C.C.; Huang, Y.N.; He, M.L.; Xu, W.W.; Li, B. Molecular mechanisms of chemo- and radiotherapy resistance and the potential implications for cancer treatment. MedComm 2021, 2, 315–340. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Stenvinkel, P.; Meyer, C.J.; Block, G.A.; Chertow, G.M.; Shiels, P.G. Understanding the role of the cytoprotective transcription factor nuclear factor erythroid 2-related factor 2-lessons from evolution, the animal kingdom and rare progeroid syndromes. Nephrol. Dial. Transplant. 2020, 35, 2036–2045. [Google Scholar] [CrossRef]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2020, 11, 629266. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Righetti, S.C.; Della Torre, G.; Pilotti, S.; Ménard, S.; Ottone, F.; Colnaghi, M.I.; Pierotti, M.A.; Lavarino, C.; Cornarotti, M.; Oriana, S.; et al. A comparative study of p53 gene mutations, protein accumulation, and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Cancer Res. 1996, 56, 689–693. [Google Scholar]
- Lavarino, C.; Pilotti, S.; Oggionni, M.; Gatti, L.; Perego, P.; Bresciani, G.; Pierotti, M.A.; Scambia, G.; Ferrandina, G.; Fagotti, A.; et al. p53 gene status and response to platinum/paclitaxel-based chemotherapy in advanced ovarian carcinoma. J. Clin. Oncol. 2000, 18, 3936–3945. [Google Scholar] [CrossRef]
- King, T.C.; Akerley, W.; Fan, A.C.; Moore, T.; Mangray, S.; Hsiu Chen, M.; Safran, H. p53 mutations do not predict response to paclitaxel in metastatic nonsmall cell lung carcinoma. Cancer 2000, 89, 769–773. [Google Scholar] [CrossRef]
- Kandioler-Eckersberger, D.; Ludwig, C.; Rudas, M.; Kappel, S.; Janschek, E.; Wenzel, C.; Schlagbauer-Wadl, H.; Mittlböck, M.; Gnant, M.; Steger, G.; et al. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin. Cancer Res. 2000, 6, 50–56. [Google Scholar] [PubMed]
- Sengeløv, L.; Horn, T.; Steven, K. p53 nuclear immunoreactivity as a predictor of response and outcome following chemotherapy for metastatic bladder cancer. J. Cancer Res. Clin. Oncol. 1997, 123, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Varna, M.; Bousquet, G.; Plassa, L.F.; Bertheau, P.; Janin, A. TP53 status and response to treatment in breast cancers. J. Biomed. Biotechnol. 2011, 2011, 284584. [Google Scholar] [CrossRef] [PubMed]
- Galeaz, C.; Totis, C.; Bisio, A. Radiation Resistance: A Matter of Transcription Factors. Front. Oncol. 2021, 11, 662840. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnaraj, J.; Yamamoto, T.; Ohki, R. p53-Dependent Cytoprotective Mechanisms behind Resistance to Chemo-Radiotherapeutic Agents Used in Cancer Treatment. Cancers 2023, 15, 3399. https://doi.org/10.3390/cancers15133399
Krishnaraj J, Yamamoto T, Ohki R. p53-Dependent Cytoprotective Mechanisms behind Resistance to Chemo-Radiotherapeutic Agents Used in Cancer Treatment. Cancers. 2023; 15(13):3399. https://doi.org/10.3390/cancers15133399
Chicago/Turabian StyleKrishnaraj, Jayaraman, Tatsuki Yamamoto, and Rieko Ohki. 2023. "p53-Dependent Cytoprotective Mechanisms behind Resistance to Chemo-Radiotherapeutic Agents Used in Cancer Treatment" Cancers 15, no. 13: 3399. https://doi.org/10.3390/cancers15133399
APA StyleKrishnaraj, J., Yamamoto, T., & Ohki, R. (2023). p53-Dependent Cytoprotective Mechanisms behind Resistance to Chemo-Radiotherapeutic Agents Used in Cancer Treatment. Cancers, 15(13), 3399. https://doi.org/10.3390/cancers15133399






