Insights into the Role of LncRNAs and miRNAs in Glioma Progression and Their Potential as Novel Therapeutic Targets
Abstract
Simple Summary
Abstract
1. Introduction
2. LncRNAs in Gliomas
2.1. LncRNAs in Adult Gliomas
2.2. LncRNAs in Pediatric Gliomas
2.3. LncRNAs as Biomarkers in Adults and Pediatric Glioma Diagnosis
2.4. LncRNAs in Adults and Pediatric Glioma Therapy
3. Micro RNAs in Gliomas
3.1. Micro RNAs in Adults Gliomas
3.2. Micro RNAs in Pediatric Gliomas
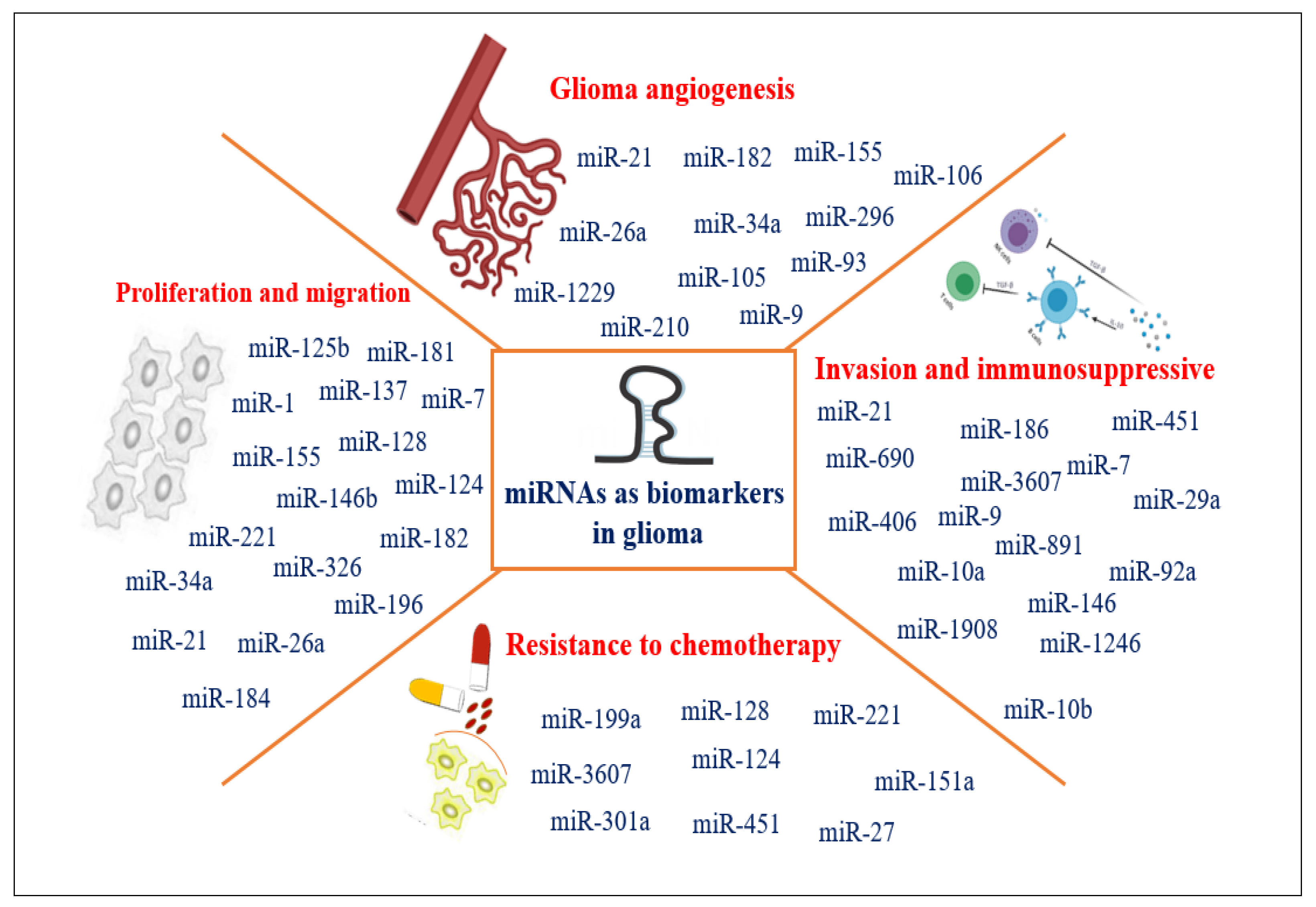
3.3. miRNAs as Biomarkers in Adults and Pediatric Glioma Diagnosis
3.4. Micro RNAs in Adults and Pediatric Glioma Therapy
4. Long and Micro RNAs Interaction
5. Future Directions and Clinical Relevance
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jin, K.-T.; Yao, J.-Y.; Fang, X.-L.; Di, H.; Ma, Y.-Y. Roles of lncRNAs in cancer: Focusing on angiogenesis. Life Sci. 2020, 252, 117647. [Google Scholar] [CrossRef]
- Santos, J.M.; Peixoto da Silva, S.; Gil da Costa, R.M.; Medeiros, R. The emerging role of microRNAs and other non-coding RNAs in cancer cachexia. Cancers 2020, 12, 1004. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Non-Coding RNAs Color. Cancer 2016, 937, 3–17. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Yang, H.; Yang, C.; Bao, Y.-l.; Yang, S.-m.; Liu, J.; Xiao, Y.-f. Translation of noncoding RNAs and cancer. Cancer Lett. 2021, 497, 89–99. [Google Scholar] [CrossRef]
- Alfonso, J.; Talkenberger, K.; Seifert, M.; Klink, B.; Hawkins-Daarud, A.; Swanson, K.; Hatzikirou, H.; Deutsch, A. The biology and mathematical modelling of glioma invasion: A review. J. R. Soc. Interface 2017, 14, 20170490. [Google Scholar] [CrossRef]
- Riemenschneider, M.J.; Jeuken, J.W.; Wesseling, P.; Reifenberger, G. Molecular diagnostics of gliomas: State of the art. Acta Neuropathol. 2010, 120, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, R.; Kurokawa, M.; Baba, A.; Ota, Y.; Pinarbasi, E.; Camelo-Piragua, S.; Capizzano, A.A.; Liao, E.; Srinivasan, A.; Moritani, T. Major Changes in 2021 World Health Organization Classification of Central Nervous System Tumors. RadioGraphics 2022, 42, 1474–1493. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Han, P.; Dahiya, S. Glioblastoma: Changing concepts in the WHO CNS5 classification. Indian J. Pathol. Microbiol. 2022, 65, 24–32. [Google Scholar]
- Reuss, D.E. Updates on the WHO diagnosis of IDH-mutant glioma. J. Neuro-Oncol. 2023, 162, 461–469. [Google Scholar] [CrossRef]
- Von Deimling, A.; Eibl, R.H.; Ohgaki, H.; Louis, D.N.; von Ammon, K.; Petersen, I.; Kleihues, P.; Chung, R.Y.; Wiestler, O.D.; Seizinger, B.R. p53 mutations are associated with 17p allelic loss in grade II and grade III astrocytoma. Cancer Res. 1992, 52, 2987–2990. [Google Scholar]
- Ohgaki, H.; Eibl, R.H.; Reichel, M.B.; Mariani, L.; Petersen, I.; Höll, T.; Wiestler, O.D.; Kleihues, P.; Schwab, M.; Gehring, M. Mutations of the p53 tumor suppressor gene in neoplasms of the human nervous system. Mol. Carcinog. 1993, 8, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma; Exon Publications: Brisbane, QLD, Australia, 2017; pp. 143–153. [Google Scholar]
- Elwood, J.M.; Win, S.S.; Aye, P.S.; Sanagou, M. Trends in brain cancers (glioma) in New Zealand from 1995 to 2020, with reference to mobile phone use. Cancer Epidemiol. 2022, 80, 102234. [Google Scholar] [CrossRef] [PubMed]
- Kreth, F.-W.; Thon, N.; Simon, M.; Westphal, M.; Schackert, G.; Nikkhah, G.; Hentschel, B.; Reifenberger, G.; Pietsch, T.; Weller, M. Gross total but not incomplete resection of glioblastoma prolongs survival in the era of radiochemotherapy. Ann. Oncol. 2013, 24, 3117–3123. [Google Scholar] [CrossRef]
- Chiang, J.C.; Ellison, D.W. Molecular pathology of paediatric central nervous system tumours. J. Pathol. 2017, 241, 159–172. [Google Scholar] [CrossRef]
- Gould, J. Breaking down the epidemiology of brain cancer. Nature 2018, 561, S40. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Suri, V.; Sahu, S.; Sharma, M.C.; Sarkar, C. World Health Organization classification of tumors of the central nervous system 5th Edition (WHO CNS5): What’s new? Indian J. Pathol. Microbiol. 2022, 65, 5. [Google Scholar]
- Machado, G.C.; Ferrer, V.P. MUC17 mutations are associated with poor prognosis in both adult low-grade glioma and glioblastoma patients. medRxiv 2023. [Google Scholar] [CrossRef]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Li, G.; Qin, Z.; Chen, Z.; Xie, L.; Wang, R.; Zhao, H. Tumor microenvironment in treatment of glioma. Open Med. 2017, 12, 247–251. [Google Scholar] [CrossRef]
- Yang, K.; Wu, Z.; Zhang, H.; Zhang, N.; Wu, W.; Wang, Z.; Dai, Z.; Zhang, X.; Zhang, L.; Peng, Y. Glioma targeted therapy: Insight into future of molecular approaches. Mol. Cancer 2022, 21, 39. [Google Scholar] [CrossRef] [PubMed]
- Brandt, B.; Németh, M.; Berta, G.; Szünstein, M.; Heffer, M.; Rauch, T.A.; Pap, M. A Promising Way to Overcome Temozolomide Resistance through Inhibition of Protein Neddylation in Glioblastoma Cell Lines. Int. J. Mol. Sci. 2023, 24, 7929. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liu, J.; Zhang, Y.; You, Q.; Chen, B.; Cheng, J.; Deng, C. Long Non-Coding RNA UBA6-AS1 Promotes the Malignant Properties of Glioblastoma by Competitively Binding to microRNA-760 and Enhancing Homeobox A2 Expression. Cancer Manag. Res. 2021, 13, 379. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Q.; Chen, Z.; Wu, M.; Zhang, C.; Su, J.; Li, Y.; Zhang, C. Hsa_circ_0110757 upregulates ITGA1 to facilitate temozolomide resistance in glioma by suppressing hsa-miR-1298-5p. Cell Death Dis. 2021, 12, 252. [Google Scholar] [CrossRef]
- Cheng, H.; Zhao, H.; Xiao, X.; Huang, Q.; Zeng, W.; Tian, B.; Ma, T.; Lu, D.; Jin, Y.; Li, Y. Long Non-coding RNA MALAT1 Upregulates ZEB2 Expression to Promote Malignant Progression of Glioma by Attenuating miR-124. Mol. Neurobiol. 2021, 58, 1006–1016. [Google Scholar] [CrossRef]
- Roux, A.; Pallud, J.; Saffroy, R.; Edjlali-Goujon, M.; Debily, M.-A.; Boddaert, N.; Sanson, M.; Puget, S.; Knafo, S.; Adam, C. High-grade gliomas in adolescents and young adults highlight histomolecular differences from their adult and pediatric counterparts. Neuro-Oncology 2020, 22, 1190–1202. [Google Scholar] [CrossRef]
- Khan, S.; Mittal, S.; McGee, K.; Alfaro-Munoz, K.D.; Majd, N.; Balasubramaniyan, V.; de Groot, J.F. Role of neutrophils and myeloid-derived suppressor cells in glioma progression and treatment resistance. Int. J. Mol. Sci. 2020, 21, 1954. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Hamilton, R.L. Molecular diagnostics of gliomas. Arch. Pathol. Lab. Med. 2011, 135, 558–568. [Google Scholar] [CrossRef]
- Kim, H.J.; Park, J.W.; Lee, J.H. Genetic architectures and cell-of-origin in glioblastoma. Front. Oncol. 2021, 10, 615400. [Google Scholar] [CrossRef]
- Muñoz-Hidalgo, L.; San-Miguel, T.; Megías, J.; Monleón, D.; Navarro, L.; Roldán, P.; Cerdá-Nicolás, M.; López-Ginés, C. Somatic copy number alterations are associated with EGFR amplification and shortened survival in patients with primary glioblastoma. Neoplasia 2020, 22, 10–21. [Google Scholar] [CrossRef]
- Joseph, G.P.; McDermott, R.; Baryshnikova, M.A.; Cobbs, C.S.; Ulasov, I.V. Cytomegalovirus as an oncomodulatory agent in the progression of glioma. Cancer Lett. 2017, 384, 79–85. [Google Scholar] [CrossRef]
- Bertout, J.A.; Patel, S.A.; Simon, M.C. The impact of O 2 availability on human cancer. Nat. Rev. Cancer 2008, 8, 967–975. [Google Scholar] [CrossRef]
- Patra, K.; Jana, S.; Sarkar, A.; Mandal, D.P.; Bhattacharjee, S. The inhibition of hypoxia-induced angiogenesis and metastasis by cinnamaldehyde is mediated by decreasing HIF-1α protein synthesis via PI3K/Akt pathway. Biofactors 2019, 45, 401–415. [Google Scholar] [CrossRef]
- Dong, L.; Li, W.; Lin, T.; Liu, B.; Hong, Y.; Zhang, X.; Li, X. PSF functions as a repressor of hypoxia-induced angiogenesis by promoting mitochondrial function. Cell Commun. Signal. 2021, 19, 14. [Google Scholar] [CrossRef]
- Wang, J.-C.; Li, G.-Y.; Li, P.-P.; Sun, X.; Li, W.-M.; Li, Y.; Lu, S.-Y.; Liu, P.-J. Suppression of hypoxia-induced excessive angiogenesis by metformin via elevating tumor blood perfusion. Oncotarget 2017, 8, 73892. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- King, J.L.; Benhabbour, S.R. Glioblastoma Multiforme—A Look at the Past and a Glance at the Future. Pharmaceutics 2021, 13, 1053. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Shi, Y.; Liu, J.-B.; Wu, T.-M.; Jia, C.-Y.; Yang, H.-Q.; Zhang, D.-D.; Yang, X.-L.; Wang, H.-M.; Ma, Y.-S. Targeting long non-coding RNA to therapeutically regulate gene expression in cancer. Mol. Ther. Nucleic Acids 2020, 21, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Zhao, M.; Wang, N.; Xu, L.; Wu, T.; Li, Z. LncRNA RGMB-AS1 promotes glioma growth and invasion through miR-1200/HOXB2 axis. OncoTargets Ther. 2019, 12, 10107. [Google Scholar] [CrossRef]
- Li, Q.; Dong, C.; Cui, J.; Wang, Y.; Hong, X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2018, 37, 265. [Google Scholar] [CrossRef] [PubMed]
- Mazurov, E.; Sizykh, A.; Medvedeva, Y.A. HiMoRNA: A Comprehensive Database of Human lncRNAs Involved in Genome-Wide Epigenetic Regulation. Non-Coding RNA 2022, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 2019, 20, 5758. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Khatami, S.H.; Khorsand, M.; Jamali, Z.; Shabaninejad, Z.; Moazamfard, M.; Majidpoor, J.; Zarch, S.M.A.; Movahedpour, A. Long non-coding RNAs (lncRNAs); roles in tumorigenesis and potentials as biomarkers in cancer diagnosis. Exp. Cell Res. 2022, 418, 113294. [Google Scholar] [CrossRef]
- Sur, S.; Ray, R.B. Emerging role of lncRNA ELDR in development and cancer. FEBS J. 2022, 289, 3011–3023. [Google Scholar] [CrossRef]
- Chen, L.; Xie, Y.; Yu, M.; Gou, Q. Long noncoding RNAs in lung cancer: From disease markers to treatment roles. Cancer Manag. Res. 2022, 14, 1771–1782. [Google Scholar] [CrossRef]
- Kumar, A.; Girisa, S.; Alqahtani, M.S.; Abbas, M.; Hegde, M.; Sethi, G.; Kunnumakkara, A.B. Targeting Autophagy Using Long Non-Coding RNAs (LncRNAs): New Landscapes in the Arena of Cancer Therapeutics. Cells 2023, 12, 810. [Google Scholar] [CrossRef]
- Sheng, S.-R.; Wu, J.-S.; Tang, Y.-L.; Liang, X.-H. Long noncoding RNAs: Emerging regulators of tumor angiogenesis. Future Oncol. 2017, 13, 1551–1562. [Google Scholar] [CrossRef]
- Teppan, J.; Barth, D.A.; Prinz, F.; Jonas, K.; Pichler, M.; Klec, C. Involvement of long non-coding RNAs (lncRNAs) in tumor angiogenesis. Non-Coding RNA 2020, 6, 42. [Google Scholar] [CrossRef]
- Song, E. The Long and Short Non-Coding RNAs in Cancer Biology; Springer: Berlin/Heidelberg, Germany, 2016; Volume 927. [Google Scholar]
- Tang, Y.; He, Y.; Zhang, P.; Wang, J.; Fan, C.; Yang, L.; Xiong, F.; Zhang, S.; Gong, Z.; Nie, S. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol. Cancer 2018, 17, 77. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Wang, H.; Chen, E.; Bian, E.; Xu, Y.; Ji, X.; Yang, Z.; Hua, X.; Zhang, Y.; Zhao, B. LncRNA-ATB promotes TGF-β-induced glioma cells invasion through NF-κB and P38/MAPK pathway. J. Cell. Physiol. 2019, 234, 23302–23314. [Google Scholar] [CrossRef] [PubMed]
- Mu, M.; Niu, W.; Zhang, X.; Hu, S.; Niu, C. LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene 2020, 39, 6879–6892. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Shields, C.E.; Haynes, K.A. Beyond the marks: Reader-effectors as drivers of epigenetics and chromatin engineering. Trends Biochem. Sci. 2022, 47, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Tan, Y.; Zhu, Y.; Cui, X.; Wang, Q.; Zhao, J.; Tian, S.; Xu, C.; Xiao, M.; Hong, B. EPIC-0307-mediated selective disruption of PRADX-EZH2 interaction and enhancement of temozolomide sensitivity to glioblastoma via inhibiting DNA repair and MGMT. Neuro-Oncology 2023, noad102. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yang, Z.; Luo, K.; Li, X. Small-molecule natural products for reversing liver fibrosis based on modulation of HSCs activation: An update. Authorea Prepr. 2022. [Google Scholar] [CrossRef]
- Melnyk, T.; Masiá, E.; Zagorodko, O.; Conejos-Sánchez, I.; Vicent, M.J. Rational design of poly-L-glutamic acid-palbociclib conjugates for pediatric glioma treatment. J. Control. Release 2023, 355, 385–394. [Google Scholar] [CrossRef]
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef]
- Gritsch, S.; Batchelor, T.T.; Gonzalez Castro, L.N. Diagnostic, therapeutic, and prognostic implications of the 2021 World Health Organization classification of tumors of the central nervous system. Cancer 2022, 128, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.S.; Smith, O.V.; Prathipati, P.; Gupta, S.C.; Challagundla, K.B. Clinical implications of noncoding RNAs in neuroblastoma patients. In Clinical Applications of Non-Coding RNAs in Cancer; Elsevier: Amsterdam, The Netherlands, 2022; pp. 409–431. [Google Scholar]
- Leung, D. Long non-coding RNA in glioblastoma invasion: Angiogenesis and mesenchymal transition via PI3K and Wnt signalling. Asia Pac. J. Mol. Biol. Biotechnol. 2023, 31, 36–52. [Google Scholar] [CrossRef]
- Roh, J.; Im, M.; Kang, J.; Youn, B.; Kim, W. Long non-coding RNA in glioma: Novel genetic players in temozolomide resistance. Anim. Cells Syst. 2023, 27, 19–28. [Google Scholar] [CrossRef]
- Teng, Y.; Mortazavi, D.; Sohrabi, B.; Mosallaei, M.; Nariman-Saleh-Fam, Z.; Bastami, M.; Mansoori, Y.; Daraei, A.; Vahed, S.Z.; Navid, S. Epi-miRNAs: Regulators of the Histone Modification Machinery in Human Cancer. J. Oncol. 2022, 2022, 4889807. [Google Scholar]
- Yin, X.; Lin, H.; Lin, L.; Miao, L.; He, J.; Zhuo, Z. LncRNAs and CircRNAs in cancer. MedComm 2022, 3, e141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Miao, L.; Lin, H.; Zhuo, Z.; He, J. The role of m6A modification in pediatric cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2022, 1877, 188691. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liao, K.; Zhuang, Z.; Chen, B.; Zhou, Z.; Zhou, S.; Lin, G.; Zhang, F.; Lin, Y.; Miao, Y. AHIF promotes glioblastoma progression and radioresistance via exosomes. Int. J. Oncol. 2019, 54, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhu, J.; Chen, H.; Qian, J.; Zhang, L.; Wan, Z.; Chen, F.; Sun, S.; Li, W.; Luo, C. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int. J. Cancer 2020, 146, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Bai, X.; Liu, C.; Hu, Z. Long noncoding RNA XIST participates hypoxia-induced angiogenesis in human brain microvascular endothelial cells through regulating miR-485/SOX7 axis. Am. J. Transl. Res. 2019, 11, 6487. [Google Scholar] [PubMed]
- Cheng, Z.; Li, Z.; Ma, K.; Li, X.; Tian, N.; Duan, J.; Xiao, X.; Wang, Y. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J. Cancer 2017, 8, 4106. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, Y.; Liu, X.; Qu, C.; Cai, H.; Wang, P.; Li, Z.; Li, Z.; Liu, Y. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol. Rep. 2017, 38, 3265–3277. [Google Scholar] [CrossRef]
- Banerjee, A.; Vest, K.E.; Pavlath, G.K.; Corbett, A.H. Nuclear poly (A) binding protein 1 (PABPN1) and Matrin3 interact in muscle cells and regulate RNA processing. Nucleic Acids Res. 2017, 45, 10706–10725. [Google Scholar] [CrossRef]
- Yang, C.; Zheng, J.; Liu, X.; Xue, Y.; He, Q.; Dong, Y.; Wang, D.; Li, Z.; Liu, L.; Ma, J. Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via staufen1-mediated mRNA decay. Mol. Ther. Nucleic Acids 2020, 20, 866–878. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Yuan, H.-Y.; Ren, X.-Y.; Huang, K.; Guo, Z.-Y. Association between expression of HOTAIR and invasiveness of gliomas, and its predictive value. Adv. Clin. Exp. Med. 2019, 28, 1179–1183. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, X.; Zhou, X.; Han, L.; Chen, L.; Shi, Z.; Zhang, A.; Ye, M.; Wang, Q.; Liu, C. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015, 6, 537. [Google Scholar] [CrossRef]
- Cai, G.; Zhu, Q.; Yuan, L.; Lan, Q. LncRNA SNHG6 acts as a prognostic factor to regulate cell proliferation in glioma through targeting p21. Biomed. Pharmacother. 2018, 102, 452–457. [Google Scholar] [CrossRef]
- Liao, K.; Lin, Y.; Gao, W.; Xiao, Z.; Medina, R.; Dmitriev, P.; Cui, J.; Zhuang, Z.; Zhao, X.; Qiu, Y. Blocking lncRNA MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and progression. Mol. Ther. Nucleic Acids 2019, 18, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, C.; Ding, B.; Gao, M.; Wei, X.; Ji, N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J. Neuro-Oncol. 2017, 134, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, Q.; Guo, Y.; Xiao, Z.; Hu, L.; Xu, Q. LncRNA LINC00689 promotes the growth, metastasis and glycolysis of glioma cells by targeting miR-338-3p/PKM2 axis. Biomed. Pharmacother. 2019, 117, 109069. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zhao, J.; Zhang, Z.-P.; Wu, M.; Li, J.; Xiao, G.-L.; Liu, B.; Liao, Y.-X.; Liu, J.-P. Long non-coding RNA GAS5, by up-regulating PRC2 and targeting the promoter methylation of miR-424, suppresses multiple malignant phenotypes of glioma. J. Neuro-Oncol. 2020, 148, 529–543. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, W.; Zhu, S.; Cheng, K.; Xu, H.; Lv, Y.; Long, X.; Ma, L.; Huang, J.; Sun, S. Long noncoding RNA GAS5 regulates the proliferation, migration, and invasion of glioma cells by negatively regulating miR-18a-5p. J. Cell. Physiol. 2019, 234, 757–768. [Google Scholar] [CrossRef]
- Yu, G.; Li, S.; Liu, P.; Shi, Y.; Liu, Y.; Yang, Z.; Fan, Z.; Zhu, W. LncRNA TUG1 functions as a ceRNA for miR-6321 to promote endothelial progenitor cell migration and differentiation. Exp. Cell Res. 2020, 388, 111839. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, R.; Liu, Q.; Liu, D.; Du, P.; Yuan, J.; Peng, G.; Liao, Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch. Biochem. Biophys. 2016, 610, 1–7. [Google Scholar] [CrossRef]
- Jia, P.; Cai, H.; Liu, X.; Chen, J.; Ma, J.; Wang, P.; Liu, Y.; Zheng, J.; Xue, Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016, 381, 359–369. [Google Scholar] [CrossRef]
- Lang, H.-L.; Hu, G.-W.; Zhang, B.; Kuang, W.; Chen, Y.; Wu, L.; Xu, G.-H. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 2017, 38, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.; Hu, G.; Chen, Y.; Liu, Y.; Tu, W.; Lu, Y.; Wu, L.; Xu, G. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharm. Sci. 2017, 21, 959–972. [Google Scholar]
- Gutschner, T.; Richtig, G.; Haemmerle, M.; Pichler, M. From biomarkers to therapeutic targets—The promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev. 2018, 37, 83–105. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.S.; Meryet-Figuiere, M.; Sabzalipoor, H.; Kashani, H.H.; Nikzad, H.; Asemi, Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol. Cancer 2017, 16, 107. [Google Scholar] [CrossRef]
- Hishii, M.; Matsumoto, T.; Arai, H. Diagnosis and treatment of early-stage glioblastoma. Asian J. Neurosurg. 2019, 14, 589. [Google Scholar] [CrossRef]
- Tykocki, T.; Eltayeb, M. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 2018, 54, 7–13. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21, v1–v100. [Google Scholar] [CrossRef]
- Hutterer, M.; Hattingen, E.; Palm, C.; Proescholdt, M.A.; Hau, P. Current standards and new concepts in MRI and PET response assessment of antiangiogenic therapies in high-grade glioma patients. Neuro-Oncology 2015, 17, 784–800. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, L.; Song, Z.; Guo, C.; Zhu, J.; Li, Z.; Zhu, S. Potential diagnostic and prognostic value of plasma circulating microRNA-182 in human glioma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 855. [Google Scholar] [CrossRef]
- Lin, J.-z.; Lin, N.; Zhao, W.-j. Identification and validation of a six-lncRNA prognostic signature with its ceRNA networks and candidate drugs in lower-grade gliomas. Genomics 2020, 112, 2990–3002. [Google Scholar] [CrossRef]
- Zhuo, H.; Tang, J.; Lin, Z.; Jiang, R.; Zhang, X.; Ji, J.; Wang, P.; Sun, B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol. Carcinog. 2016, 55, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Bin, X.; Hongjian, Y.; Xiping, Z.; Bo, C.; Shifeng, Y.; Binbin, T. Research progresses in roles of LncRNA and its relationships with breast cancer. Cancer Cell Int. 2018, 18, 179. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.; Zambalde, E.; Mathias, C.; Barazetti, J.; Gradia, D.; Oliveira, J. lncRNAs in Hallmarks of Cancer and Clinical Applications. In Non-Coding RNAs; IntechOpen: London, UK, 2019. [Google Scholar]
- Lai, H.; Li, Y.; Zhang, H.; Hu, J.; Liao, J.; Su, Y.; Li, Q.; Chen, B.; Li, C.; Wang, Z. exoRBase 2.0: An atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022, 50, D118–D128. [Google Scholar] [CrossRef] [PubMed]
- Parasramka, M.A.; Maji, S.; Matsuda, A.; Yan, I.K.; Patel, T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol. Ther. 2016, 161, 67–78. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, X.; Qi, L.; Cai, Y.; Yang, P.; Xuan, G.; Jiang, Y. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget 2016, 7, 14429. [Google Scholar] [CrossRef]
- Wang, Z.; Katsaros, D.; Biglia, N.; Shen, Y.; Fu, Y.; Loo, L.W.; Jia, W.; Obata, Y.; Yu, H. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res. Treat. 2018, 171, 261–271. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, C.; Cai, J.; Yang, F.; Liang, T.; Yan, X.; Wang, H.; Wang, W.; Chen, J.; Jiang, T. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget 2017, 8, 53110. [Google Scholar] [CrossRef]
- Wang, W.; Wu, F.; Zhao, Z.; Wang, K.Y.; Huang, R.Y.; Wang, H.Y.; Lan, Q.; Wang, J.F.; Zhao, J.Z. Long noncoding RNA LINC00152 is a potential prognostic biomarker in patients with high-grade glioma. CNS Neurosci. Ther. 2018, 24, 957–966. [Google Scholar] [CrossRef]
- Peng, Z.; Liu, C.; Wu, M. New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer 2018, 17, 61. [Google Scholar] [CrossRef]
- Wang, S.; Wang, H.; Zhao, C.; Li, R. Long noncoding RNA LINC01426 promotes glioma progression through PI3K/AKT signaling pathway and serves as a prognostic biomarker. Eur. Rev. Med. Pharm. Sci. 2018, 22, 6358–6368. [Google Scholar]
- Yang, X.; Wang, C.C.; Lee, W.Y.W.; Trovik, J.; Chung, T.K.H.; Kwong, J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018, 413, 23–34. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Liu, Y.; Zhang, W.; Zhou, J.; Duan, R.; Pu, P.; Kang, C.; Han, L. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016, 373, 251–259. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, Z.; Chai, R.; Liu, Y.; Wang, K.; Wang, Z.; Li, G.; Huang, R.; Jiang, H.; Zhang, K. Expression profile analysis of antisense long non-coding RNA identifies WDFY3-AS2 as a prognostic biomarker in diffuse glioma. Cancer Cell Int. 2018, 18, 107. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, F.; Zhang, L.; Chen, J.; Zhao, Z.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget 2016, 7, 77225. [Google Scholar] [CrossRef] [PubMed]
- Abbastabar, M.; Kheyrollah, M.; Azizian, K.; Bagherlou, N.; Tehrani, S.S.; Maniati, M.; Karimian, A. Multiple functions of p27 in cell cycle, apoptosis, epigenetic modification and transcriptional regulation for the control of cell growth: A double-edged sword protein. DNA Repair 2018, 69, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J. Anim. Sci. Technol. 2018, 60, 25. [Google Scholar] [CrossRef]
- Chen, H.; Li, Q.; Liang, J.; Jin, M.; Lu, A. LncRNA CPS1-IT1 serves as anti-oncogenic role in glioma. Biomed. Pharmacother. 2019, 118, 109277. [Google Scholar] [CrossRef]
- Liu, S.J.; Malatesta, M.; Lien, B.V.; Saha, P.; Thombare, S.S.; Hong, S.J.; Pedraza, L.; Koontz, M.; Seo, K.; Horlbeck, M.A. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 2020, 21, 83. [Google Scholar] [CrossRef]
- Ji, J.; Xu, R.; Ding, K.; Bao, G.; Zhang, X.; Huang, B.; Wang, X.; Martinez, A.; Wang, X.; Li, G. Long noncoding RNA SChLAP1 forms a growth-promoting complex with HNRNPL in human glioblastoma through stabilization of ACTN4 and activation of NF-κB signaling. Clin. Cancer Res. 2019, 25, 6868–6881. [Google Scholar] [CrossRef]
- He, Z.; Wang, Y.; Huang, G.; Wang, Q.; Zhao, D.; Chen, L. The lncRNA UCA1 interacts with miR-182 to modulate glioma proliferation and migration by targeting iASPP. Arch. Biochem. Biophys. 2017, 623, 1–8. [Google Scholar] [CrossRef]
- Pandya, G.; Kirtonia, A.; Sethi, G.; Pandey, A.K.; Garg, M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1874, 188423. [Google Scholar] [CrossRef] [PubMed]
- Kciuk, M.; Yahya, E.B.; Mohamed Ibrahim Mohamed, M.; Rashid, S.; Iqbal, M.O.; Kontek, R.; Abdulsamad, M.A.; Allaq, A.A. Recent Advances in Molecular Mechanisms of Cancer Immunotherapy. Cancers 2023, 15, 2721. [Google Scholar] [CrossRef]
- Ghani, A.R.I.; Yahya, E.B.; Allaq, A.A.; Khalil, A.S.A. Novel insights into genetic approaches in glioblastoma multiforme therapy. Biomed. Res. Ther. 2022, 9, 4851–4864. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, X.; Wu, X.; Xiang, L.; Yuan, Y.; Zhou, S.; Yu, W. LncRNA UCA1 facilitated cell growth and invasion through the miR-206/CLOCK axis in glioma. Cancer Cell Int. 2019, 19, 316. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. LncRNA structural characteristics in epigenetic regulation. Int. J. Mol. Sci. 2017, 18, 2659. [Google Scholar] [CrossRef]
- Luan, W.; Zhou, Z.; Ni, X.; Xia, Y.; Wang, J.; Yan, Y.; Xu, B. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J. Cancer Res. Clin. Oncol. 2018, 144, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, Y.; She, Q.; Li, X.; Peng, L.; Wang, X.; Liu, S.; Shen, X.; Zhang, W.; Dong, Y. Long noncoding RNA H19/miR-675 axis promotes gastric cancer via FADD/Caspase 8/Caspase 3 signaling pathway. Cell. Physiol. Biochem. 2017, 42, 2364–2376. [Google Scholar] [CrossRef]
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452. [Google Scholar] [CrossRef]
- Conigliaro, A.; Costa, V.; Dico, A.L.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef]
- Zhao, W.; Lin, X.; Han, H.; Zhang, H.; Li, X.; Jiang, C.; Feng, M. Long noncoding RNA H19 contributes to the proliferation and autophagy of glioma cells through mTOR/ULK1 pathway. Neuroreport 2021, 32, 352–358. [Google Scholar] [CrossRef]
- Sheng, J.; He, X.; Yu, W.; Chen, Y.; Long, Y.; Wang, K.; Zhu, S.; Liu, Q. p53-targeted lncRNA ST7-AS1 acts as a tumour suppressor by interacting with PTBP1 to suppress the Wnt/β-catenin signalling pathway in glioma. Cancer Lett. 2021, 503, 54–68. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Agabalazadeh, A.; Abak, A.; Shoorei, H.; Hassanzadeh Taheri, M.M.; Taheri, M.; Sharifi, G. Role of long non-coding RNAs in conferring resistance in tumors of the nervous system. Front. Oncol. 2021, 11, 670917. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Li, Z.; Song, S.; Xu, L.; Tong, X.; Yan, H. Silencing of lncRNA LBX2-AS1 suppresses glioma cell proliferation and metastasis through the Akt/GSK3β pathway in vitro. Acta Biochim. Biophys. Sin. 2021, 53, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yao, J.; Geng, P.; Fu, X.; Xue, J.; Zhang, Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014, 7, 3065. [Google Scholar] [PubMed]
- Xu, L.-M.; Chen, L.; Li, F.; Zhang, R.; Li, Z.-y.; Chen, F.-F.; Jiang, X.-D. Over-expression of the long non-coding RNA HOTTIP inhibits glioma cell growth by BRE. J. Exp. Clin. Cancer Res. 2016, 35, 162. [Google Scholar] [CrossRef]
- Li, H.; Yan, R.; Chen, W.; Ding, X.; Liu, J.; Chen, G.; Zhao, Q.; Tang, Y.; Lv, S.; Liu, S. Long non coding RNA SLC26A4-AS1 exerts antiangiogenic effects in human glioma by upregulating NPTX1 via NFKB1 transcriptional factor. FEBS J. 2021, 288, 212–228. [Google Scholar] [CrossRef]
- Bi, C.-L.; Liu, J.-F.; Zhang, M.-Y.; Lan, S.; Yang, Z.-Y.; Fang, J.-S. LncRNA NEAT1 promotes malignant phenotypes and TMZ resistance in glioblastoma stem cells by regulating let-7g-5p/MAP3K1 axis. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Wang, A.; Meng, M.; Zhao, X.; Kong, L. Long non-coding RNA ENST00462717 suppresses the proliferation, survival, and migration by inhibiting MDM2/MAPK pathway in glioma. Biochem. Biophys. Res. Commun. 2017, 485, 513–521. [Google Scholar] [CrossRef]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7, e2123. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; An, G.; Ma, Q. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp. Biol. Med. 2016, 241, 644–649. [Google Scholar] [CrossRef]
- Luo, Q.; Zhang, F.; Li, W.; Wang, F.; Wu, L.; Huang, B. Overexpression of lncRNA MEG3 inhibits proliferation and invasion of glioblastoma U251 cells in vitro by suppressing HIF1α expression. Nan Fang Yi Ke Da Xue Xue Bao = J. South Med. Univ. 2021, 41, 141–145. [Google Scholar]
- Li, J.; Liao, T.; Liu, H.; Yuan, H.; Ouyang, T.; Wang, J.; Chai, S.; Li, J.; Chen, J.; Li, X. Hypoxic Glioma Stem Cell–Derived Exosomes Containing Linc01060 Promote Progression of Glioma by Regulating the MZF1/c-Myc/HIF1α Axis. Cancer Res. 2021, 81, 114–128. [Google Scholar] [CrossRef]
- Ebrahimpour, A.; Sarfi, M.; Rezatabar, S.; Tehrani, S.S. Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma. Mol. Cell. Biochem. 2021, 476, 2317–2335. [Google Scholar] [CrossRef]
- Adams, B.D.; Kasinski, A.L.; Slack, F.J. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 2014, 24, R762–R776. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Avsar, B.; Zhao, Y.; Li, W.; Lukiw, W.J. Atropa belladonna expresses a microRNA (aba-miRNA-9497) highly homologous to Homo sapiens miRNA-378 (hsa-miRNA-378); both miRNAs target the 3′-Untranslated region (3′-UTR) of the mRNA encoding the neurologically relevant, zinc-finger transcription factor ZNF-691. Cell. Mol. Neurobiol. 2020, 40, 179–188. [Google Scholar]
- Rezaei, R.; Baghaei, K.; Amani, D.; Piccin, A.; Hashemi, S.M.; Aghdaei, H.A.; Zali, M.R. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci. 2021, 269, 119035. [Google Scholar] [CrossRef]
- Elshaer, S.S.; Abulsoud, A.I.; Fathi, D.; Abdelmaksoud, N.M.; Zaki, M.B.; El-Mahdy, H.A.; Ismail, A.; Elsakka, E.G.; Abd-Elmawla, M.A.; Abulsoud, L.A. miRNAs role in glioblastoma pathogenesis and targeted therapy: Signaling pathways interplay. Pathol. Res. Pract. 2023, 246, 154511. [Google Scholar] [CrossRef]
- Nurzadeh, M.; Naemi, M.; Sheikh Hasani, S. A comprehensive review on oncogenic miRNAs in breast cancer. J. Genet. 2021, 100, 15. [Google Scholar] [CrossRef]
- Deng, S.-Z.; Lai, M.-F.; Li, Y.-P.; Xu, C.-H.; Zhang, H.-R.; Kuang, J.-G. Human marrow stromal cells secrete microRNA-375-containing exosomes to regulate glioma progression. Cancer Gene Ther. 2020, 27, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Qi, Y.; Ng, S.S.; Chen, X.; Chen, S.; Fang, M.; Li, D.; Zhao, Y.; Ge, R.; Li, G. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem. Biophys. Res. Commun. 2009, 380, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Setlai, B.P.; Hull, R.; Reis, R.M.; Agbor, C.; Ambele, M.A.; Mulaudzi, T.V.; Dlamini, Z. MicroRNA interrelated epithelial mesenchymal transition (EMT) in glioblastoma. Genes 2022, 13, 244. [Google Scholar] [CrossRef]
- Xia, Z.; Liu, F.; Zhang, J.; Liu, L. Decreased expression of MiRNA-204-5p contributes to glioma progression and promotes glioma cell growth, migration and invasion. PLoS ONE 2015, 10, e0132399. [Google Scholar] [CrossRef]
- Lin, J.; Ding, S.; Xie, C.; Yi, R.; Wu, Z.; Luo, J.; Huang, T.; Zeng, Y.; Wang, X.; Xu, A. MicroRNA-4476 promotes glioma progression through a miR-4476/APC/β-catenin/c-Jun positive feedback loop. Cell Death Dis. 2020, 11, 269. [Google Scholar] [CrossRef]
- Reddy, K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Chadda, K.R.; Blakey, E.E.; Coleman, N.; Murray, M.J. The clinical utility of dysregulated microRNA expression in paediatric solid tumours. Eur. J. Cancer 2022, 176, 133–154. [Google Scholar] [CrossRef]
- Dayakar, A.; Shanmukha, K.D.; Kalangi, S.K. Spectrum of microRNAs and their target genes in cancer: Intervention in diagnosis and therapy. Mol. Biol. Rep. 2022, 49, 6827–6846. [Google Scholar] [CrossRef]
- Schulte, J.H.; Schowe, B.; Mestdagh, P.; Kaderali, L.; Kalaghatgi, P.; Schlierf, S.; Vermeulen, J.; Brockmeyer, B.; Pajtler, K.; Thor, T. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int. J. Cancer 2010, 127, 2374–2385. [Google Scholar] [CrossRef]
- Schulte, J.H.; Horn, S.; Otto, T.; Samans, B.; Heukamp, L.C.; Eilers, U.C.; Krause, M.; Astrahantseff, K.; Klein-Hitpass, L.; Buettner, R. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer 2008, 122, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.S.; Johansson, P.; Chen, Q.-R.; Song, Y.K.; Durinck, S.; Wen, X.; Cheuk, A.T.; Smith, M.A.; Houghton, P.; Morton, C. microRNA Profiling Identifies Cancer-Specific and Prognostic Signatures in Pediatric MalignanciesmicroRNA Profiling of Pediatric Tumors. Clin. Cancer Res. 2009, 15, 5560–5568. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Fiori, M.E.; Albini, S.; Cifaldi, L.; Giovinazzi, S.; Forloni, M.; Boldrini, R.; Donfrancesco, A.; Federici, V.; Giacomini, P. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE 2008, 3, e2236. [Google Scholar] [CrossRef]
- De Preter, K.; Mestdagh, P.; Vermeulen, J.; Zeka, F.; Naranjo, A.; Bray, I.; Castel, V.; Chen, C.; Drozynska, E.; Eggert, A. miRNA Expression Profiling Enables Risk Stratification in Archived and Fresh Neuroblastoma Tumor SamplesPrognostic MicroRNA Signature for Neuroblastoma Patients. Clin. Cancer Res. 2011, 17, 7684–7692. [Google Scholar] [CrossRef]
- Balachandran, A.A.; Larcher, L.M.; Chen, S.; Veedu, R.N. Therapeutically significant microRNAs in primary and metastatic brain malignancies. Cancers 2020, 12, 2534. [Google Scholar] [CrossRef]
- Yang, J.-K.; Yang, J.-P.; Tong, J.; Jing, S.-Y.; Fan, B.; Wang, F.; Sun, G.-Z.; Jiao, B.-H. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. Neuro-Oncol. 2017, 131, 255–265. [Google Scholar] [CrossRef]
- Li, Z.; Ye, L.; Wang, L.; Quan, R.; Zhou, Y.; Li, X. Identification of miRNA signatures in serum exosomes as a potential biomarker after radiotherapy treatment in glioma patients. Ann. Diagn. Pathol. 2020, 44, 151436. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Zhu, C.; Ren, C.; Han, J.; Ding, Y.; Du, J.; Dai, N.; Dai, J.; Ma, H.; Hu, Z.; Shen, H. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer 2014, 110, 2291–2299. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Sabahi, A.; Salahandish, R.; Ghaffarinejad, A.; Omidinia, E. Electrochemical nano-genosensor for highly sensitive detection of miR-21 biomarker based on SWCNT-grafted dendritic Au nanostructure for early detection of prostate cancer. Talanta 2020, 209, 120595. [Google Scholar] [CrossRef]
- Hermansen, S.K.; Dahlrot, R.H.; Nielsen, B.S.; Hansen, S.; Kristensen, B.W. MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J. Neuro-Oncol. 2013, 111, 71–81. [Google Scholar] [CrossRef]
- Berthois, Y.; Delfino, C.; Metellus, P.; Fina, F.; Nanni-Metellus, I.; Al Aswy, H.; Pirisi, V.; Ouafik, L.H.; Boudouresque, F. Differential expression of miR200a-3p and miR21 in grade II–III and grade IV gliomas: Evidence that miR200a-3p is regulated by O6-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol. Ther. 2014, 15, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, X.; Wang, H.; Li, Y.; Yan, W.; Han, L.; Zhang, K.; Zhang, J.; Wang, Y.; Feng, Y. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur. J. Cancer 2012, 48, 3104–3111. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhao, W.; Zhao, H.; Zhang, J. Diagnostic and prognostic value of microRNA-193b in patients with glioma and its effect on tumor progression. Oncol. Lett. 2019, 18, 4882–4890. [Google Scholar] [CrossRef]
- Ma, C.; Nguyen, H.P.; Luwor, R.B.; Stylli, S.S.; Gogos, A.; Paradiso, L.; Kaye, A.H.; Morokoff, A.P. A comprehensive meta-analysis of circulation miRNAs in glioma as potential diagnostic biomarker. PLoS ONE 2018, 13, e0189452. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Shi, C.-J.; Lu, J.-X.; Wang, Y.-P.; Yuan, T.; Wang, X.-P. miR-124-3p inhibits the viability and motility of glioblastoma multiforme by targeting RhoG. Int. J. Mol. Med. 2021, 47, 69. [Google Scholar] [CrossRef]
- Cates, K.; McCoy, M.J.; Kwon, J.-S.; Liu, Y.; Abernathy, D.G.; Zhang, B.; Liu, S.; Gontarz, P.; Kim, W.K.; Chen, S. Deconstructing Stepwise Fate Conversion of Human Fibroblasts to Neurons by MicroRNAs. Cell Stem Cell 2021, 28, 127–140.e9. [Google Scholar] [CrossRef]
- Sun, J.; Jia, Z.; Li, B.; Zhang, A.; Wang, G.; Pu, P.; Chen, Z.; Wang, Z.; Yang, W. MiR-19 regulates the proliferation and invasion of glioma by RUNX3 via β-catenin/Tcf-4 signaling. Oncotarget 2017, 8, 110785. [Google Scholar] [CrossRef] [PubMed]
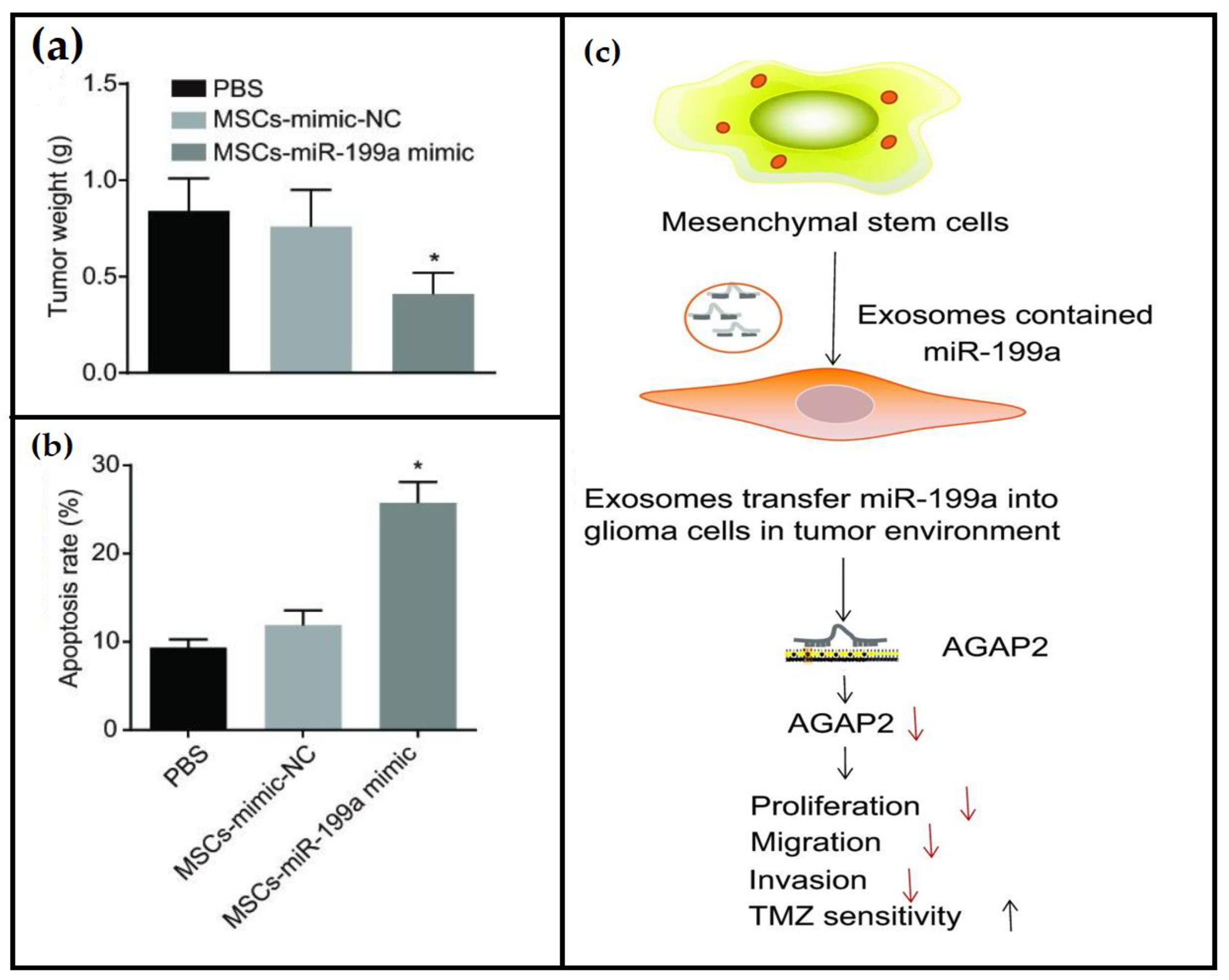
- Yu, L.; Gui, S.; Liu, Y.; Qiu, X.; Zhang, G.; Zhang, X.a.; Pan, J.; Fan, J.; Qi, S.; Qiu, B. Exosomes derived from microRNA-199a-overexpressing mesenchymal stem cells inhibit glioma progression by down-regulating AGAP2. Aging 2019, 11, 5300. [Google Scholar] [CrossRef]
- Shi, Y.; Tao, T.; Liu, N.; Luan, W.; Qian, J.; Li, R.; Hu, Q.; Wei, Y.; Zhang, J.; You, Y. PPARα, a predictor of patient survival in glioma, inhibits cell growth through the E2F1/miR-19a feedback loop. Oncotarget 2016, 7, 84623. [Google Scholar] [CrossRef]
- Shu, Y.; Yao, S.; Cai, S.; Li, J.; He, L.; Zou, J.; Zhang, Q.; Fan, H.; Zhou, L.; Yu, S. MiR-34c inhibits proliferation of glioma by targeting PTP1B. Acta Biochim. Biophys. Sin. 2021, 53, 325–332. [Google Scholar] [CrossRef]
- Xi, Z.; Wang, P.; Xue, Y.; Shang, C.; Liu, X.; Ma, J.; Li, Z.; Li, Z.; Bao, M.; Liu, Y. Overexpression of miR-29a reduces the oncogenic properties of glioblastoma stem cells by downregulating Quaking gene isoform 6. Oncotarget 2017, 8, 24949. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, K.; Zhang, A.; Wang, G.; Kang, C.; Han, L.; Pu, P. miR-19a and miR-19b overexpression in gliomas. Pathol. Oncol. Res. 2013, 19, 847–853. [Google Scholar] [CrossRef]
- Ma, C.; Zheng, C.; Bai, E.; Yang, K. miR-101 inhibits glioma cell invasion via the downregulation of COX-2. Oncol. Lett. 2016, 12, 2538–2544. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.B.; Rathod, S.S.; Karthik, S.; Kaur, N.; Muzumdar, D.; Shiras, A.S. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro-Oncology 2013, 15, 1302–1316. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Liu, X.; Chen, L.; Dou, Z.; Lei, X.; Chang, L.; Cai, J.; Cui, Y.; Yang, D.; Sun, Y. Targeting the SMO oncogene by miR-326 inhibits glioma biological behaviors and stemness. Neuro-Oncology 2015, 17, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y.; Xue, Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget 2016, 7, 62208. [Google Scholar] [CrossRef]
- Yang, C.H.; Wang, Y.; Sims, M.; Cai, C.; He, P.; Häcker, H.; Yue, J.; Cheng, J.; Boop, F.A.; Pfeffer, L.M. MicroRNA203a suppresses glioma tumorigenesis through an ATM-dependent interferon response pathway. Oncotarget 2017, 8, 112980. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, J.; Wang, Z.; Wu, G.; Liu, F. TFAM is directly regulated by miR-23b in glioma. Oncol. Rep. 2013, 30, 2105–2110. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, A.; Wu, Y.; Morris, S.-A.; Lin, K.; Amin, S.; Verhaak, R.; Fuller, G.; Xie, K.; Heimberger, A.B. miR-182-5p induced by STAT3 activation promotes glioma tumorigenesis. Cancer Res. 2016, 76, 4293–4304. [Google Scholar] [CrossRef]
- Sun, G.; Cao, Y.; Shi, L.; Sun, L.; Wang, Y.; Chen, C.; Wan, Z.; Fu, L.; You, Y. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother. Radiopharm. 2013, 28, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, J.; Hu, J.; Xue, K. MiR-21 enhanced glioma cells resistance to carmustine via decreasing Spry2 expression. Eur. Rev. Med. Pharm. Sci. 2017, 21, 5065–5071. [Google Scholar]
- Chen, L.; Li, Z.-y.; Xu, S.-y.; Zhang, X.-j.; Zhang, Y.; Luo, K.; Li, W.-p. Upregulation of miR-107 inhibits glioma angiogenesis and VEGF expression. Cell. Mol. Neurobiol. 2016, 36, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Rong, X.; Dong, J.; Yu, C.; Yang, J. miR-142 inhibits the migration and invasion of glioma by targeting Rac1. Oncol. Rep. 2017, 38, 1543–1550. [Google Scholar] [CrossRef]
- Liu, X.; Wang, S.; Yuan, A.; Yuan, X.; Liu, B. MicroRNA-140 represses glioma growth and metastasis by directly targeting ADAM9. Oncol. Rep. 2016, 36, 2329–2338. [Google Scholar] [CrossRef]
- Lan, F.; Yu, H.; Hu, M.; Xia, T.; Yue, X. miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J. Neurochem. 2015, 135, 274–286. [Google Scholar] [CrossRef]
- Yan, Z.; Che, S.; Wang, J.; Jiao, Y.; Wang, C.; Meng, Q. miR-155 contributes to the progression of glioma by enhancing Wnt/β-catenin pathway. Tumor Biol. 2015, 36, 5323–5331. [Google Scholar] [CrossRef]
- Sun, J.; Tian, X.; Zhang, J.; Huang, Y.; Lin, X.; Chen, L.; Zhang, S. Regulation of human glioma cell apoptosis and invasion by miR-152-3p through targeting DNMT1 and regulating NF2. J. Exp. Clin. Cancer Res. 2017, 36, 100. [Google Scholar] [CrossRef]
- Dong, H.; Hao, X.; Cui, B.; Guo, M. MiR-429 suppresses glioblastoma multiforme by targeting SOX2. Cell Biochem. Funct. 2017, 35, 260–268. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, Y.; Liu, M.; Jiang, Y. MicroRNA-181b inhibits cellular proliferation and invasion of glioma cells via targeting Sal-like protein 4. Oncol. Res. 2017, 25, 947. [Google Scholar] [CrossRef]
- Zhao, J.; Li, L.; Han, Z.-Y.; Wang, Z.-X.; Qin, L.-X. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am. J. Cancer Res. 2019, 9, 1367. [Google Scholar]
- Cai, H.; Liu, X.; Zheng, J.; Xue, Y.; Ma, J.; Li, Z.; Xi, Z.; Bao, M.; Liu, Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene 2017, 36, 318–331. [Google Scholar] [CrossRef]
- Gong, C.; Fang, J.; Li, G.; Liu, H.-H.; Liu, Z.-S. Effects of microRNA-126 on cell proliferation, apoptosis and tumor angiogenesis via the down-regulating ERK signaling pathway by targeting EGFL7 in hepatocellular carcinoma. Oncotarget 2017, 8, 52527. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, B.; Ren, H.; Chen, W. miR-9-5p inhibits pancreatic cancer cell proliferation, invasion and glutamine metabolism by targeting GOT1. Biochem. Biophys. Res. Commun. 2019, 509, 241–248. [Google Scholar] [CrossRef]
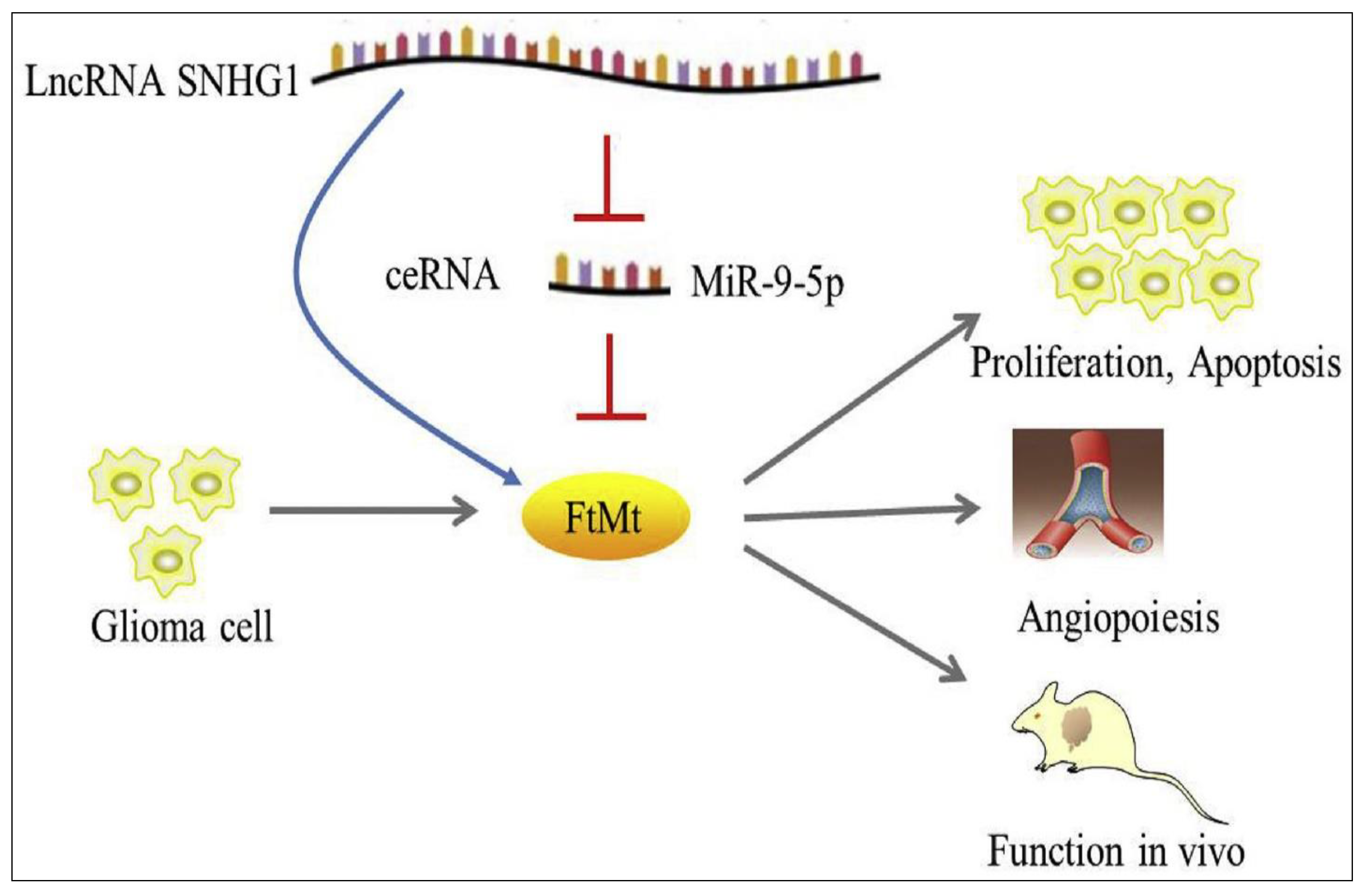
- Mi, S.; Du, J.; Liu, J.; Hou, K.; Ji, H.; Ma, S.; Ba, Y.; Chen, L.; Xie, R.; Hu, S. FtMt promotes glioma tumorigenesis and angiogenesis via lncRNA SNHG1/miR-9-5p axis. Cell. Signal. 2020, 75, 109749. [Google Scholar] [CrossRef] [PubMed]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrøm, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013, 3, 2535. [Google Scholar] [CrossRef]
- Chen, S.; Liang, H.; Yang, H.; Zhou, K.; Xu, L.; Liu, J.; Lai, B.; Song, L.; Luo, H.; Peng, J. LincRNa-p21: Function and mechanism in cancer. Med. Oncol. 2017, 34, 98. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Yu, B.; Wang, S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics 2018, 8, 3654. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Y.; Li, L.; Liu, Y.; Geng, P.; Li, G.; Song, H. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem. Cell Biol. 2018, 96, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shi, Y.; Shi, J.; Wang, H.; Sheng, Y.; Jiang, Q.; Chen, H.; Li, X.; Dong, J. The long non-coding RNA SNHG1 promotes glioma progression by competitively binding to miR-194 to regulate PHLDA1 expression. Cell Death Dis. 2019, 10, 463. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Gao, H.; Cheng, S.; Wang, B.; Guan, H. LncRNA HOXA-AS2 promotes glioblastoma carcinogenesis by targeting miR-885-5p/RBBP4 axis. Cancer Cell Int. 2021, 21, 39. [Google Scholar] [CrossRef]
- Wu, Y.; Qian, Z. Long non-coding RNAs (lncRNAs) and microRNAs regulatory pathways in the tumorigenesis and pathogenesis of glioma. Discov. Med. 2019, 28, 129–138. [Google Scholar] [PubMed]
- Guo, C.; Gong, M.; Li, Z. Knockdown of lncRNA MCM3AP-AS1 Attenuates Chemoresistance of Burkitt Lymphoma to Doxorubicin Treatment via Targeting the miR-15a/EIF4E Axis. Cancer Manag. Res. 2020, 12, 5845. [Google Scholar] [CrossRef]
- He, J.; Huang, Z.; He, M.; Liao, J.; Zhang, Q.; Wang, S.; Xie, L.; Ouyang, L.; Koeffler, H.P.; Yin, D. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer 2020, 19, 17. [Google Scholar] [CrossRef]
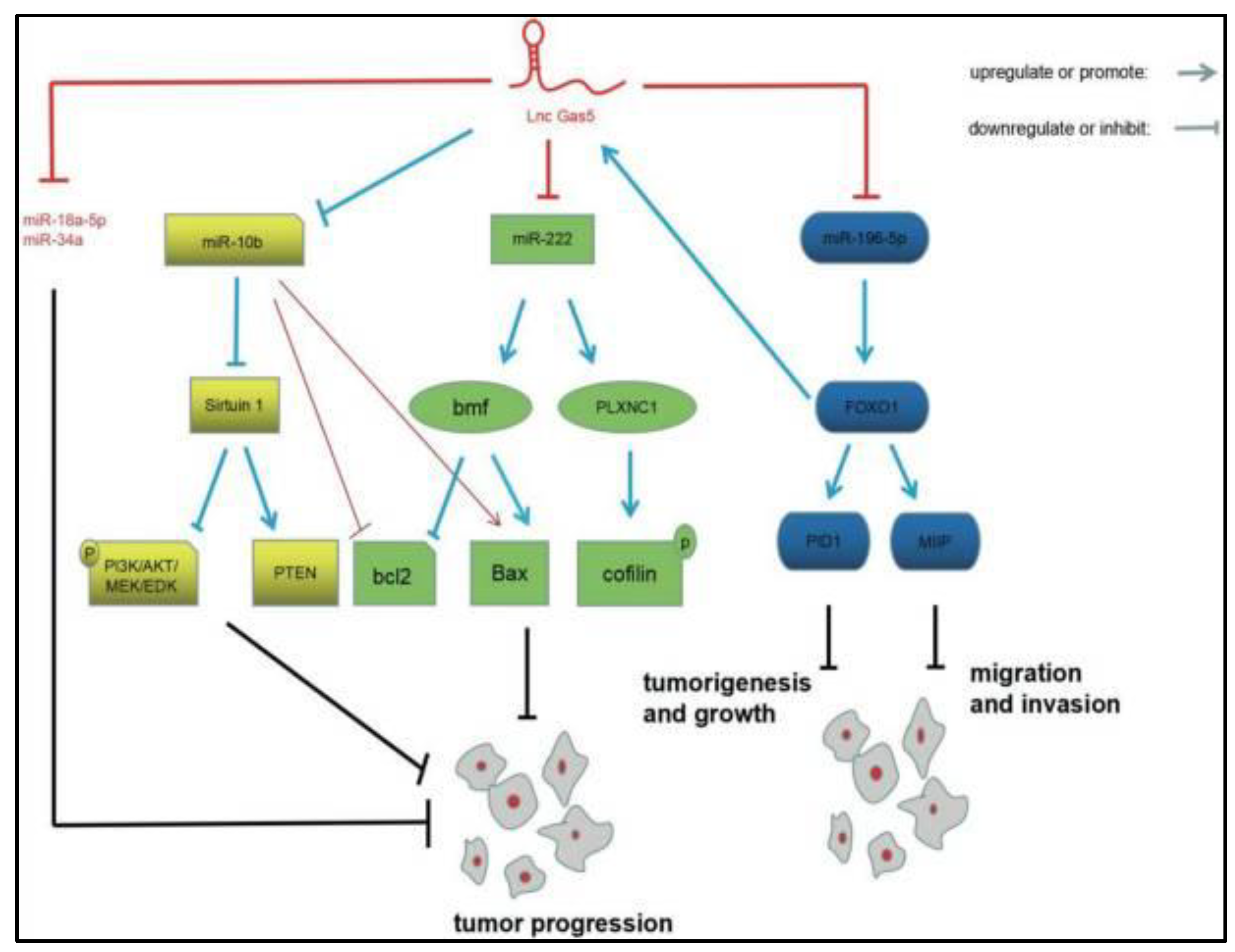
- Ding, Y.; Wang, J.; Zhang, H.; Li, H. Long noncoding RNA-GAS5 attenuates progression of glioma by eliminating microRNA-10b and Sirtuin 1 in U251 and A172 cells. Biofactors 2020, 46, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Jiang, H.; Fang, Y.; Han, D.; Guo, Y.; Wang, X.; Gong, X.; Hong, W.; Tu, J.; Wei, W. The essential role of long non-coding RNA GAS5 in glioma: Interaction with microRNAs, chemosensitivity and potential as a biomarker. J. Cancer 2021, 12, 224. [Google Scholar] [CrossRef]
- Nair, L.; Chung, H.; Basu, U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat. Rev. Mol. Cell Biol. 2020, 21, 123–136. [Google Scholar] [CrossRef]
- Cai, B.; Song, X.; Cai, J.; Zhang, S. HOTAIR: A cancer-related long non-coding RNA. Neoplasma 2014, 61, 379–391. [Google Scholar] [CrossRef]
- Chen, Y.; Bao, C.; Zhang, X.; Lin, X.; Huang, H.; Wang, Z. Long non-coding RNA HCG11 modulates glioma progression through cooperating with miR-496/CPEB3 axis. Cell Prolif. 2019, 52, e12615. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.-C.; Xiong, Z.; Zhu, G.-N.; Wang, C.; Zong, G.; Wang, H.-L.; Bian, E.-B.; Zhao, B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J. Exp. Clin. Cancer Res. 2016, 35, 90. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xiao, Z.; Du, X.; Huang, L.; Du, G. Silencing of the long non-coding RNA NEAT1 suppresses glioma stem-like properties through modulation of the miR-107/CDK6 pathway. Oncol. Rep. 2017, 37, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.; Li, B.; Liu, Q.; Cui, Y. lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J. Cell. Biochem. 2017, 118, 4548–4557. [Google Scholar] [CrossRef]
- Qin, N.; Tong, G.-F.; Sun, L.-W.; Xu, X.-L. Long noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol. Res. 2017, 25, 1471. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, J.-G.; Mi, W.-Y. Long noncoding RNA UCA1 targets miR-122 to promote proliferation, migration, and invasion of glioma cells. Oncol. Res. 2018, 26, 103. [Google Scholar] [CrossRef]
- Wang, P.; Liu, Y.-h.; Yao, Y.-l.; Li, Z.; Li, Z.-q.; Ma, J.; Xue, Y.-x. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell. Signal. 2015, 27, 275–282. [Google Scholar] [CrossRef]
- Shang, C.; Guo, Y.; Hong, Y.; Xue, Y.-x. Long non-coding RNA TUSC7, a target of miR-23b, plays tumor-suppressing roles in human gliomas. Front. Cell. Neurosci. 2016, 10, 235. [Google Scholar] [CrossRef]
- Zheng, J.; Li, X.-d.; Wang, P.; Liu, X.-b.; Xue, Y.-x.; Hu, Y.; Li, Z.; Li, Z.-q.; Wang, Z.-h.; Liu, Y.-h. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget 2015, 6, 25339. [Google Scholar] [CrossRef]
- Su, R.; Cao, S.; Ma, J.; Liu, Y.; Liu, X.; Zheng, J.; Chen, J.; Liu, L.; Cai, H.; Li, Z. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer 2017, 16, 171. [Google Scholar] [CrossRef]
- Ma, J.; Wang, P.; Yao, Y.; Liu, Y.; Li, Z.; Liu, X.; Li, Z.; Zhao, X.; Xi, Z.; Teng, H. Knockdown of long non-coding RNA MALAT1 increases the blood–tumor barrier permeability by up-regulating miR-140. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Xue, Y.; Wang, P.; Wang, Z.; Li, Z.; Hu, Y.; Li, Z.; Shang, X.; Liu, Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget 2015, 6, 19759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jin, H.; Lou, F. The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J. Cell. Biochem. 2018, 119, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, P.; Xue, Y.; Qu, C.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumor Biol. 2017, 39, 1010428317694326. [Google Scholar] [CrossRef]
- Ke, J.; Yao, Y.-l.; Zheng, J.; Wang, P.; Liu, Y.-h.; Ma, J.; Li, Z.; Liu, X.-b.; Li, Z.-q.; Wang, Z.-h. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 2015, 6, 21934. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, X.; Wang, P.; Xue, Y.; Ma, J.; Qu, C.; Liu, Y. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol. Ther. 2016, 24, 1199–1215. [Google Scholar] [CrossRef]
- Liu, C.; Sun, Y.; She, X.; Tu, C.; Cheng, X.; Wang, L.; Yu, Z.; Li, P.; Liu, Q.; Yang, H. CASC2c as an unfavorable prognosis factor interacts with miR-101 to mediate astrocytoma tumorigenesis. Cell Death Dis. 2017, 8, e2639. [Google Scholar] [CrossRef]
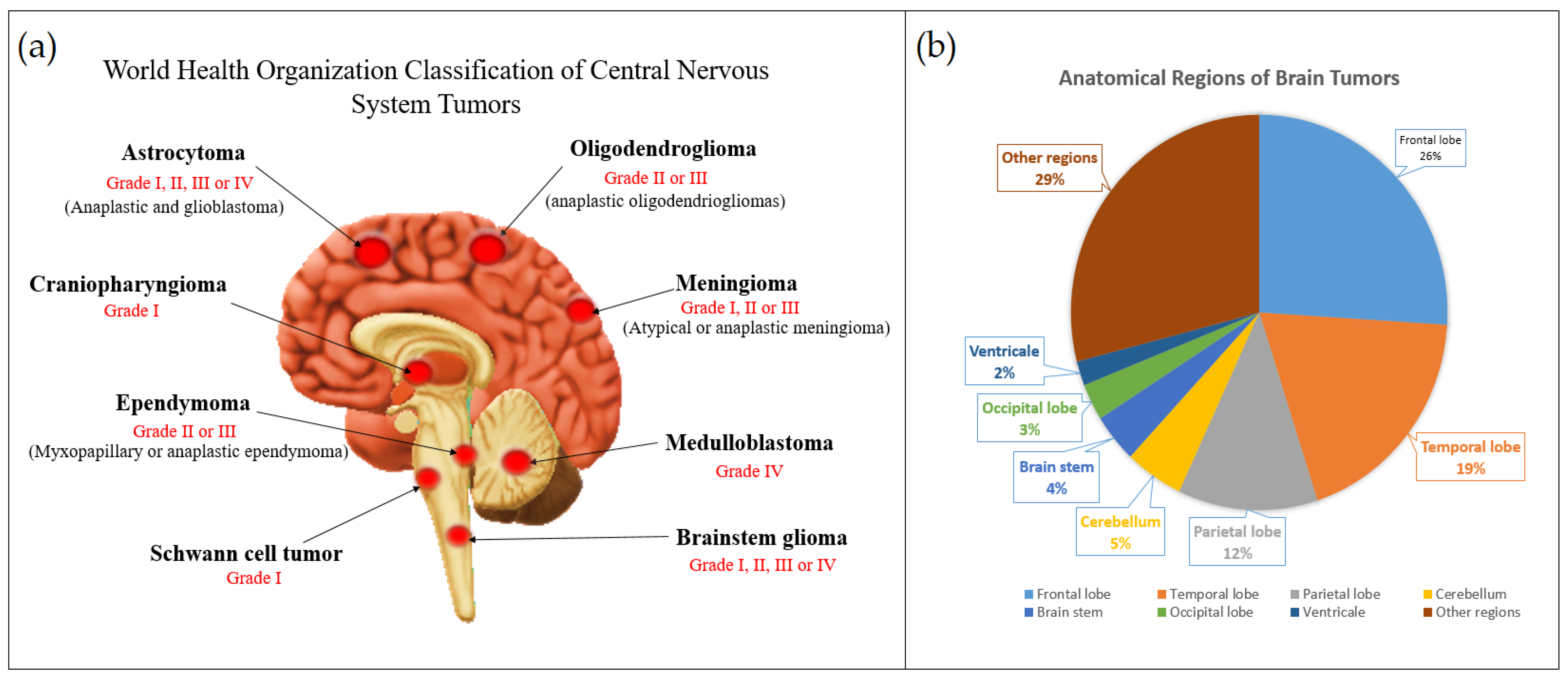
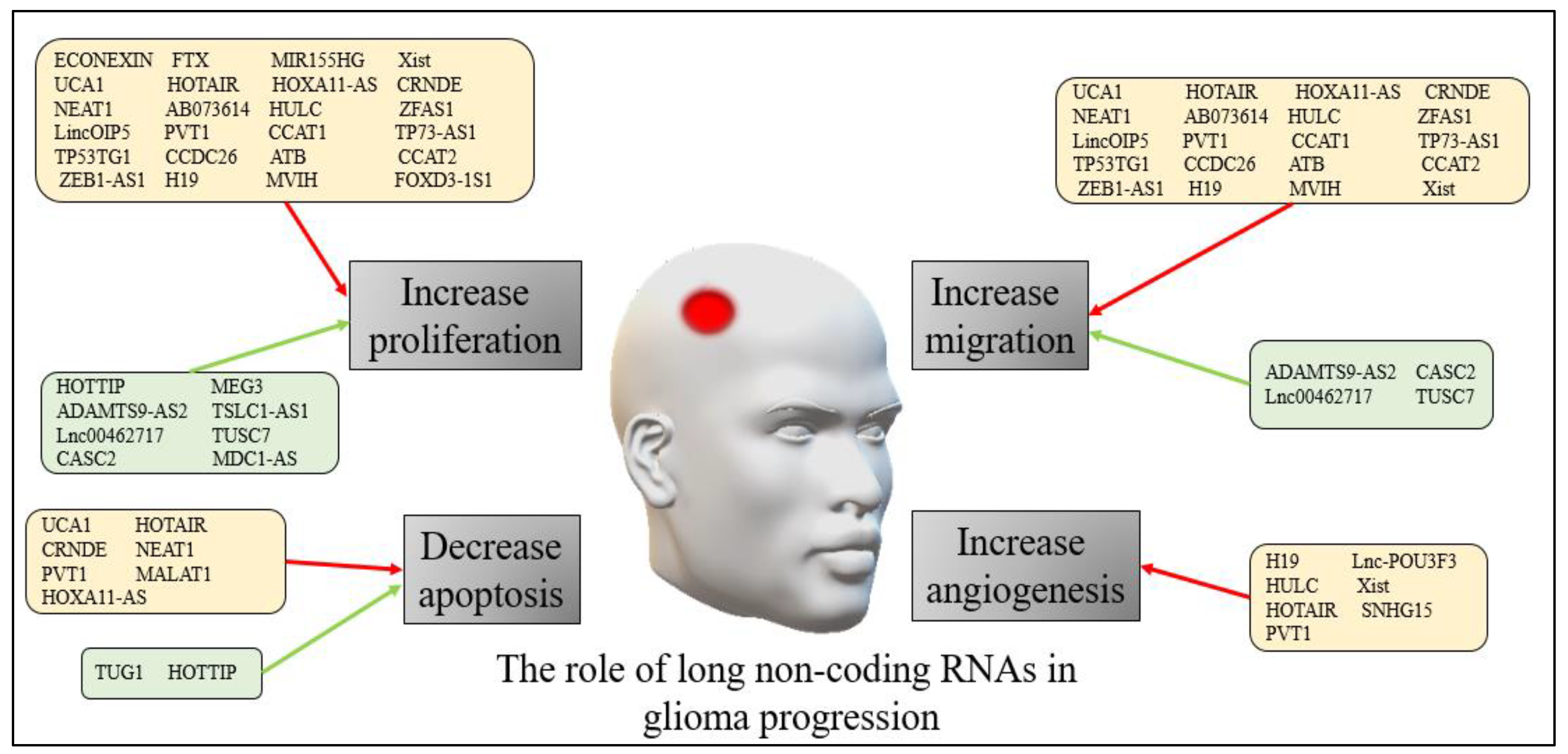
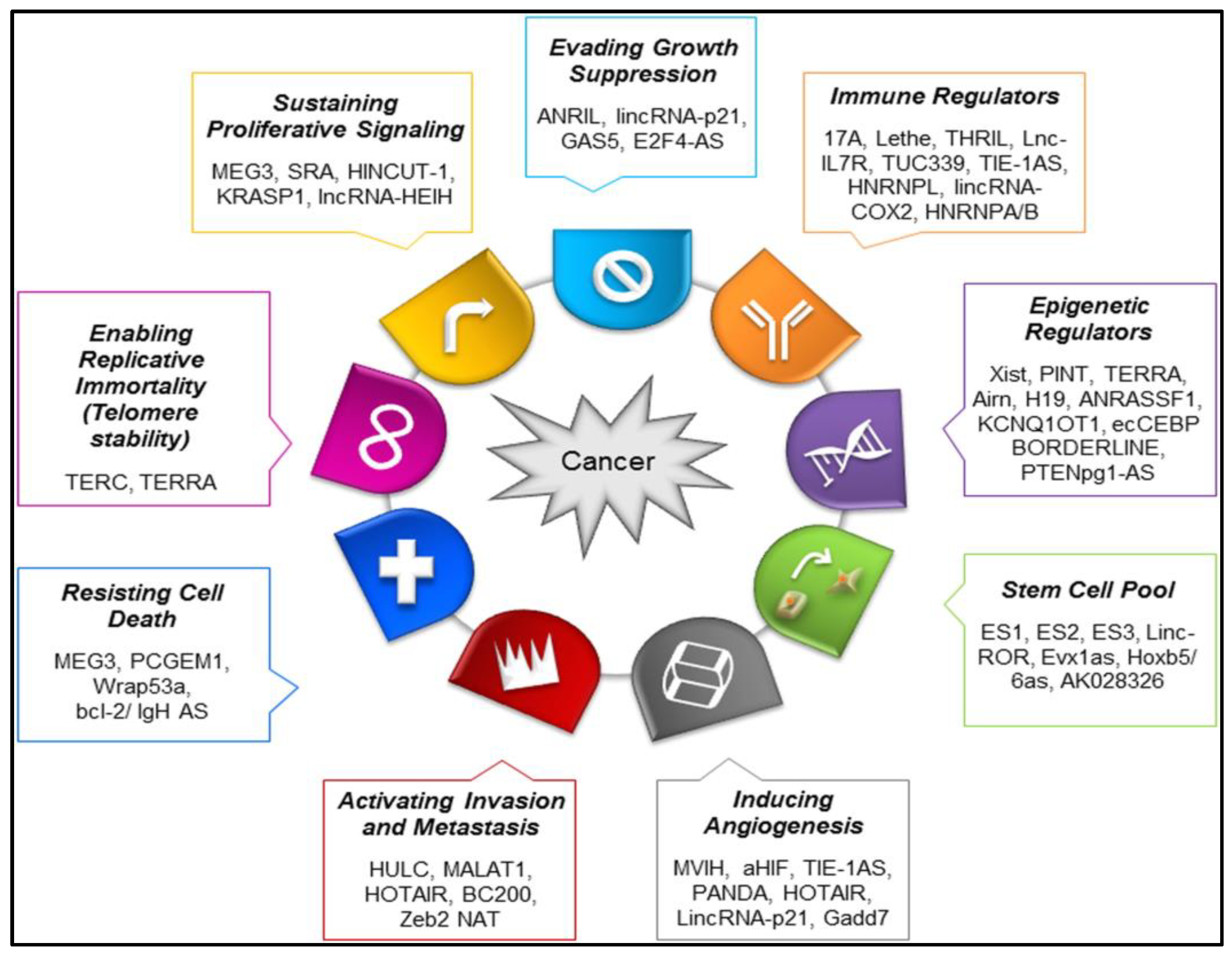
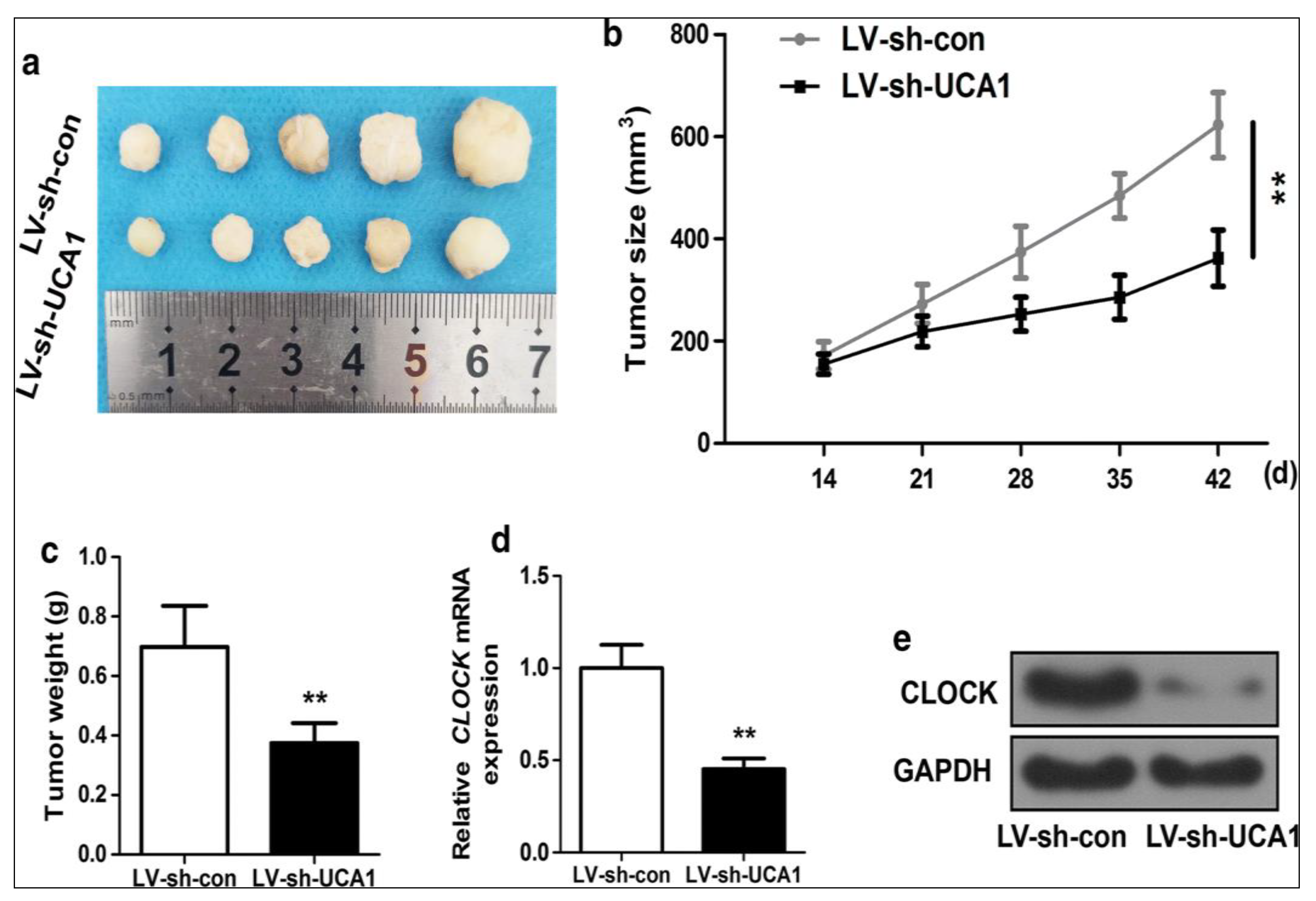
| No. | Major Category | Type | Grade |
|---|---|---|---|
| 1 | Diffuse Astrocytic and Oligodendroglial Tumors | Diffuse astrocytoma | (grade II) |
| Anaplastic astrocytoma | (grade III) | ||
| Glioblastoma | (grade IV) | ||
| 2 | Oligodendroglial Tumors | Oligodendroglioma | (grade II) |
| Anaplastic oligodendroglioma | (grade III) | ||
| 3 | Mixed Oligoastrocytic Tumors | Oligoastrocytoma | (grade II) |
| Anaplastic oligoastrocytoma | (grade III) | ||
| 4 | Ependymal Tumors | Subependymoma | (grade I) |
| Myxopapillary ependymoma | (grade I) | ||
| Ependymoma | (grade II-III) | ||
| Anaplastic ependymoma | (grade III) | ||
| 5 | Choroid Plexus Tumors | Choroid plexus papilloma | (grade I) |
| Atypical choroid plexus papilloma | (grade II) | ||
| Choroid plexus carcinoma | (grade III) | ||
| 6 | Other Rare Gliomas | Pleomorphic xanthoastrocytoma | (grade II) |
| Rosette-forming glioneuronal tumor of the fourth ventricle | (grade II) | ||
| Angiocentric glioma | (grade I) | ||
| Papillary glioneuronal tumor | (grade I) | ||
| Dysembryoplastic neuroepithelial tumor | (grade I) |
| LncRNA | The Effect on Glioma Progression | Ref. |
|---|---|---|
| HOTAIR | Cell cycle progression and promotion of adult glioma invasion by targeting enhancer of zeste homolog 2 (EZH2), vascular endothelial growth factor (VEGF), and matrix metalloproteinases (MMP-7 and MMP-9). | [73,74] |
| SNHG6 | Regulate glioma cell proliferation in adults through targeting of P21. | [75] |
| MALAT1 | Promotes progression and proliferation through miR-199a-zinc fingers and homeoboxes protein 1 (ZHX1) regulation and sponging of the miR-101. | [76,77] |
| LINC00689 | Promote the proliferation of adult glioma cells through targeting of miR-338-3p/pyruvate kinase M2 (PKM2) axis. | [78] |
| GAS5 | Reduction of adult glioma proliferation, and invasion and induces apoptosis by sponging miR-18a-5p, miR-222, and miR-424. | [79,80] |
| TUG1 | Promote migration and differentiation of adult glioma cells by functioning as a competing endogenous RNA for miR-6321. | [81] |
| H-19 | Induces proliferation, invasion, and metastasis of adult glioma cells in adults through the regulation of miR-140 and miR-29a. | [82,83] |
| CCAT2 | Enhance angiogenesis of glioma in adults and inhibit endothelial cell apoptosis through the stimulation of VEGFA expression and secretion. | [84] |
| POU3F3 | Promote glioma angiogenesis in adults by up-regulating fibroblast growth factor/receptor β (bFGF/bFGFR) and VEGFA expression. | [85] |
| Study | Type of LncRNA | Target | Remark |
|---|---|---|---|
| Wen et al. [128] | LBX2-AS1 | AKT/GSK3β pathway | Silencing of LBX2 antisense RNA 1 (LBX2-AS1) suppressed glioma cells proliferation in adults and pediatric and increased apoptosis by the protein kinase B (AKT)- glycogen synthase kinase 3β (GSK3β) pathway |
| Qin et al. [129] | TSLC1-AS1 | B-RAF proto-oncogene (BRAF) oncogene | Tumor suppressor in lung cancer 1 antisense RNA 1 (TSLC1-AS1) up-regulation resulted in a significant inhibition in the proliferation of adults and pediatric glioma cells. |
| Xu et al. [130] | HOTTIP | BRE gene | Overexpression of HOXA distal transcript antisense RNA (HOTTIP) promoted adult glioma cells apoptosis and inhibited their growth by down-regulating brain and reproductive organ-expressed protein expression. |
| Li et al. [131] | SLC26A4-AS1 | NPTX1 and NFKB1 | Solute carrier family 26 member 4 antisense RNA 1 (SLC26A4-AS1) overexpression promoted neuronal pentraxin 1 (NPTX1) expression by recruiting NFKB1 leading to an anti-angiogenic effect. |
| Bi et al. [132] | NEAT1 | let-7g-5p/MAP3K1 axis | Nuclear-Enriched Abundant Transcript 1 (NEAT1) down-regulation restrained malignant behaviors of glioma stem cells in adults and pediatric. |
| Wang et al. [133] | ENST00462717 | MDM2/MAPK | LncRNA ENST00462717 suppressed the cell’s activities by inhibiting Mouse double minute 2 homolog (MDM2)/MAPK pathway. |
| Han et al. [134] | MALAT1 | MMP2 gene | Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) overexpression suppressed cancer cells proliferation by down-regulation of matrix metalloproteinase 2 (MMP2) gene and extracellular signal-regulated kinase (ERK)/MAPK signaling inactivation. |
| Li et al. [135] | TUG1 | gene 1 | TUG1 expression up-regulated gene 1 in glioma tissues in adults and acted as a tumor suppressor by promoting cell apoptosis. |
| Luo et al. [136] | MEG3 | HIF1α | Overexpression of maternally expressed gene 3 (MEG3) inhibited the proliferation and invasion of tumor cells by suppressing the expression of hypoxia-inducible factor 1α (HIF1α). |
| Li et al. [137] | Linc01060 | MZF1/c-Myc | Long intergenic non-protein coding RNA 1060 (Linc01060) promoted myeloid zinc finger 1 (MZF1)-mediated c-MYC protooncogene transcriptional activities and thus promoted glioma progression in adults and pediatric. |
| Study | Type of miRNA | Target | Remark |
|---|---|---|---|
| Xi et al. [177] | miR-29a | Quaking gene isoform 6 | Up-regulation of miR-29a inhibited the expression of QKI-6 gene and thus inhibited the malignant behavior of adult glioma. |
| Jia et al. [178] | miR-19a | Akt protein | miR-19a/b knockdown inhibited glioma cells proliferation and invasion in adult and pediatric by down-regulating Akt expression. |
| Ma et al. [179] | miR-101 | COX-2 enzyme | miR-101 significantly down-regulated the expression of COX-2 leading to inhibition of tumor development. |
| Rani et al. [180] | miR-145 | Sox9 and ADD3 proteins | miR-145 up-regulation has a tumor-suppress the activity of oncogenic proteins Sox9 and ADD3 leading to reduction in glioma activities both in adults and pediatrics. |
| Du et al. [181] | miR-326 | SMO oncogene | miR-326 overexpression repressed SMO oncogene and inhibited adult glioma biological behaviors. |
| Gong et al. [182] | let-7e | NRAS protein | let-7e up-regulation led to inhibition proliferation and migration of adult glioma cells by reducing the expression of NRAS oncoprotein. |
| Yang et al. [183] | miR-203a | IFN-stimulated gene | Up-regulation of miR-203a promoted the interferon response, leading to suppression of adult glioma proliferation. |
| Jiang et al. [184] | miR-23b | PI3K/Akt signaling | miRNA-23b up-regulation suppressed PI3K/Akt signaling pathway leading to inhibition of glioma cells activities. |
| Xue et al. [185] | miR-182-5p | PCDH8 gene | miR-182-5p negatively regulated the expression of tumor suppressor gene PCDH8, leading to enhance adult glioma progression. |
| Sun et al. [186] | miR-137 | Rac1 gene | Overexpressed miRNA-137 Inhibited the growth of glioma cells by directly targeting Rac1 gene. |
| Wang et al. [187] | miR-21 | SPRY2 gene | miR-21 significantly increased glioma cells resistance to carmustine by decreasing the expression of SPRY2 protein. |
| Chen et al. [188] | miRNA-107 | VEGF | Up-regulation of miR-107 significantly inhibited the angiogenesis of glioma cells and the expression of VEGF. |
| Qin et al. [189] | miR-142 | Rac1 gene | miR-142 inhibited glioma migration and invasion by direct targeting Rac1. |
| Liu et al. [190] | miR-140 | ADAM9 protein | miR-140 up-regulation inhibited glioma cells proliferation and invasion via suppressing the expression of ADAM9 protein. |
| Lan et al. [191] | miR-144-3p | c-Met protein | Up-regulation of miR-144-3p inhibited survival capability in glioma cells and increased apoptosis by targeting c-Met. |
| Yan et al. [192] | miR-155 | Wnt signaling pathways | miR-155 promoted glioma cells progression by promoting Wnt signaling pathways. |
| Sun et al. [193] | miR-152-3p | DNMT1 gene | miR-152-3p up-regulation directly knocked down DNMT1 and led to significant induction of glioma cells apoptosis. |
| Dong et al. [194] | miR-429 | SOX2 gene | MiR-429 suppressed the proliferation of human glioma cells by targeting SOX2. |
| Zhou et al. [195] | miR-181b | Sal-Like Protein 4 | miR-181b up-regulation significantly decreased glioma cells proliferation and Invasion in adults via Targeting SALL4. |
| Study | lncRNA/miRNAInteraction | Remark |
|---|---|---|
| Ma et al. [216] | ATB/miR-200a | LncRNA activated by TGF-beta (ATB) up-regulation inhibits miR-200a in adult glioma cells and facilitates the transforming growth factor β2 (TGF-β2) signaling, while its knockdown significantly inhibits glioma cell growth/proliferation. |
| [53] | BCYRN1/miR-619-5p | Brain cytoplasmic RNA 1 (BCYRN1) acted as competing endogenous RNA, which inhibits glioma progression by sponging miR-619-5p and regulating the expression of the CUE domain-containing protein 2 (CUEDC2) gene. |
| Yang et al. [217] | NEAT1/miR-107 miR-7e-5p | This binding prevented the repression of cell division protein kinase 6, resulting in increased cancer cells growth. |
| Cui et al. [218] | CCAT1/miR-181b | lncRNA colon cancer-associated transcript 1 (CCAT1) knockdown notably suppressed glioma cell proliferation and promoted apoptosis by acting as a competing endogenous RNA for miR-181b. |
| Qin et al. [219] | MEG3/miR-19a | Suppression of glioma cells proliferation and invasion in adults by acting as a competing endogenous RNA for miR-19a. |
| Sun et al. [220] | UCA1/miR-122 | UCA1 acted as an endogenous sponge for miR-122. The binding led to miR-122 downregulation and significant suppression in all glioma cell activities. |
| Gong et al. [182] | NEAT1/let-7e | NEAT1 up-regulation reduced let-7e expression, which seemed to suppress tumor function. Thus, NEAT1 knockdown led to let-7e overexpression and reduced the oncoprotein NRAS expression. |
| Wang et al. [221] | CASC2/miR-21 | Cancer susceptibility candidate 2 (CASC2) suppresses glioma cell proliferation via the negative regulation of miR-21. |
| Shang et al. [222] | TUSC7/miR-23b | Tumor suppressor candidate 7 (TUSC7) up-regulation suppressed the activities of glioma cells in adult and led to the down-regulation of miR-23b. |
| Zheng et al. [223] | CRNDE/miR-186 | Colorectal neoplasia differentially expressed (CRNDE) knockdown positively regulated miR-186 expression and suppressed the activity of glioma cells. |
| Su et al. [224] | SOX2OT/miR-122 and miR-194-5p | Knocking down of SOX2 overlapping transcript (SOX2OT) inhibited the malignant biological behaviors of glioma cells via miR-122 and miR-194-5p up-regulation. |
| Ma et al. [225] | MALAT/miR-140 | Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) downregulation increased the blood–tumor barrier and the permeability by up-regulating miR-140. |
| Cai et al. [226] | TUG1/miR-144 | Taurine upregulated gene 1 (TUG1) regulated blood-tumor barrier permeability in adult and pediatric glioma by targeting miR-144. |
| Zhang et al. [227] | TP73-AS1/miR-142 | TP73 antisense RNA 1 (TP73-AS1) promoted the growth of adult glioma via sponging miR-142 and acting as a competing endogenous RNA to promote high mobility group box 1 (HMGB1) expression. |
| Cui et al. [218] | CCAT1/miR-181b | Knockdown of colon cancer-associated transcript 1 (CCAT1) inhibited glioma proliferation by sponging miR-181b, resulting in de-repression of its endogenous targets platelet-derived growth factor receptor (PDGFRα) and fibroblast growth factor receptor 3 (FGFR3). |
| Ma et al. [228] | PVT1/miR-186 | The overexpression of plasmacytoma variant translocation 1 (PVT1) increased autophagy related 7 (ATG7) and Beclin1 expression by targeting miR-186, thus promoting the proliferation of glioma cells. |
| Ke et al. [229] | HOTAIR/miR-326 | HOX transcript antisense intergenic RNA (HOTAIR) knockdown inhibited the malignant behaviors of glioma cells in adults through the modulation of miR-326. |
| Zheng et al. [230] | CRNDE/miR-384 | Knockdown of colorectal neoplasia differentially expressed (CRNDE) and miR-384 overexpression significantly led to tumor regression. |
| Liu et al. [231] | CASC2c/miR-101 | Overexpression of cancer susceptibility 2 (CASC2c) promoted the malignant behavior of adult glioma cells by directly bounding with miR-101. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kciuk, M.; Yahya, E.B.; Mohamed, M.M.I.; Abdulsamad, M.A.; Allaq, A.A.; Gielecińska, A.; Kontek, R. Insights into the Role of LncRNAs and miRNAs in Glioma Progression and Their Potential as Novel Therapeutic Targets. Cancers 2023, 15, 3298. https://doi.org/10.3390/cancers15133298
Kciuk M, Yahya EB, Mohamed MMI, Abdulsamad MA, Allaq AA, Gielecińska A, Kontek R. Insights into the Role of LncRNAs and miRNAs in Glioma Progression and Their Potential as Novel Therapeutic Targets. Cancers. 2023; 15(13):3298. https://doi.org/10.3390/cancers15133298
Chicago/Turabian StyleKciuk, Mateusz, Esam Bashir Yahya, Montaha Mohamed Ibrahim Mohamed, Muhanad A. Abdulsamad, Abdulmutalib A. Allaq, Adrianna Gielecińska, and Renata Kontek. 2023. "Insights into the Role of LncRNAs and miRNAs in Glioma Progression and Their Potential as Novel Therapeutic Targets" Cancers 15, no. 13: 3298. https://doi.org/10.3390/cancers15133298
APA StyleKciuk, M., Yahya, E. B., Mohamed, M. M. I., Abdulsamad, M. A., Allaq, A. A., Gielecińska, A., & Kontek, R. (2023). Insights into the Role of LncRNAs and miRNAs in Glioma Progression and Their Potential as Novel Therapeutic Targets. Cancers, 15(13), 3298. https://doi.org/10.3390/cancers15133298










