Genomic Mapping of Epidermal Growth Factor Receptor and Mesenchymal–Epithelial Transition-Up-Regulated Tumors Identifies Novel Therapeutic Opportunities
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression and Genomic Alteration Analysis
2.2. Correlation between Gene Expression and Immune Cell Infiltration
2.3. Outcome and Prognosis Analysis
3. Results
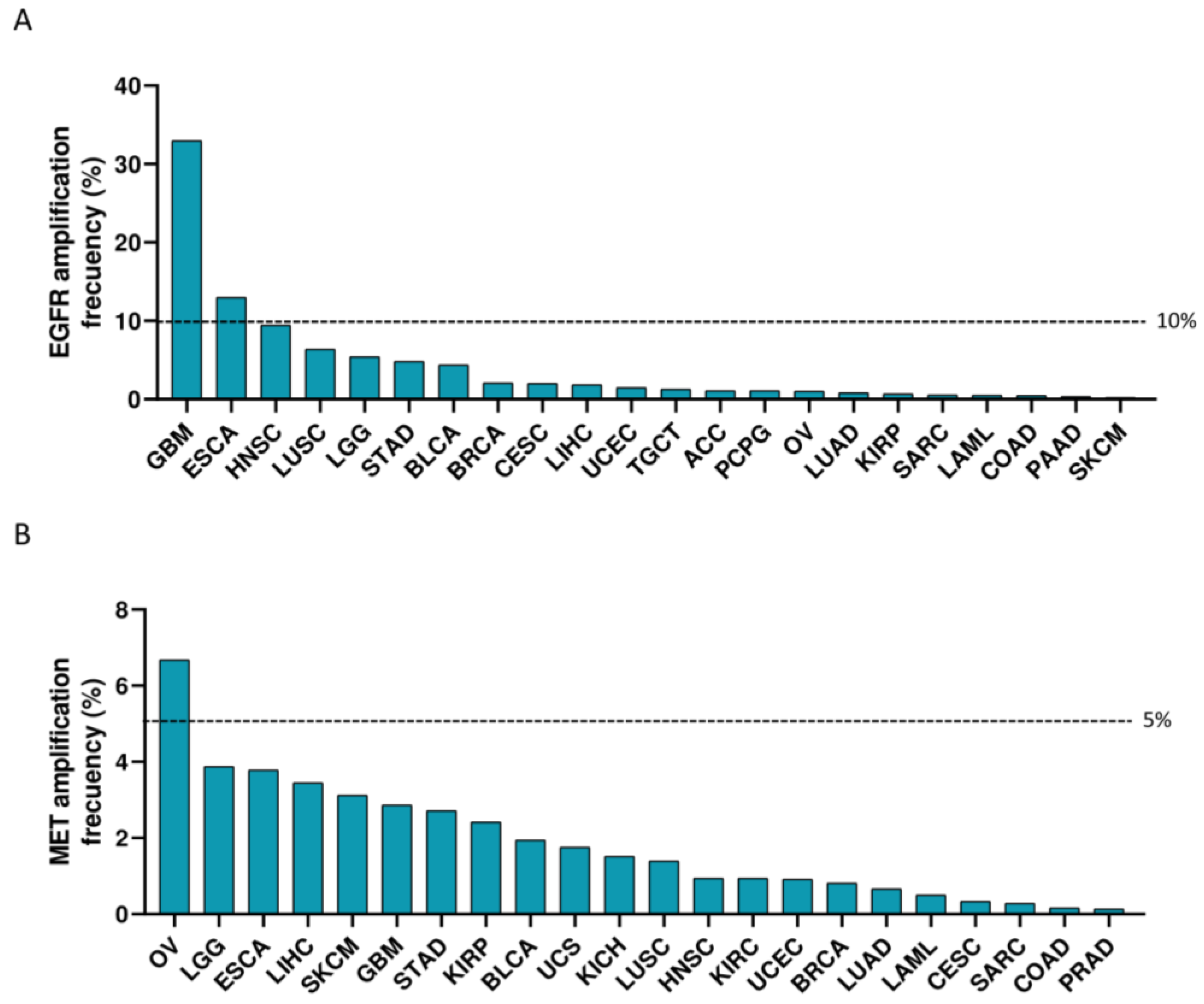
3.1. Expression of EGFR and MET in Cancer
3.2. Co-Occurrent Expression of EGFR and MET with PD-L1 in Solid Tumors
3.3. Association of EGFR and MET Expression with Clinical Outcome in PD(L)1-Treated Population
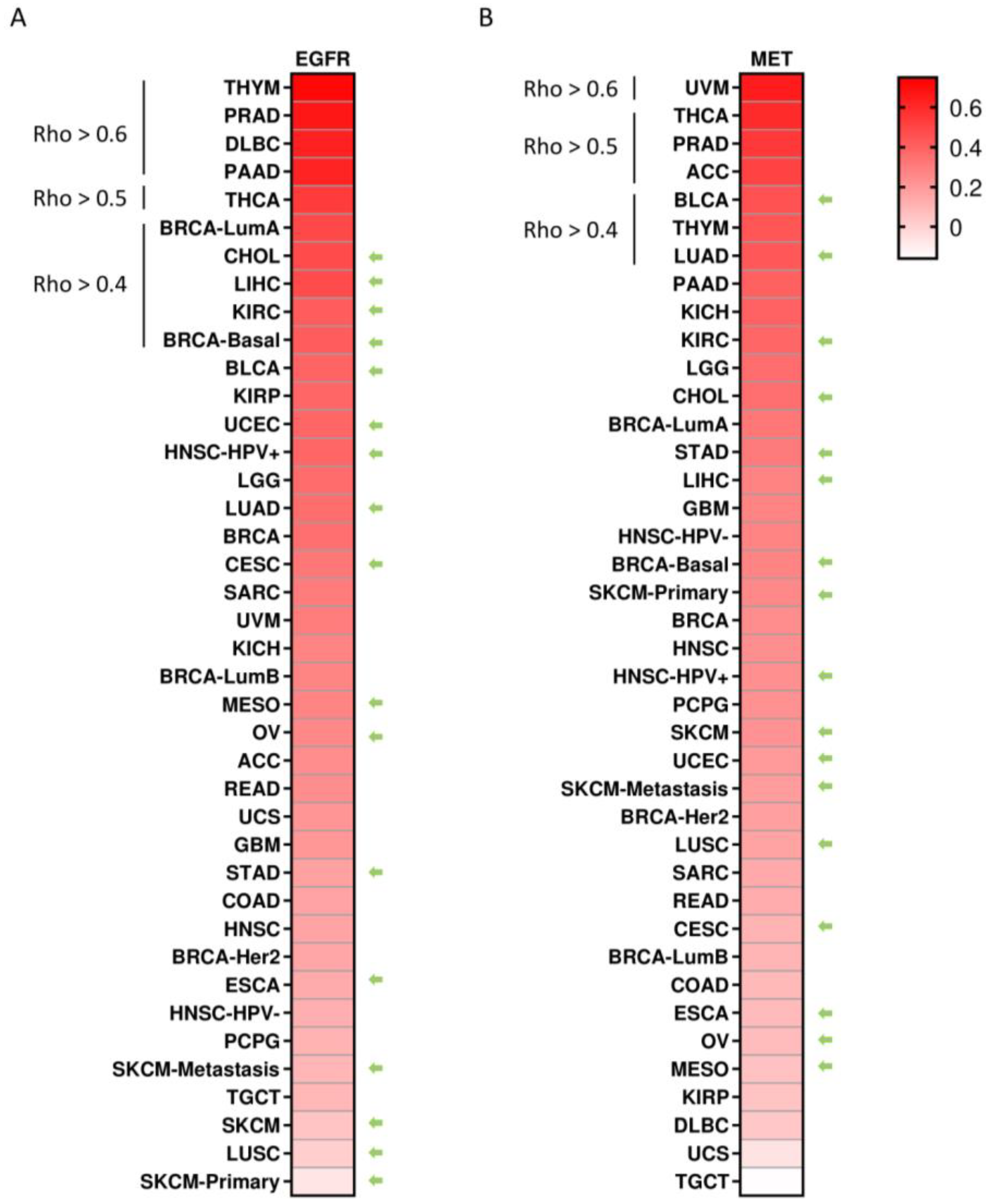
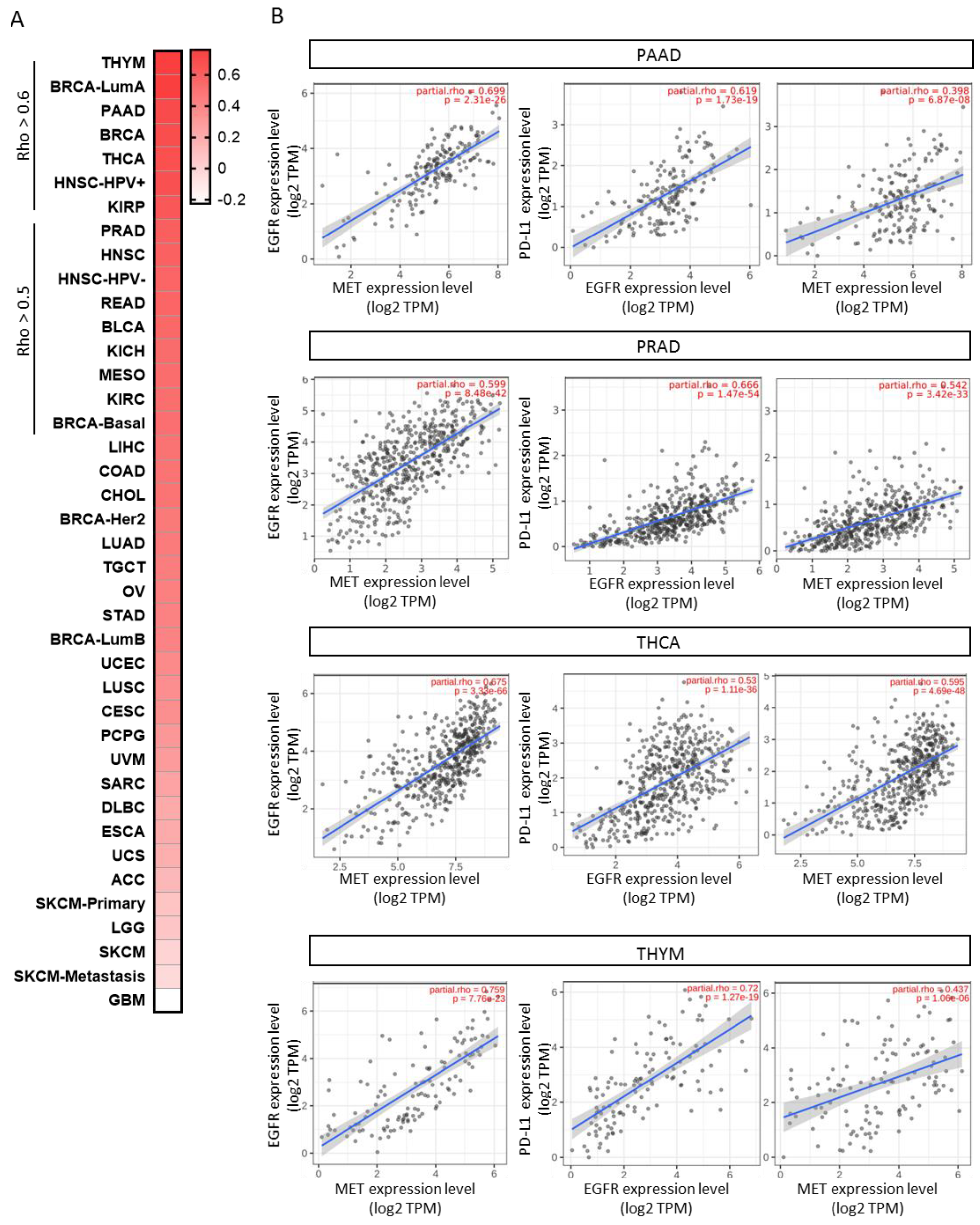
3.4. Co-Occurrent Expression of EGFR and MET in Solid Tumors
3.5. Therapeutic Opportunities to Exploit Anti-EGFR/MET with Anti-PD-(L)1 Antibodies
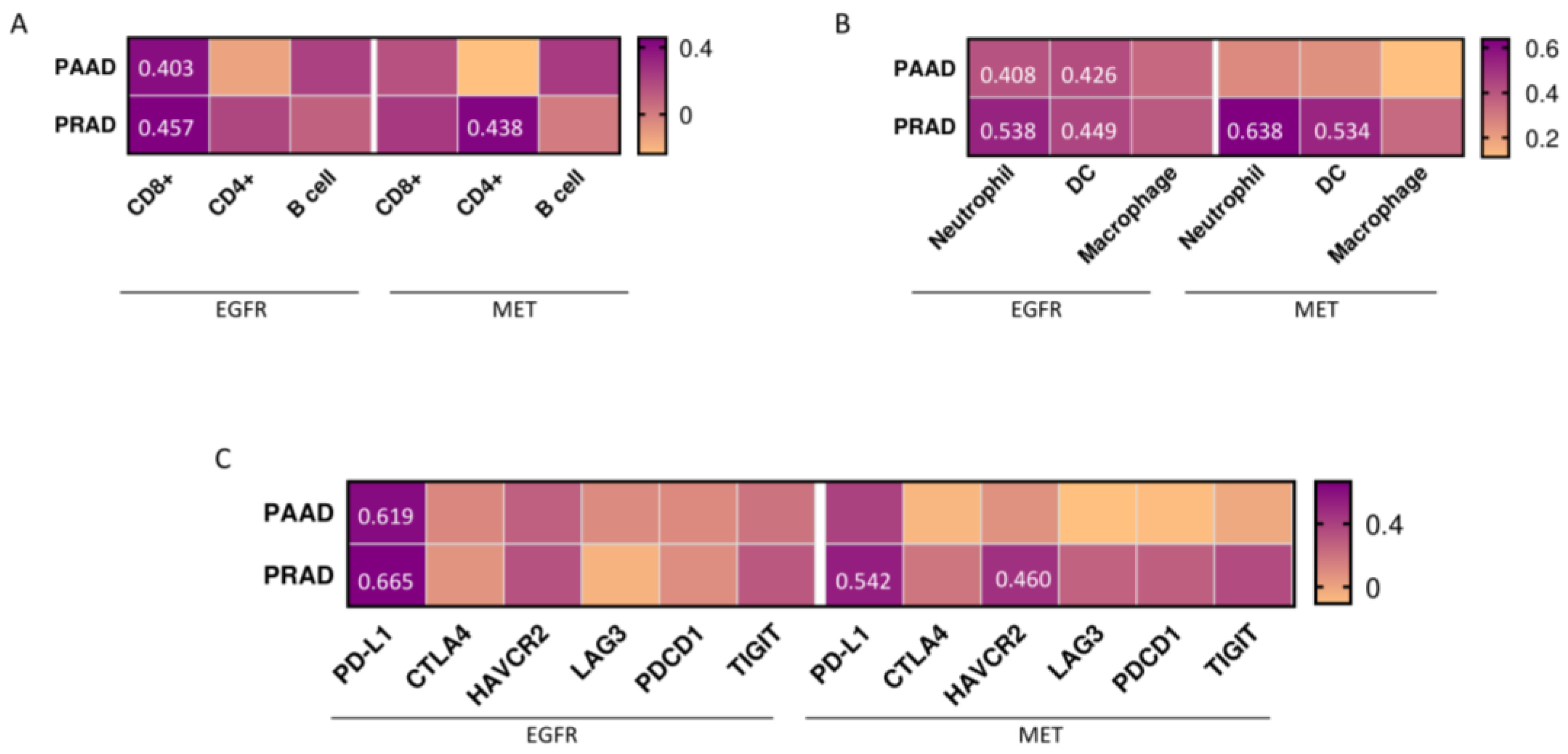
3.6. Expression of Immune Populations and Co-Inhibitors in PAAD and PRAD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ocana, A.; Pandiella, A.; Siu, L.L.; Tannock, I.F. Preclinical development of molecular-targeted agents for cancer. Nat. Rev. Clin. Oncol. 2010, 8, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, J.; Settleman, J.; Malek, S. Emerging Trends in Cancer Drug Discovery-From Drugging the “Undruggable” to Overcoming Resistance. Cancer Discov. 2021, 11, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Gandullo-Sanchez, L.; Ocana, A.; Pandiella, A. HER3 in cancer: From the bench to the bedside. J. Exp. Clin. Cancer Res. 2022, 41, 310. [Google Scholar] [CrossRef] [PubMed]
- Esparis-Ogando, A.; Montero, J.C.; Arribas, J.; Ocana, A.; Pandiella, A. Targeting the EGF/HER Ligand-Receptor System in Cancer. Curr. Pharm. Des. 2016, 22, 5887–5898. [Google Scholar] [CrossRef]
- Ocana, A.; Amir, E. Irreversible pan-ErbB tyrosine kinase inhibitors and breast cancer: Current status and future directions. Cancer Treat. Rev. 2009, 35, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Ocana, A.; Pandiella, A. Targeting HER receptors in cancer. Curr. Pharm. Des. 2013, 19, 808–817. [Google Scholar] [CrossRef]
- Hynes, N.E. ErbB2: From an EGFR Relative to a Central Target for Cancer Therapy. Cancer Res. 2016, 76, 3659–3662. [Google Scholar] [CrossRef]
- Musolino, A.; Gradishar, W.J.; Rugo, H.S.; Nordstrom, J.L.; Rock, E.P.; Arnaldez, F.; Pegram, M.D. Role of Fcγ receptors in HER2-targeted breast cancer therapy. J. Immunother. Cancer 2022, 10, e003171. [Google Scholar] [CrossRef]
- Lin, N.U.; Murthy, R.K.; Abramson, V.; Anders, C.; Bachelot, T.; Bedard, P.L.; Borges, V.; Cameron, D.; Carey, L.A.; Chien, A.J.; et al. Tucatinib vs. Placebo, Both in Combination with Trastuzumab and Capecitabine, for Previously Treated ERBB2 (HER2) Positive Metastatic Breast Cancer in Patients with Brain Metastases: Updated Exploratory Analysis of the HER2CLIMB Randomized Clinical Trial. JAMA Oncol. 2023, 9, 197–205. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Kaplon, H.; Crescioli, S.; Chenoweth, A.; Visweswaraiah, J.; Reichert, J.M. Antibodies to watch in 2023. MAbs 2023, 15, 2153410. [Google Scholar] [CrossRef]
- Ocana, A.; Garcia-Alonso, S.; Amir, E.; Pandiella, A. Refining Early Antitumoral Drug Development. Trends Pharmacol. Sci. 2018, 39, 922–925. [Google Scholar] [CrossRef]
- Runcie, K.; Budman, D.R.; John, V.; Seetharamu, N. Bi-specific and tri-specific antibodies—The next big thing in solid tumor therapeutics. Mol. Med. 2018, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Haura, E.B.; Leighl, N.B.; Mitchell, P.; Shu, C.A.; Girard, N.; Viteri, S.; Han, J.Y.; Kim, S.W.; Lee, C.K.; et al. Amivantamab in EGFR Exon 20 Insertion-Mutated Non-Small-Cell Lung Cancer Progressing on Platinum Chemotherapy: Initial Results from the CHRYSALIS Phase I Study. J. Clin. Oncol. 2021, 39, 3391–3402. [Google Scholar] [CrossRef]
- Yun, J.; Lee, S.H.; Kim, S.Y.; Jeong, S.Y.; Kim, J.H.; Pyo, K.H.; Park, C.W.; Heo, S.G.; Yun, M.R.; Lim, S.; et al. Antitumor Activity of Amivantamab (JNJ-61186372), an EGFR-MET Bispecific Antibody, in Diverse Models of EGFR Exon 20 Insertion-Driven NSCLC. Cancer Discov. 2020, 10, 1194–1209. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H.; Moreno Garcia, V.; Gil Bazo, I.; Prenen, H.; Moreno, I.; Johnson, M.; Castañón Álvarez, E.; Nagasaka, M.; Adeyemi, S.; Barasa, B.; et al. MCLA-129, a human anti-EGFR and anti-c-MET bispecific antibody, in patients with advanced NSCLC and other solid tumors: An ongoing phase 1/2 study. Eur. J. Cancer 2022, 174, S122. [Google Scholar] [CrossRef]
- Andre, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Blery, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-Tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e1713. [Google Scholar] [CrossRef]
- La Corte, C.M.; Fasano, M.; Ciaramella, V.; Cimmino, F.; Cardnell, R.; Gay, C.M.; Ramkumar, K.; Diao, L.; Di Liello, R.; Viscardi, G.; et al. Anti-tumor activity of cetuximab plus avelumab in non-small cell lung cancer patients involves innate immunity activation: Findings from the CAVE-Lung trial. J. Exp. Clin. Cancer Res. 2022, 41, 109. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Zhang, Z. Available online: http://gepia2.cancer-pku.cn/#index (accessed on 6 February 2023).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. Available online: https://www.cbioportal.org (accessed on 20 February 2023).
- Nguyen, B.; Mota, J.M.; Nandakumar, S.; Stopsack, K.H.; Weg, E.; Rathkopf, D.; Morris, M.J.; Scher, H.I.; Kantoff, P.W.; Gopalan, A.; et al. Pan-cancer Analysis of CDK12 Alterations Identifies a Subset of Prostate Cancers with Distinct Genomic and Clinical Characteristics. Eur. Urol. 2020, 78, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Jusakul, A.; Cutcutache, I.; Yong, C.H.; Lim, J.Q.; Huang, M.N.; Padmanabhan, N.; Nellore, V.; Kongpetch, S.; Ng, A.W.T.; Ng, L.M.; et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017, 7, 1116–1135. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Nacev, B.A.; Sanchez-Vega, F.; Smith, S.A.; Antonescu, C.R.; Rosenbaum, E.; Shi, H.; Tang, C.; Socci, N.D.; Rana, S.; Gularte-Mérida, R.; et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat. Commun. 2022, 13, 3405. [Google Scholar] [CrossRef]
- Caso, R.; Sanchez-Vega, F.; Tan, K.S.; Mastrogiacomo, B.; Zhou, J.; Jones, G.D.; Nguyen, B.; Schultz, N.; Connolly, J.G.; Brandt, W.S.; et al. The Underlying Tumor Genomics of Predominant Histologic Subtypes in Lung Adenocarcinoma. J. Thorac. Oncol. 2020, 15, 1844–1856. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Taiwen, L. Available online: http:/timer.cistrome.org/ (accessed on 8 March 2023).
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef]
- Kovacs, S.A.; Gyorffy, B. Transcriptomic datasets of cancer patients treated with immune-checkpoint inhibitors: A systematic review. J. Transl. Med. 2022, 20, 249. [Google Scholar] [CrossRef]
- Gyorffy, B. Available online: https://kmplot.com/analysis/ (accessed on 24 March 2023).
- To, K.K.W.; Fong, W.; Cho, W.C.S. Immunotherapy in Treating EGFR-Mutant Lung Cancer: Current Challenges and New Strategies. Front. Oncol. 2021, 11, 635007. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Baas, P.; Kim, D.W.; Felip, E.; Perez-Gracia, J.L.; Han, J.Y.; Molina, J.; Kim, J.H.; Arvis, C.D.; Ahn, M.J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Man, J.; Lord, S.; Links, M.; Gebski, V.; Mok, T.; Yang, J.C. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non Small Cell Lung Cancer—A Meta-Analysis. J. Thorac. Oncol. 2017, 12, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Dantoing, E.; Piton, N.; Salaun, M.; Thiberville, L.; Guisier, F. Anti-PD1/PD-L1 Immunotherapy for Non-Small Cell Lung Cancer with Actionable Oncogenic Driver Mutations. Int. J. Mol. Sci. 2021, 22, 6288. [Google Scholar] [CrossRef]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the immunotarget registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef]
- Esteva, F.J.; Hubbard-Lucey, V.M.; Tang, J.; Pusztai, L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019, 20, e175–e186. [Google Scholar] [CrossRef]
- Kumagai, S.; Koyama, S.; Nishikawa, H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 2021, 21, 181–197. [Google Scholar] [CrossRef]
- Dong, Z.Y.; Zhang, J.T.; Liu, S.Y.; Su, J.; Zhang, C.; Xie, Z.; Zhou, Q.; Tu, H.Y.; Xu, C.R.; Yan, L.X.; et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 2017, 6, e1356145. [Google Scholar] [CrossRef]
- Petrylak, D.P.; Loriot, Y.; Shaffer, D.R.; Braiteh, F.; Powderly, J.; Harshman, L.C.; Conkling, P.; Delord, J.P.; Gordon, M.; Kim, J.W.; et al. Safety and Clinical Activity of Atezolizumab in Patients with Metastatic Castration-Resistant Prostate Cancer: A Phase I Study. Clin. Cancer Res. 2021, 27, 3360–3369. [Google Scholar] [CrossRef]
- Powles, T.; Yuen, K.C.; Gillessen, S.; Kadel, E.E., III; Rathkopf, D.; Matsubara, N.; Drake, C.G.; Fizazi, K.; Piulats, J.M.; Wysocki, P.J.; et al. Atezolizumab with enzalutamide versus enzalutamide alone in metastatic castration-resistant prostate cancer: A randomized phase 3 trial. Nat. Med. 2022, 28, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Wiesehöfer, M.; Raczinski, B.B.G.; Wiesehöfer, C.; Dankert, J.T.; Czyrnik, E.D.; Spahn, M.; Kruithof-de Julio, M.; Wennemuth, G. Epiregulin expression and secretion is increased in castration-resistant prostate cancer. Front. Oncol. 2023, 13, 1107021. [Google Scholar] [CrossRef]
- Verras, M.; Lee, J.; Xue, H.; Li, T.H.; Wang, Y.; Sun, Z. The androgen receptor negatively regulates the expression of c-Met: Implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007, 67, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Gong, Y.; Fan, Z.; Luo, G.; Huang, Q.; Deng, S.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2020, 13, 130. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gulati, M.; Larson, A.C.; Solheim, J.C.; Jain, M.; Kumar, S.; Batra, S.K. Immune checkpoint blockade in pancreatic cancer: Trudging through the immune desert. Semin. Cancer Biol. 2022, 86, 14–27. [Google Scholar] [CrossRef]
- Shi, X.; Wang, M.; Zhang, Y.; Guo, X.; Liu, M.; Zhou, Z.; Zhao, Y.; He, R.; Gao, Y.; Liu, Y.; et al. Hypoxia activated HGF expression in pancreatic stellate cells confers resistance of pancreatic cancer cells to EGFR inhibition. EBioMedicine 2022, 86, 104352. [Google Scholar] [CrossRef]
- Salles, G.; Duell, J.; Gonzalez Barca, E.; Tournilhac, O.; Jurczak, W.; Liberati, A.M.; Nagy, Z.; Obr, A.; Gaidano, G.; Andre, M.; et al. Tafasitamab plus lenalidomide in relapsed or refractory diffuse large B-cell lymphoma (L-MIND): A multicentre, prospective, single-arm, phase 2 study. Lancet Oncol. 2020, 21, 978–988. [Google Scholar] [CrossRef]
- Rubio-Perez, L.; Lazaro-Gorines, R.; Harwood, S.L.; Compte, M.; Navarro, R.; Tapia-Galisteo, A.; Bonet, J.; Blanco, B.; Lykkemark, S.; Ramirez-Fernandez, A.; et al. A PD-L1/EGFR bispecific antibody combines immune checkpoint blockade and direct anti-cancer action for an enhanced anti-tumor response. Oncoimmunology 2023, 12, 2205336. [Google Scholar] [CrossRef]
- Yuan, Q.; Liang, Q.; Sun, Z.; Yuan, X.; Hou, W.; Wang, Y.; Wang, H.; Yu, M. Development of bispecific anti-c-Met/PD-1 diabodies for the treatment of solid tumors and the effect of c-Met binding affinity on efficacy. Oncoimmunology 2021, 10, 1914954. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paniagua-Herranz, L.; Doger, B.; Díaz-Tejeiro, C.; Sanvicente, A.; Nieto-Jiménez, C.; Moreno, V.; Pérez Segura, P.; Gyorffy, B.; Calvo, E.; Ocana, A. Genomic Mapping of Epidermal Growth Factor Receptor and Mesenchymal–Epithelial Transition-Up-Regulated Tumors Identifies Novel Therapeutic Opportunities. Cancers 2023, 15, 3250. https://doi.org/10.3390/cancers15123250
Paniagua-Herranz L, Doger B, Díaz-Tejeiro C, Sanvicente A, Nieto-Jiménez C, Moreno V, Pérez Segura P, Gyorffy B, Calvo E, Ocana A. Genomic Mapping of Epidermal Growth Factor Receptor and Mesenchymal–Epithelial Transition-Up-Regulated Tumors Identifies Novel Therapeutic Opportunities. Cancers. 2023; 15(12):3250. https://doi.org/10.3390/cancers15123250
Chicago/Turabian StylePaniagua-Herranz, Lucía, Bernard Doger, Cristina Díaz-Tejeiro, Adrián Sanvicente, Cristina Nieto-Jiménez, Víctor Moreno, Pedro Pérez Segura, Balazs Gyorffy, Emiliano Calvo, and Alberto Ocana. 2023. "Genomic Mapping of Epidermal Growth Factor Receptor and Mesenchymal–Epithelial Transition-Up-Regulated Tumors Identifies Novel Therapeutic Opportunities" Cancers 15, no. 12: 3250. https://doi.org/10.3390/cancers15123250
APA StylePaniagua-Herranz, L., Doger, B., Díaz-Tejeiro, C., Sanvicente, A., Nieto-Jiménez, C., Moreno, V., Pérez Segura, P., Gyorffy, B., Calvo, E., & Ocana, A. (2023). Genomic Mapping of Epidermal Growth Factor Receptor and Mesenchymal–Epithelial Transition-Up-Regulated Tumors Identifies Novel Therapeutic Opportunities. Cancers, 15(12), 3250. https://doi.org/10.3390/cancers15123250





