Implementation of Nurse Navigation Improves Rate of Molecular Tumor Testing for Ovarian Cancer in a Gynecologic Oncology Practice
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Study Population
3.2. Clinical Impact
3.3. Timeliness of Testing
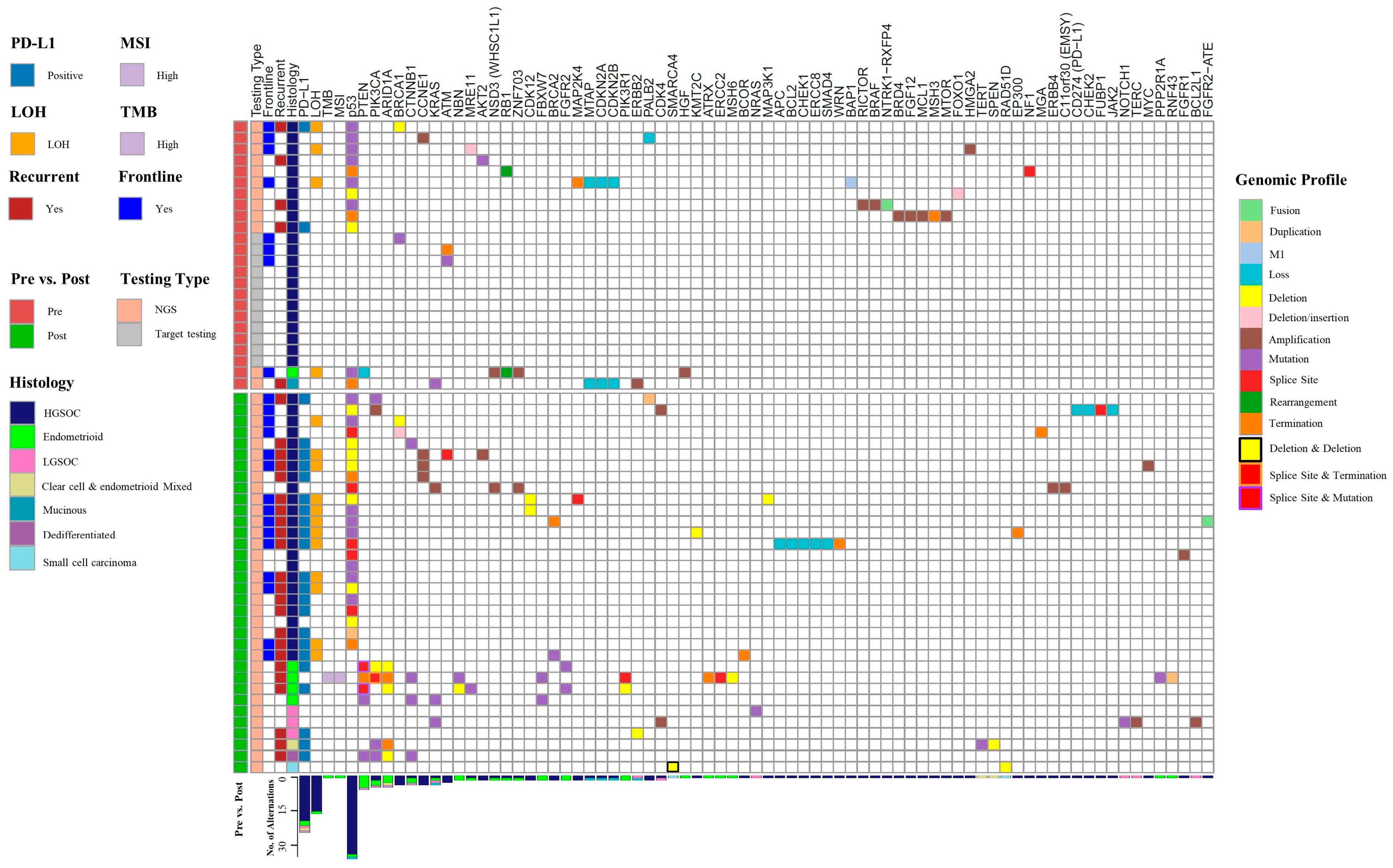
3.4. Mutational Analysis
3.5. Germline Testing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited Mutations in Women With Ovarian Carcinoma. JAMA Oncol. 2016, 2, 482–490. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Norquist, B.; Lacchetti, C.; Armstrong, D.; Grisham, R.N.; Goodfellow, P.J.; Kohn, E.C.; Levine, D.A.; Liu, J.F.; Lu, K.H.; et al. Germline and Somatic Tumor Testing in Epithelial Ovarian Cancer: ASCO Guideline. J. Clin. Oncol. 2020, 38, 1222–1245. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic (Version 1.2023). Available online: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf (accessed on 12 November 2022).
- National Comprehensive Cancer Network. Ovarian Cancer Including Fallopian Tube Cancer and Primary Peritoneal Cancer (Version 5.2022). Available online: https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf (accessed on 9 November 2022).
- Yamamoto, H.; Hirasawa, A. Homologous Recombination Deficiencies and Hereditary Tumors. Int. J. Mol. Sci. 2021, 23, 348. [Google Scholar] [CrossRef]
- Harbin, L.M.; Gallion, H.H.; Allison, D.B.; Kolesar, J.M. Next Generation Sequencing and Molecular Biomarkers in Ovarian Cancer—An Opportunity for Targeted Therapy. Diagnostics 2022, 12, 842. [Google Scholar] [CrossRef] [PubMed]
- Gamble, C.R.; Huang, Y.; Wright, J.D.; Hou, J.Y. Precision medicine testing in ovarian cancer: The growing inequity between patients with commercial vs medicaid insurance. Gynecol. Oncol. 2021, 162, 18–23. [Google Scholar] [CrossRef]
- Lin, J.; Sharaf, R.N.; Saganty, R.; Ahsan, D.; Feit, J.; Khoury, A.; Bergeron, H.; Chapman-Davis, E.; Cantillo, E.; Holcomb, K.; et al. Achieving universal genetic assessment for women with ovarian cancer: Are we there yet? A systematic review and meta-analysis. Gynecol. Oncol. 2021, 162, 506–516. [Google Scholar] [CrossRef]
- Kurian, A.W.; Ward, K.C.; Howlader, N.; Deapen, D.; Hamilton, A.S.; Mariotto, A.; Miller, D.; Penberthy, L.S.; Katz, S.J. Genetic Testing and Results in a Population-Based Cohort of Breast Cancer Patients and Ovarian Cancer Patients. J. Clin. Oncol. 2019, 37, 1305–1315. [Google Scholar] [CrossRef]
- Aversano, M.J.; Boehmer, L.M.; Spira, A. Improving Cancer Care Delivery: Learnings for Oncology Nurses and Patient Navigation From a National Quality Survey. J. Adv. Pract. Oncol. 2022, 13, 484–493. [Google Scholar] [CrossRef]
- McAllister, K.; Schmitt, M. Impact of a Nurse Navigator on Genomic Testing and Timely Treatment Decision Making in Patients With Breast Cancer. Clin. J. Oncol. Nurs. 2015, 19, 510–512. [Google Scholar] [CrossRef]
- Basu, M.; Linebarger, J.; Gabram, S.G.A.; Patterson, S.G.; Amin, M.; Ward, K.C. The effect of nurse navigation on timeliness of breast cancer care at an academic comprehensive cancer center. Cancer 2013, 119, 2524–2531. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Boehmer, L.M.; Celestin, C.; Yu, H.; Spigel, D.R. Improving Care for Patients With Stage III or IV NSCLC: Learnings for Multidisciplinary Teams From the ACCC National Quality Survey. JCO Oncol. Pract. 2021, 17, e1120–e1130. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Chen, Q.; Allison, D.; El Khouli, R.; Peh, K.H.; Mobley, J.; Anderson, A.; Durbin, E.B.; Goodin, D.; Villano, J.L.; et al. Molecular Tumor Board Review and Improved Overall Survival in Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2021, 5, 1530–1539. [Google Scholar] [CrossRef]
- Miller, R.W.; Hutchcraft, M.L.; Weiss, H.L.; Wu, J.; Wang, C.; Liu, J.; Jayswal, R.; Buchanan, M.; Anderson, A.; Allison, D.B.; et al. Molecular Tumor Board–Assisted Care in an Advanced Cancer Population: Results of a Phase II Clinical Trial. JCO Precis. Oncol. 2022, 6, e2100524. [Google Scholar] [CrossRef]
- Larson, K.L.; Huang, B.; Weiss, H.L.; Hull, P.; Westgate, P.M.; Miller, R.W.; Arnold, S.M.; Kolesar, J.M. Clinical Outcomes of Molecular Tumor Boards: A Systematic Review. JCO Precis. Oncol. 2021, 5, 1122–1132. [Google Scholar] [CrossRef]
- Miller, R.; Leary, A.; Scott, C.; Serra, V.; Lord, C.; Bowtell, D.; Chang, D.; Garsed, D.; Jonkers, J.; Ledermann, J.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef]
- Hodgson, D.R.; Dougherty, B.A.; Lai, Z.; Fielding, A.; Grinsted, L.; Spencer, S.; O’connor, M.J.; Ho, T.W.; Robertson, J.D.; Lanchbury, J.S.; et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br. J. Cancer 2018, 119, 1401–1409. [Google Scholar] [CrossRef]
- Kondrashova, O.; Nguyen, M.; Shield-Artin, K.; Tinker, A.V.; Teng, N.N.H.; Harrell, M.I.; Kuiper, M.J.; Ho, G.Y.; Barker, H.; Jasin, M.; et al. Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2017, 7, 984–998. [Google Scholar] [CrossRef]
- McCabe, N.; Turner, N.C.; Lord, C.J.; Kluzek, K.; Białkowska, A.; Swift, S.; Giavara, S.; O’Connor, M.J.; Tutt, A.N.; Zdzienicka, M.Z.; et al. Deficiency in the Repair of DNA Damage by Homologous Recombination and Sensitivity to Poly(ADP-Ribose) Polymerase Inhibition. Cancer Res. 2006, 66, 8109–8115. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Harter, P.; Mouret-Reynier, M.A.; Pignata, S.; Cropet, C.; González-Martín, A.; Bogner, G.; Fujiwara, K.; Vergote, I.; Colombo, N.; Nøttrup, T.J.; et al. Efficacy of maintenance olaparib plus bevacizumab according to clinical risk in patients with newly diagnosed, advanced ovarian cancer in the phase III PAOLA-1/ENGOT-ov25 trial. Gynecol. Oncol. 2021, 164, 254–264. [Google Scholar] [CrossRef]
- Matulonis, U.A.; Shapira-Frommer, R.; Santin, A.D.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Provencher, D.M.; et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase II KEYNOTE-100 study. Ann. Oncol. 2019, 30, 1080–1087. [Google Scholar] [CrossRef]
- Varga, A.; Piha-Paul, S.; Ott, P.A.; Mehnert, J.M.; Berton-Rigaud, D.; Morosky, A.; Yang, P.; Ruman, J.; Matei, D. Pembrolizumab in patients with programmed death ligand 1–positive advanced ovarian cancer: Analysis of KEYNOTE-028. Gynecol. Oncol. 2018, 152, 243–250. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Subbiah, V.; Wolf, J.; Konda, B.; Kang, H.; Spira, A.; Weiss, J.; Takeda, M.; Ohe, Y.; Khan, S.; Ohashi, K.; et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): A phase 1/2, open-label, basket trial. Lancet Oncol. 2022, 23, 1261–1273. [Google Scholar] [CrossRef]
- Salama, A.K.S.; Li, S.; Macrae, E.R.; Park, J.-I.; Mitchell, E.P.; Zwiebel, J.A.; Chen, H.X.; Gray, R.J.; McShane, L.M.; Rubinstein, L.V.; et al. Dabrafenib and Trametinib in Patients With Tumors With BRAFV600E Mutations: Results of the NCI-MATCH Trial Subprotocol H. J. Clin. Oncol. 2020, 38, 3895–3904. [Google Scholar] [CrossRef]
- Jardim, D.L.F.; Schwaederle, M.; Wei, C.; Lee, J.J.; Hong, D.S.; Eggermont, A.M.; Schilsky, R.L.; Mendelsohn, J.; Lazar, V.; Kurzrock, R. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. Gynecol. Oncol. 2015, 107, djv253. [Google Scholar] [CrossRef]
- Schwaederle, M.; Zhao, M.; Lee, J.J.; Lazar, V.; Leyland-Jones, B.; Schilsky, R.L.; Mendelsohn, J.; Kurzrock, R. Association of Biomarker-Based Treatment Strategies With Response Rates and Progression-Free Survival in Refractory Malignant Neoplasms A Meta-analysis. JAMA Oncol. 2016, 2, 1452–1459. [Google Scholar] [CrossRef]
- Da Cunha Colombo Bonadio, R.R.; Fogace, R.N.; Miranda, V.C.; Diz, M.D.P.E. Homologous recombination deficiency in ovarian cancer: A review of its epidemiology and management. Clinics 2018, 73 (Suppl. 1), e450s. [Google Scholar] [CrossRef]
| All | Pre NN | Post NN | p-Value * | |
|---|---|---|---|---|
| n = 78 | n = 34 | n = 44 | ||
| Age at diagnosis, year, mean (range) | 61.67 (23–89) | 63.76 (32–89) | 60.05 (23–82) | 0.22 |
| Age at diagnosis, n (%) | 0.14 | |||
| <50 years old | 15 (19.23) | 4 (11.76) | 11 (25) | |
| ≥50 years old | 63 (80.77) | 30 (88.24) | 33 (75) | |
| BMI, mean (IQR) | 30.43 (12.7) | 29.78 (11.6) | 30.93 (14.8) | 0.57 |
| Race/Ethnicity, n (%) | 0.43 | |||
| Non-Hispanic White | 73 (93.59) | 31 (91.81) | 42 (95.45) | |
| Black | 2 (2.56) | 1 (2.94) | 1 (2.27) | |
| Hispanic | 2 (2.56) | 2 (5.88) | 0 | |
| Unknown | 1 (1.28) | 0 | 1 (2.27) | |
| Primary, n (%) | 0.81 | |||
| Ovary | 62 (79.49) | 26 (76.47) | 36 (81.82) | |
| Fallopian | 13 (16.67) | 7 (20.59) | 6 (13.64) | |
| Peritoneal | 3 (3.85) | 1 (2.94) | 2 (4.55) | |
| Histology, n (%) | 0.07 | |||
| HGSOC | 59 (75.64) | 30 (88.24) | 29 (65.91) | |
| LGSOC | 4 (5.13) | 0 | 4 (9.09) | |
| Endometrioid | 10 (12.82) | 2 (5.88) | 8 (18.18) | |
| Mucinous | 1 (1.28) | 1 (2.94) | 0 | |
| Clear cell and endometrioid mixed | 2 (2.56) | 1 (2.94) | 1 (2.27) | |
| Dedifferentiated | 1 (1.28) | 0 | 1 (2.27) | |
| Small cell carcinoma | 1 (1.28) | 0 | 1 (2.27) | |
| Initial Stage, n (%) | 0.40 | |||
| IA | 14 (17.95) | 4 (11.76) | 10 (22.73) | |
| IB | 1 (1.28) | 0 | 1 (2.27) | |
| IC1 | 3 (3.85) | 2 (5.88) | 1 (2.27) | |
| IC2 | 2 (2.56) | 1 (2.94) | 1 (2.27) | |
| IC3 | 2 (2.56) | 2 (5.88) | 0 | |
| IIA | 2 (2.56) | 2 (5.88) | 0 | |
| IIB | 6 (7.69) | 1 (2.94) | 5 (11.36) | |
| IIIA1i | 1 (1.28) | 1 (2.94) | 0 | |
| IIIA1ii | 1 (1.28) | 0 | 1 (2.27) | |
| IIIA2 | 1 (1.28) | 0 | 1 (2.27) | |
| IIIB | 1 (1.28) | 0 | 1 (2.27) | |
| IIIC | 28 (35.90) | 13 (38.24) | 15 (34.09) | |
| IVA | 8 (10.26) | 4 (11.76) | 4 (9.09) | |
| IVB | 7 (8.97) | 4 (11.76) | 3 (6.82) | |
| Specimen, n (%) | 0.40 | |||
| Pretreatment biopsy | 6 (7.89) | 2 (5.88) | 4 (9.52) | |
| Primary surgery | 34 (44.74) | 17 (50) | 17 (40.48) | |
| Interval debulking | 13 (17.11) | 3 (8.82) | 10 (23.81) | |
| Recurrence | 5 (6.58) | 2 (5.88) | 3 (7.14) | |
| Unknown | 18 (23.68) | 10 (29.41) | 8 (19.05) | |
| Current Disease Status, n (%) | 0.0002 | |||
| Initial therapy | 34 (43.59) | 6 (17.65) | 28 (63.64) | |
| Recurrent | 11 (14.10) | 7 (20.59) | 4 (9.09) | |
| Persistent | 7 (8.97) | 6 (17.65) | 1 (2.27) | |
| No evidence of disease | 25 (32.05) | 14 (41.18) | 11 (25) | |
| MD, n (%) | 0.77 | |||
| 1 | 17 (21.79) | 8 (23.53) | 9 (20.45) | |
| 2 | 8 (10.26) | 3 (8.82) | 5 (11.36) | |
| 3 | 8 (10.26) | 4 (11.76) | 4 (9.09) | |
| 4 | 22 (28.21) | 9 (26.47) | 13 (29.55) | |
| 5 | 11 (14.10) | 3 (8.82) | 8 (18.18) | |
| 6 | 12 (15.38) | 7 (20.59) | 5 (11.36) | |
| Hypertension, n (%) | 0.24 | |||
| No | 31 (39.74) | 11 (32.35) | 20 (45.45) | |
| Yes | 47 (60.26) | 23 (67.65) | 24 (54.55) | |
| Diabetes, n (%) | 0.50 | |||
| No | 59 (75.64) | 27 (79.41) | 32 (72.73) | |
| Yes | 19 (24.36) | 7 (20.59) | 12 (27.27) | |
| Cardiac, n (%) | 0.51 | |||
| No | 64 (82.05) | 29 (85.29) | 35 (79.55) | |
| Yes | 14 (17.95) | 5 (14.71) | 9 (20.45) | |
| Respiratory, n (%) | 0.56 | |||
| No | 62 (79.49) | 26 (76.47) | 36 (81.82) | |
| Yes | 16 (20.51) | 8 (23.53) | 8 (18.18) | |
| Neuro, n (%) | 1.00 | |||
| No | 74 (94.87) | 32 (94.12) | 42 (95.45) | |
| Yes | 4 (5.13) | 2 (5.88) | 2 (4.55) | |
| Renal, n (%) | 0.65 | |||
| No | 73 (93.59) | 31 (91.18) | 42 (95.45) | |
| Yes | 5 (6.41) | 3 (8.82) | 2 (4.55) | |
| Total Comorbidities, n (%) | 0.88 | |||
| 0 | 20 (25.64) | 7 (20.59) | 13 (29.55) | |
| 1 | 26 (33.33) | 13 (38.24) | 13 (29.55) | |
| 2 | 19 (24.36) | 8 (23.53) | 11 (25) | |
| 3 | 11 (14.10) | 5 (14.71) | 6 (13.64) | |
| 4 | 2 (2.56) | 1 (2.94) | 1 (2.27) |
| Pre-NN (n = 34) | Post-NN (n = 44) | p-Value * | |
|---|---|---|---|
| NGS, n (%) | 12 (35.29) | 34 (77.27) | 0.0002 |
| No NGS, n (%) | 22 (64.71) | 10 (22.72) | |
| No testing | 10 (29.41) | 10 (22.72) | |
| Targeted panel testing | 12 (35.29) | 0 |
| Demographic | Pre NN N = 34 | Odds Ratio # | 95% CI | p-Value | Post NN N = 44 | Odds Ratio # | 95% CI | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| NGS-No n = 22 | NGS-Yes n = 12 | NGS-No n = 10 | NGS-Yes n = 34 | |||||||
| Age, mean (SD) | 62.27 (12.94) | 66.5 (9.46) | 1.03 | 0.97–1.10 | 0.32 | 61.4 (11.12) | 59.65 (15.15) | 0.99 | 0.94–1.04 | 0.73 |
| Age, n | 6.08 | 0.30–123.2 | 0.24 | 0.70 | 0.12–3.90 | 0.68 | ||||
| <50 yo | 4 | 0 * | 2 | 9 | ||||||
| ≥50 yo | 18 | 12 | 8 | 25 | ||||||
| BMI, mean (SD) | 29.49 (7.93) | 30.32 (8.97) | 1.01 | 0.93–1.10 | 0.78 | 31.75 (9.49) | 30.69 (9.33) | 0.99 | 0.92–1.07 | 0.75 |
| MD | 0.71 | 0.09 | ||||||||
| 1 (reference) | 5 | 3 | 2 | 7 | ||||||
| 2 | 2 | 1 | 0.83 | 0.05–13.63 | 3 | 2 | 0.24 * | 0.03–2.07 | ||
| 3 | 3 | 1 | 0.56 | 0.04–8.09 | 2 | 2 | 0.33 * | 0.04–3.21 | ||
| 4 | 4 | 5 | 2.08 | 0.30–14.55 | 0 | 13 | 9.00 * | 0.38–213.17 | ||
| 5 | 2 | 1 | 0.83 | 0.05–13.63 | 0 | 8 | 5.67 * | 0.23–137.80 | ||
| 6 | 6 | 1 | 0.28 | 0.02–3.58 | 3 | 2 | 0.24 * | 0.03–2.07 | ||
| Total Comorbidities | 0.41 | 0.03 | ||||||||
| 0–2 (reference) | 19 | 9 | 6 | 31 | ||||||
| >2 | 3 | 3 | 2.11 | 0.35–12.59 | 4 | 3 | 0.16 | 0.03–0.90 | ||
| Primary | 0.88 | 0.46 | ||||||||
| Ovary (reference) | 17 | 9 | 9 | 27 | ||||||
| Fallopian/ Peritoneal | 5 | 3 | 1.13 | 0.22–5.86 | 1 | 7 | 2.33 | 0.25–21.63 | ||
| Histology | 0.52 | 0.23 | ||||||||
| HGSOC (reference) | 20 | 10 | 5 | 24 | ||||||
| Other | 2 | 2 | 2.00 | 0.24–16.36 | 5 | 10 | 0.42 | 0.10–1.76 | ||
| Initial Stage | 0.11 | 0.03 | ||||||||
| Early (reference) | 10 | 2 | 7 | 11 | ||||||
| Late | 12 | 10 | 4.18 | 0.74–23.61 | 2 | 23 | 6.13 | 1.20–31.33 | ||
| Specimen | 0.30 | 0.95 | ||||||||
| Pre-treatment biopsy (reference) | 0 | 2 | 0 | 4 | ||||||
| Primary surgery | 11 | 6 | 0.11 * | 0.01–2.73 | 0 | 17 | 3.89 * | 0.07–224.22 | ||
| Interval debulking | 1 | 2 | 0.33 * | 0.01–12.82 | 0 | 10 | 2.33 * | 0.04–136.98 | ||
| Recurrence | 0 | 2 | 1.00 * | NA | 0 | 3 | 0.78 * | 0.01–49.9 | ||
| Unknown | 10 | 0 | NA | NA | 8 | 0 | NA | NA | ||
| Current Disease Status | 0.32 | 0.64 | ||||||||
| Initial therapy (reference) | 3 | 3 | 7 | 21 | ||||||
| NED | 11 | 3 | 0.27 | 0.04–2.11 | 3 | 8 | 0.85 * | 0.19–3.79 | ||
| Recurrent/Persistent | 7 | 6 | 0.86 | 0.12–5.94 | 0 | 5 | 3.84 * | 0.19–78.00 | ||
| Reason | Pre-NN | Post-NN | p-Value * |
|---|---|---|---|
| n = 22 | n = 10 | ||
| Reason for no NGS, n (%) | 0.0004 | ||
| Targeted screening | 13 (59.08) | 0 | |
| Unknown | 3 (13.64) | 0 | |
| Cancer characteristics (e.g., stage/grade/type) | 2 (9.09) | 5 (50) | |
| Community provider primary | 2 (9.09) | 1 (10) | |
| Not enough tissue | 1 (4.55) | 3 (30) | |
| Wait for recurrence | 1 (4.55) | 0 | |
| Sent on second primary malignancy | 0 | 1 (10) | |
| Not ordered (appropriate reason) **, n (%) | 3 (13.64) | 5 (50) | 0.0722 |
| Not ordered (inappropriate reason) ***, n (%) | 19 (86.36) | 5 (50) |
| Time Interval | Pre NN (n = 24) | Post NN (n = 34) | p-Value * |
|---|---|---|---|
| n = 23 | n = 34 | <0.0001 | |
| Specimen collection date to date resulted ***, mean number days (95% CI) | 145.2 (103.5–187.0) | 42.8 (31.6–54.0) | |
| n = 22 | n = 33 | <0.0001 | |
| Specimen collected date to physician order date, mean number days (95% CI) | 102.6 (69.3–136.0) | 24.7 (13.4–35.9) | |
| n = 21 | n = 33 | 0.48 | |
| Physician order date to date received by testing company **, mean number days (95% CI) | 6.1 (2.3–10.0) | 4.7 (4.7–6.1) | |
| n = 22 | n = 34 | 0.0003 | |
| Date received by testing company ** to date resulted ***, mean number days (95% CI) | 23.1 (16.0–30.1) | 12.3 (11.3–13.2) |
| Pre NN n = 24 | Post NN n = 34 | p-Value * | |
|---|---|---|---|
| Tumor mutation, n (%) | 0.0001 | ||
| Yes | 15 (62.5) | 34 (100) | |
| No | 9 (37.5) | 0 | |
| Frontline actionable mutations **, n (%) | 0.41 | ||
| Yes | 8 (33.3) | 15 (44.1) | |
| No | 16 (66.6) | 19 (55.8) | |
| Recurrence actionable mutations ***, n (%) | 0.0005 | ||
| Yes | 5 (20.8) | 23 (67.6) | |
| No | 19 (79.2) | 11 (32.4) | |
| LOH results, n (%) | <0.0001 | ||
| Yes | 9 (37.5) | 33 (97.1) | |
| No | 15 (62.5) | 1 (2.9) | |
| LOH High (score ≥ 16), n (%) | n = 9 | n = 33 | 0.71 |
| Yes | 4 (44.4) | 12 (36.4) | |
| No | 5 (55.6) | 21 (63.6) |
| Pre NN | Post NN | p-Value * | |
| n = 34 | n = 44 | ||
| Physician recommended germline testing, n (%) | 1.00 | ||
| Yes | 31 (91.2) | 41 (93.2) | |
| No | 3 (8.8) | 3 (6.8) | |
| Germline testing completed, n (%) | 0.008 | ||
| Yes | 27 (79.4) | 22 (50) | |
| No | 7 (20.6) | 22 (50) | |
| n = 27 | n = 22 | ||
| Germline testing results, n (%) | 0.31 | ||
| Negative/VUS | 23 (85.2) | 16 (72.7) | |
| Positive | 4 (14.8) | 6 (27.3) | |
| Germline BRCA or HRD mutated ** | 1.00 | ||
| Yes | 4 (14.8) | 4 (18.2) | |
| No | 23 (85.2) | 18 (81.8) | |
| n = 4 | n = 6 | ||
| Positive germline mutation results, n (%) | 0.30 | ||
| ATM | 1 (25) | 0 | |
| BRCA1 | 3 (75) | 1 (16.7) | |
| BRCA2 | 0 | 2 (33.3) | |
| NF1 | 0 | 1 (16.7) | |
| PALB2 | 0 | 1 (16.7) | |
| WRN | 0 | 1 (16.7) | |
| n = 6 | n = 4 | ||
| Germline VUS, n (%) | 0.22 | ||
| ATM VUS | 0 | 2 (50) | |
| CDKN1B VUS | 1 (16.7) | 0 | |
| MSH6 VUS | 1 (16.7) | 0 | |
| PMS2 VUS | 0 | 1 (25) | |
| DICER1 VUS | 2 (33.3) | 0 | |
| PALB2 VUS | 1 (16.7) | 0 | |
| POLD1 VUS | 1 (16.7) | 0 | |
| SMARCA4 VUS | 0 | 1 (25) | |
| Time Interval Date physician recommended genetics to germline test results, mean number days (95% CI) | n = 19 135.4 (88.4–182.5) | n = 16 69.1 (34.8–103.5) | 0.03 |
| Date physician recommended genetics to genetic counseling, mean number days (95% CI) | n = 19 82.5 (43.4–121.7) | n = 18 48.4 (17.0–79.8) | 0.16 |
| Date of genetic counseling to germline test results, mean number days (95% CI) | n = 19 52.9 (12.3–93.5) | n = 15 24.2 (11.9–36.5) | 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rives, T.A.; Pavlik, H.; Li, N.; Qasrawi, L.; Yan, D.; Pickarski, J.; Dietrich, C.S.; Miller, R.W.; Ueland, F.R.; Kolesar, J.M. Implementation of Nurse Navigation Improves Rate of Molecular Tumor Testing for Ovarian Cancer in a Gynecologic Oncology Practice. Cancers 2023, 15, 3192. https://doi.org/10.3390/cancers15123192
Rives TA, Pavlik H, Li N, Qasrawi L, Yan D, Pickarski J, Dietrich CS, Miller RW, Ueland FR, Kolesar JM. Implementation of Nurse Navigation Improves Rate of Molecular Tumor Testing for Ovarian Cancer in a Gynecologic Oncology Practice. Cancers. 2023; 15(12):3192. https://doi.org/10.3390/cancers15123192
Chicago/Turabian StyleRives, Taylor A., Heather Pavlik, Ning Li, Lien Qasrawi, Donglin Yan, Justine Pickarski, Charles S. Dietrich, Rachel W. Miller, Frederick R. Ueland, and Jill M. Kolesar. 2023. "Implementation of Nurse Navigation Improves Rate of Molecular Tumor Testing for Ovarian Cancer in a Gynecologic Oncology Practice" Cancers 15, no. 12: 3192. https://doi.org/10.3390/cancers15123192
APA StyleRives, T. A., Pavlik, H., Li, N., Qasrawi, L., Yan, D., Pickarski, J., Dietrich, C. S., Miller, R. W., Ueland, F. R., & Kolesar, J. M. (2023). Implementation of Nurse Navigation Improves Rate of Molecular Tumor Testing for Ovarian Cancer in a Gynecologic Oncology Practice. Cancers, 15(12), 3192. https://doi.org/10.3390/cancers15123192








