The Role of Microbiota in Pancreatic Cancer
Abstract
Simple Summary
Abstract
1. Introduction
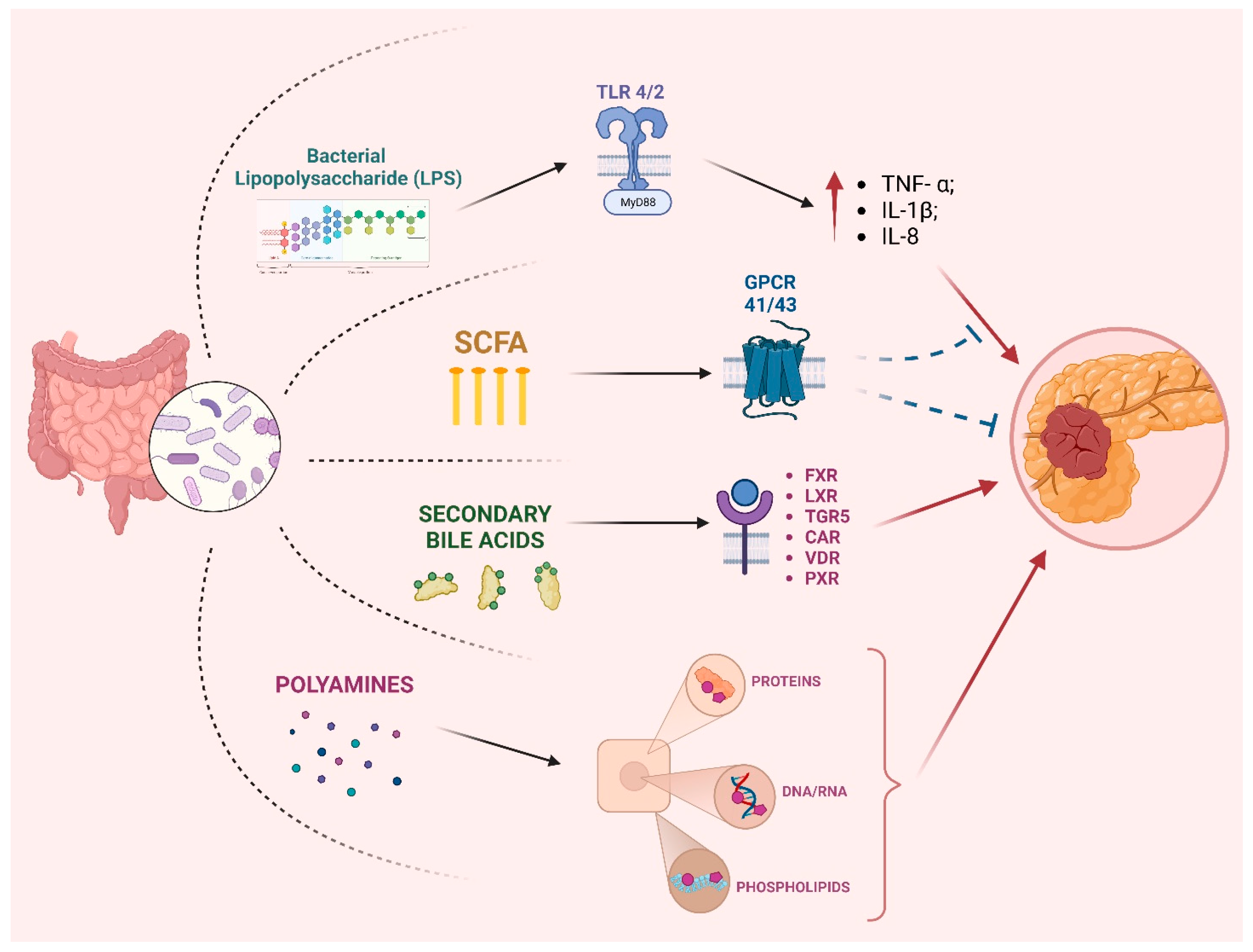
2. Microbiota and Pancreatic Cancer Pathogenesis
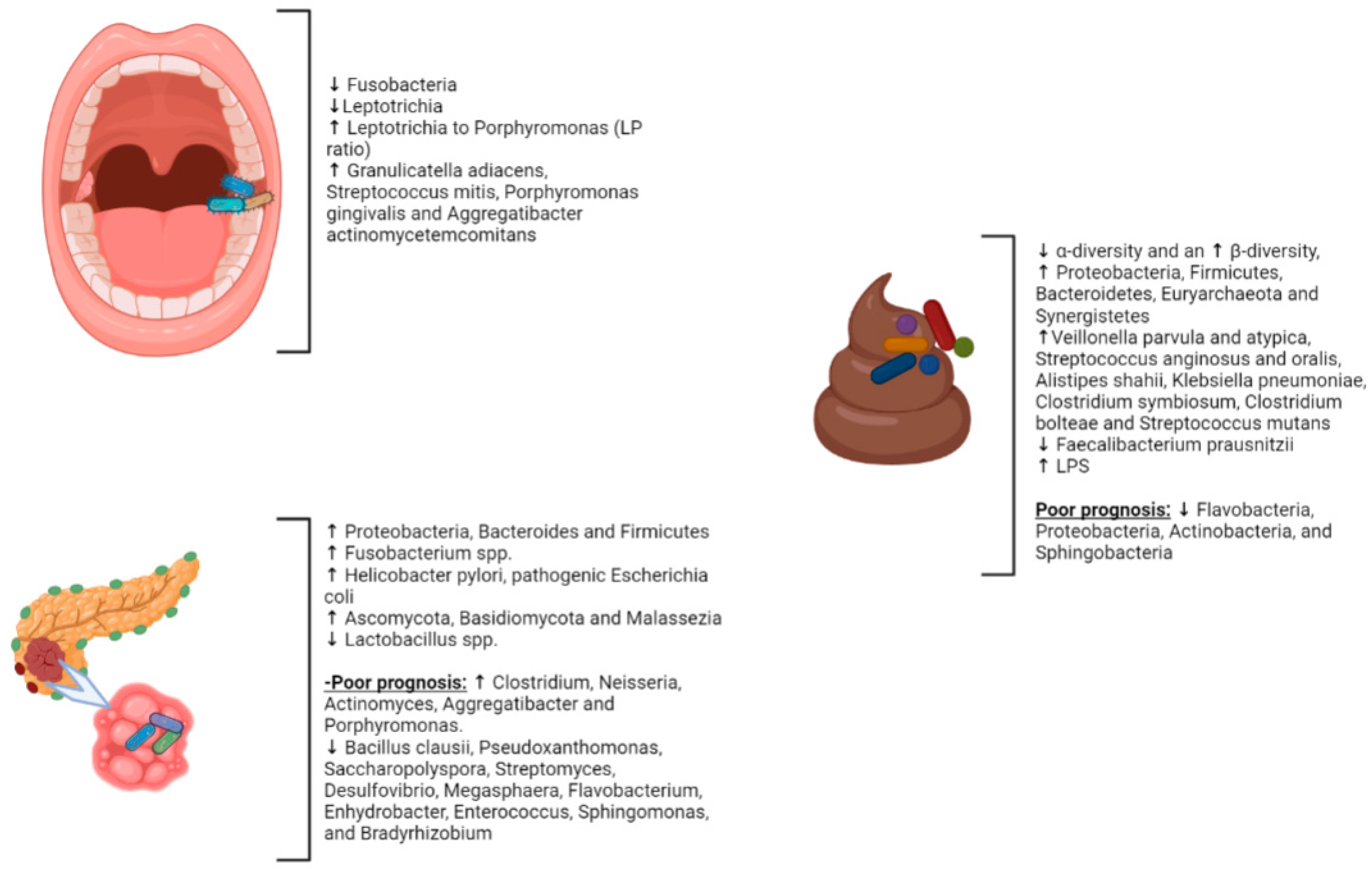
2.1. Gut Microbiota and PC
2.2. Intrapancreatic Microbiota in PC
2.3. Periodontal Diseases and PC
2.4. The Oral Microbiota and PC
3. Role of Microbiota in PC Progression
4. Microbiota Involvement in Therapy Response
4.1. The Microbiome as a Non-Invasive Biomarker in PDAC Management
4.2. Cytotoxic and Immune Therapy
5. Modulation of the Gut Microbiota
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ramakrishna, B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef]
- Flandroy, L.; Poutahidis, T.; Berg, G.; Clarke, G.; Dao, M.C.; Decaestecker, E.; Furman, E.; Haahtela, T.; Massart, S.; Plovier, H.; et al. Human activities and lifestyles impact the interlinked microbiota and health of humans and ecosystems. Sci. Total Environ. 2018, 627, 1018–1038. [Google Scholar] [CrossRef]
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell Mol. Life Sci. 2019, 76, 473–493. [Google Scholar] [CrossRef]
- Thomas, R.M.; Jobin, C. Microbiota in pancreatic health and disease: The next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 53–64. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo, E.; Meier, R.; Chung, M.; Koestler, D.C.; Chen, T.; Paster, B.J.; Charpentier, K.P.; Kelsey, K.T.; Izard, J.; Michaud, D.S. The microbiomes of pancreatic and duodenum tissue overlap and are highly subject-specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol. Biomark. Prev. 2019, 28, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Diehl, G.E.; Longman, R.S.; Zhang, J.X.; Breart, B.; Galan, C.; Cuesta, A.; Schwab, S.R.; Littman, D.R. Microbiota restricts the trafficking of bacteria to mesenteric lymph nodes by CX 3 CR1 hi cells. Nature 2013, 494, 116–120. [Google Scholar] [CrossRef]
- Pietzner, M.; Budde, K.; Rühlemann, M.; Völzke, H.; Homuth, G.; Weiss, F.U.; Lerch, M.M.; Frost, F. Exocrine Pancreatic Function Modulates Plasma Metabolites Through Changes in Gut Microbiota Composition. J. Clin. Endocrinol. Metab. 2021, 106, e2290–e2298. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jin, M.; Liu, Y.; Jin, L. Gut Microbiota: Its Potential Roles in Pancreatic Cancer. Front. Cell. Infect. Microbiol. 2020, 10, 572492. [Google Scholar] [CrossRef]
- Yuan, M.; Xu, Y.; Guo, Z. Association of oral microbiome and pancreatic cancer: A systematic review and meta-analysis. Ther. Adv. Gastroenterol. 2022, 15, 17562848221123980. [Google Scholar] [CrossRef]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef]
- Chang, J.S.; Tsai, C.R.; Chen, L.T.; Shan, Y.S. Investigating the Association Between Periodontal Disease and Risk of Pancreatic Cancer. Pancreas 2016, 45, 134–141. [Google Scholar] [CrossRef]
- Nagata, N.; Nishijima, S.; Kojima, Y.; Hisada, Y.; Imbe, K.; Miyoshi-Akiyama, T.; Suda, W.; Kimura, M.; Aoki, R.; Sekine, K.; et al. Metagenomic Identification of Microbial Signatures Predicting Pancreatic Cancer From a Multinational Study. Gastroenterology 2022, 163, 222–238. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, J.; Zhu, Y. Potential Roles of the Gut Microbiota in Pancreatic Carcinogenesis and Therapeutics. Front. Cell. Infect. Microbiol. 2022, 12, 872019. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, H.; Iida, N.; Kitamura, K.; Terashima, T.; Seishima, J.; Makino, I.; Kannon, T.; Hosomichi, K.; Yamashita, T.; Sakai, Y.; et al. Dysbiotic gut microbiota in pancreatic cancer patients form correlation networks with the oral microbiota and prognostic factors. Am. J. Cancer Res. 2021, 11, 3163–3175. [Google Scholar]
- Zhou, W.; Zhang, D.; Li, Z.; Jiang, H.; Li, J.; Ren, R.; Gao, X.; Li, J.; Wang, X.; Wang, W.; et al. The faecal microbiota of patients with pancreatic ductal adenocarcinoma and autoimmune pancreatitis characterised by metagenomic sequencing. J. Transl. Med. 2021, 19, 215. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, W.; Wu, J. Helicobacter pylori infection and pancreatic cancer risk: A meta-analysis. J. Cancer Res. Ther. 2016, 12, C229–C232. [Google Scholar] [CrossRef] [PubMed]
- Sammallahti, H.; Sarhadi, V.K.; Kokkola, A.; Ghanbari, R.; Rezasoltani, S.; Aghdaei, H.A.; Puolakkainen, P.; Knuutila, S. Oncogenomic Changes in Pancreatic Cancer and Their Detection in Stool. Biomolecules 2022, 12, 652. [Google Scholar] [CrossRef]
- Di Magliano, M.P.; Logsdon, C.D. Roles for KRAS in pancreatic tumour development and progression. Gastroenterology 2013, 144, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Vujasinovic, M.; Dugic, A.; Maisonneuve, P.; Aljic, A.; Berggren, R.; Panic, N.; Valente, R.; Mucelli, R.P.; Waldthaler, A.; Ghorbani, P.; et al. Risk of developing pancreatic cancer in patients with chronic pancreatitis. Pancreatology 2021, 21, S38. [Google Scholar] [CrossRef]
- Van DIjk, S.M.; Hallensleben, N.D.L.; Van Santvoort, H.C.; Fockens, P.; Van Goor, H.; Bruno, M.J.; Besselink, M.G. Acute pancreatitis: Recent advances through randomised trials. Gut 2017, 66, 2024–2032. [Google Scholar] [CrossRef]
- Yadav, D.; Lowenfels, A.B. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013, 144, 1252–1261. [Google Scholar] [CrossRef]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef]
- Elaskandrany, M.; Patel, R.; Patel, M.; Miller, G.; Saxena, D.; Saxena, A. Fungi, host immune response, and tumorigenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G213–G222. [Google Scholar] [CrossRef]
- Conche, C.; Greten, F.R. Fungi Enter the Stage of Colon Carcinogenesis. Immunity 2018, 49, 384–386. [Google Scholar] [CrossRef]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef] [PubMed]
- Sobocki, B.K.; Kaźmierczak-Siedlecka, K.; Folwarski, M.; Hawryłkowicz, V.; Makarewicz, W.; Stachowska, E. Pancreatic cancer and gut microbiome-related aspects: A comprehensive review and dietary recommendations. Nutrients 2021, 13, 4425. [Google Scholar] [CrossRef]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K.; et al. The host microbiome regulates and maintains human health: A primer and perspective for non-microbiologists. Cancer Res. 2017, 77, 1783–1812. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef]
- Djuric, Z. Obesity-associated cancer risk: The role of intestinal microbiota in the aetiology of the host pro-inflammatory state. Transl. Res. 2017, 179, 155–167. [Google Scholar] [CrossRef]
- Sharif, R.; Dawra, R.; Wasiluk, K.; Phillips, P.; Dudeja, V.; Kurt-Jones, E.; Finberg, R.; Saluja, A. Impact of toll-like receptor 4 on the severity of acute pancreatitis and pancreatitis-associated lung injury in mice. Gut 2009, 58, 813–819. [Google Scholar] [CrossRef]
- Ren, Z.; Jiang, J.; Xie, H.; Li, A.; Lu, H.; Xu, S.; Zhou, L.; Zhang, H.; Cui, G.; Chen, X.; et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget 2017, 8, 95176–95191. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumour resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Mirji, G.; Worth, A.; Bhat, S.A.; El Sayed, M.; Kannan, T.; Goldman, A.R.; Tang, H.Y.; Liu, Q.; Auslander, N.; Dang, C.V.; et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci. Immunol. 2022, 7, eabn0704. [Google Scholar] [CrossRef]
- Corrales, L.; Matson, V.; Flood, B.; Spranger, S.; Gajewski, T.F. Innate immune signalling and regulation in cancer immunotherapy. Cell Res. 2017, 27, 96–108. [Google Scholar] [CrossRef]
- Santoni, M.; Andrikou, K.; Sotte, V.; Bittoni, A.; Lanese, A.; Pellei, C.; Piva, F.; Conti, A.; Nabissi, M.; Santoni, G.; et al. Toll-like receptors and pancreatic diseases: From a pathogenetic mechanism to a therapeutic target. Cancer Treat. Rev. 2015, 41, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, D.; Wang, Q.; Zheng, D.; Jiang, X.; Xu, L. LPS induced miR-181a promotes pancreatic cancer cell migration via targeting PTEN and MAP2K4. Dig. Dis. Sci. 2014, 59, 1452–1460. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Pu, N.; Chen, Q.; Zhang, J.; Zhao, G.; Xu, X.; Wang, D.; Kuang, T.; Jin, D.; Lou, W.; et al. Gut-derived lipopolysaccharide remodels tumoral microenvironment and synergises with PD-L1 checkpoint blockade via TLR4/MyD88/AKT/NF-κB pathway in pancreatic cancer. Cell Death Dis. 2021, 12, 1033. [Google Scholar] [CrossRef] [PubMed]
- Leung, P.S.; Chan, Y.C. Role of oxidative stress in pancreatic inflammation. Antioxid. Redox Signal. 2009, 11, 135–166. [Google Scholar] [CrossRef] [PubMed]
- Noureldein, M.H.; Eid, A.A. Gut microbiota and mTOR signalling: Insight on a new pathophysiological interaction. Microb. Pathog. 2018, 118, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Hao, J. Dedifferentiation of Epithelial Cells Incorporates Immune Reprogramming. Trends Cell Biol. 2021, 31, 237–240. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, S.; Dong, S.; Xu, H.; Zhou, W. Association of the Microbiota and Pancreatic Cancer: Opportunities and Limitations. Front. Immunol. 2022, 13, 844401. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef] [PubMed]
- Kartal, E.; Schmidt, T.S.B.; Molina-Montes, E.; Rodríguez-Perales, S.; Wirbel, J.; Maistrenko, O.M.; Akanni, W.A.; Alashkar Alhamwe, B.; Alves, R.J.; Carrato, A.; et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut 2022, 71, 1359–1372. [Google Scholar] [CrossRef] [PubMed]
- Chakladar, J.; Kuo, S.Z.; Castaneda, G.; Li, W.T.; Gnanasekar, A.; Yu, M.A.; Chang, E.Y.; Wang, X.Q.; Ongkeko, W.M. The pancreatic microbiome is associated with carcinogenesis and worse prognosis in males and smokers. Cancers 2020, 12, 2672. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Chu, C.S.; Yang, C.Y.; Yeh, C.C.; Lin, R.T.; Chen, C.C.; Bai, L.Y.; Hung, M.C.; Lin, C.C.; Wu, C.Y.; Lin, J.T. Endoscopic ultrasound-guided fine-needle biopsy as a tool for studying the intra-tumoral microbiome in pancreatic ductal adenocarcinoma: A pilot study. Sci. Rep. 2022, 12, 107. [Google Scholar] [CrossRef]
- Kurihara, H.; Nosho, K.; Mitsuhashi, K.; Sukawa, Y.; Matsunaga, Y.; Ito, M.; Kanno, S.; Igarashi, H.; Yamamoto, I.; Adachi, Y.; et al. Association of tumour fusobacterium status in pancreatic cancers with molecular alterations and patient survival. Gastroenterology 2015, 148, S589. [Google Scholar] [CrossRef]
- Yu, D.; Wang, T.; Liang, D.; Mei, Y.; Zou, W.; Guo, S. The Landscape of Microbial Composition and Associated Factors in Pancreatic Ductal Adenocarcinoma Using RNA-Seq Data. Front. Oncol. 2021, 11, 651350. [Google Scholar] [CrossRef]
- Lee, A.A.; Wang, Q.L.; Kim, J.; Babic, A.; Zhang, X.; Perez, K.; Ng, K.; Nowak, J.; Rifai, N.; Sesso, H.D.; et al. Helicobacter pylori Seropositivity, ABO Blood Type, and Pancreatic Cancer Risk from 5 Prospective Cohorts. Clin. Transl. Gastroenterol. 2023, 14, e00573. [Google Scholar] [CrossRef]
- Nilsson, H.O.; Wadström, T.; Stenram, U.; Ihse, I. Helicobacter species ribosomal DNA in the pancreas, stomach and duodenum of pancreatic cancer patients. World J. Gastroenterol. 2006, 12, 3038–3043. [Google Scholar] [CrossRef] [PubMed]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef] [PubMed]
- Sendler, M.; Maertin, S.; John, D.; Persike, M.; Weiss, F.U.; Krüger, B.; Wartmann, T.; Wagh, P.; Halangk, W.; Schaschke, N.; et al. Cathepsin B activity initiates apoptosis via digestive protease activation in pancreatic acinar cells and experimental pancreatitis. J. Biol. Chem. 2016, 291, 14717–14731. [Google Scholar] [CrossRef]
- Lerch, M.M.; Saluja, A.K.; Dawra, R.; Saluja, M.; Steer, M.L. The effect of chloroquine administration on two experimental models of acute pancreatitis. Gastroenterology 1993, 104, 1768–1779. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Nandy, S.; Dudeja, M.; Das, A.K.; Tiwari, R. Community-acquired bacteremia by Sphingomonas paucimobilis: Two rare case reports. J. Clin. Diagn. Res. 2013, 7, 2947–2949. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, Y.; Guo, S.; Mei, Z.; Liao, H.; Dong, H.; Wu, K.; Ye, H.; Zhang, Y.; Zhu, Y.; et al. The tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun. Biol. 2021, 4, 1019. [Google Scholar] [CrossRef]
- Charlebois, A.; Jacques, M.; Archambault, M. Biofilm formation of Clostridium perfringens and its exposure to low-dose antimicrobials. Front. Microbiol. 2014, 5, 183. [Google Scholar] [CrossRef]
- Yu, J.; Ploner, A.; Chen, M.S.; Zhang, J.; Sandborgh-Englund, G.; Ye, W. Poor dental health and risk of pancreatic cancer: A nationwide registry-based cohort study in Sweden, 2009–2016. Br. J. Cancer 2022, 127, 2133–2140. [Google Scholar] [CrossRef]
- Gerlovin, H.; Michaud, D.S.; Cozier, Y.C.; Palmer, J.R. Oral health in relation to pancreatic cancer risk in African American women. Cancer Epidemiol. Biomark. Prev. 2019, 28, 675–679. [Google Scholar] [CrossRef]
- Michaud, D.S.; Joshipura, K.; Giovannucci, E.; Fuchs, C.S. A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J. Natl. Cancer Inst. 2007, 99, 171–175. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Amar, S.; Lowenfels, A.B. Periodontal disease, edentulism, and pancreatic cancer: A meta-analysis. Ann. Oncol. 2017, 28, 985–995. [Google Scholar] [CrossRef]
- Heikkilä, P.; But, A.; Sorsa, T.; Haukka, J. Periodontitis and cancer mortality: Register-based cohort study of 68,273 adults in a 10-year follow-up. Int. J. Cancer 2018, 142, 2244–2253. [Google Scholar] [CrossRef]
- Ahn, J.; Segers, S.; Hayes, R.B. Periodontal disease, Porphyromonas gingivalis serum antibody levels and or digestive cancer mortality. Carcinogenesis 2012, 33, 1055–1058. [Google Scholar] [CrossRef]
- Tan, Q.; Ma, X.; Yang, B.; Liu, Y.; Xie, Y.; Wang, X.; Yuan, W.; Ma, J. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes 2022, 14, 2073785. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T.W. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef]
- Torres, P.J.; Fletcher, E.M.; Gibbons, S.M.; Bouvet, M.; Doran, K.S.; Kelley, S.T. Characterisation of the salivary microbiome in patients with pancreatic cancer. PeerJ 2015, 3, e1373. [Google Scholar] [CrossRef]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.H.; Grote, V.A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Jobin, C.; Thomas, R.M. Implications of the microbiome in the development and treatment of pancreatic cancer: Thinking outside of the box by looking inside the gut. Neoplasia 2021, 23, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Sexton, R.E.; Uddin, M.H.; Bannoura, S.; Khan, H.Y.; Mzannar, Y.; Li, Y.; Aboukameel, A.; Al-Hallak, M.N.; Al-Share, B.; Mohamed, A.; et al. Connecting the Human Microbiome and Pancreatic Cancer. Cancer Metastasis Rev. 2022, 41, 317–331. [Google Scholar] [CrossRef]
- Swidnicka-Siergiejko, A.K.; Gomez-Chou, S.B.; Cruz-Monserrate, Z.; Deng, D.; Liu, Y.; Huang, H.; Ji, B.; Azizian, N.; Daniluk, J.; Lu, W.; et al. Chronic inflammation initiates multiple forms of K-Ras-independent mouse pancreatic cancer in the absence of TP53. Oncogene 2017, 36, 3149–3158. [Google Scholar] [CrossRef]
- Tjomsland, V.; Bojmar, L.; Sandström, P.; Bratthäll, C.; Messmer, D.; Spångeus, A.; Larsson, M. IL-1α Expression in Pancreatic Ductal Adenocarcinoma Affects the Tumor Cell Migration and Is Regulated by the p38MAPK Signaling Pathway. PLoS ONE 2013, 8, e70874. [Google Scholar] [CrossRef] [PubMed]
- Zambirinis, C.P.; Pushalkar, S.; Saxena, D.; Miller, G. Pancreatic cancer, inflammation, and microbiome. Cancer J. 2014, 20, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.L. Primers on molecular pathways: Lipopolysaccharide signalling—Potential role in pancreatitis and pancreatic cancer. Pancreatology 2010, 10, 114–118. [Google Scholar] [CrossRef]
- Ikebe, M.; Kitaura, Y.; Nakamura, M.; Tanaka, H.; Yamasaki, A.; Nagai, S.; Wada, J.; Yanai, K.; Koga, K.; Sato, N.; et al. Lipopolysaccharide (LPS) increases the invasive ability of pancreatic cancer cells through the TLR4/MyD88 signalling pathway. J. Surg. Oncol. 2009, 100, 725–731. [Google Scholar] [CrossRef]
- Sethi, V.; Kurtom, S.; Tarique, M.; Lavania, S.; Malchiodi, Z.; Hellmund, L.; Zhang, L.; Sharma, U.; Giri, B.; Garg, B.; et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018, 155, 33–37.e6. [Google Scholar] [CrossRef]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Mullins, T.D.; Kern, H.F.; Metzgar, R.S. Ultrastructural differentiation of sodium butyrate-treated human pancreatic adenocarcinoma cell lines. Pancreas 1991, 6, 578–587. [Google Scholar] [CrossRef]
- Bülow, R.; Fitzner, B.; Sparmann, G.; Emmrich, J.; Liebe, S.; Jaster, R. Anti-fibrogenic effects of histone deacetylase inhibitors on pancreatic stellate cells. Biochem. Pharmacol. 2007, 74, 1747–1757. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, T.D.; Von Ahgrens, D.; Dawlaty, M.; Zou, Y.; Baddour, J.; Achreja, A.; Zhao, H.; Yang, L.; Patel, B.; Kwak, C.; et al. Lactate-mediated epigenetic reprogramming regulates the formation of human pancreatic cancer-associated fibroblasts. eLife 2019, 8, e50663. [Google Scholar] [CrossRef]
- Kiss, B.; Mikó, E.; Sebő, É.; Toth, J.; Ujlaki, G.; Szabó, J.; Uray, K.; Bai, P.; Árkosy, P. Oncobiosis and microbial metabolite signaling in pancreatic adenocarcinoma. Cancers 2020, 12, 1068. [Google Scholar] [CrossRef]
- Nagathihalli, N.S.; Beesetty, Y.; Lee, W.; Washington, M.K.; Chen, X.; Lockhart, A.C.; Merchant, N.B. Novel mechanistic insights into ectodomain shedding of egfr ligands amphiregulin and TGF-α: Impact on gastrointestinal cancers driven by secondary bile acids. Cancer Res. 2014, 74, 2062–2072. [Google Scholar] [CrossRef] [PubMed]
- Tucker, O.N.; Dannenberg, A.J.; Yang, E.K.; Fahey, T.J. Bile acids induce cyclooxygenase-2 expression in human pancreatic cancer cell lines. Carcinogenesis 2004, 25, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signalling pathways in cancer. Cancer Cell Int. 2020, 20, 539. [Google Scholar] [CrossRef]
- Wortham, B.W.; Oliveira, M.A.; Patel, C.N. Polyamines in bacteria: Pleiotropic effects yet specific mechanisms. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2007. [Google Scholar] [CrossRef]
- Michael, A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018, 293, 18693–18701. [Google Scholar] [CrossRef]
- Mendez, R.; Kesh, K.; Arora, N.; Di Martino, L.; McAllister, F.; Merchant, N.; Banerjee, S.; Banerjee, S. Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis 2020, 41, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering the gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; DeLuca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptors and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Kim, C.H. Immune regulation by microbiome metabolites. Immunology 2018, 154, 220–229. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Jin, U.H.; Kim, S.B.; Safe, S. Omeprazole Inhibits Pancreatic Cancer Cell Invasion through a Nongenomic Aryl Hydrocarbon Receptor Pathway. Chem. Res. Toxicol. 2015, 28, 907–918. [Google Scholar] [CrossRef]
- Gabarrini, G.; Grasso, S.; van Winkelhoff, A.J.; van Dijl, J.M. Gingimaps: Protein Localization in the Oral Pathogen Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 2020, 84, e00032-19. [Google Scholar] [CrossRef]
- Öğrendik, M. Oral bacteria in pancreatic cancer: Mutagenesis of the p53 tumour suppressor gene. Int. J. Clin. Exp. Pathol. 2015, 8, 11835. [Google Scholar]
- Kunovsky, L.; Dite, P.; Jabandziev, P.; Dolina, J.; Vaculova, J.; Blaho, M.; Bojkova, M.; Dvorackova, J.; Uvirova, M.; Kala, Z.; et al. Helicobacter pylori infection and other bacteria in pancreatic cancer and autoimmune pancreatitis. World J. Gastrointest. Oncol. 2021, 13, 835–844. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, X.; Yin, M.; Gao, J.; Weng, Z.; Xu, C. The relationship between Helicobacter pylori and pancreatic cancer: A meta-analysis. Transl. Cancer Res. 2022, 11, 2810–2822. [Google Scholar] [CrossRef]
- Zhou, B.G.; Mei, Y.Z.; Wang, J.S.; Xia, J.L.; Jiang, X.; Ju, S.Y.; Ding, Y.B. Is Helicobacter pylori infection associated with pancreatic cancer? A systematic review and meta-analysis of observational studies. Ther. Adv. Chronic Dis. 2023, 14, 20406223231155119. [Google Scholar] [CrossRef]
- Kolbeinsson, H.M.; Chandana, S.; Wright, G.P.; Chung, M. Pancreatic Cancer: A Review of Current Treatment and Novel Therapies. J. Investig. Surg. 2023, 36, 2129884. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, K.; Nosho, K.; Sukawa, Y.; Matsunaga, Y.; Ito, M.; Kurihara, H.; Kanno, S.; Igarashi, H.; Naito, T.; Adachi, Y.; et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015, 6, 7209–7220. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies the association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and mycobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Udayasuryan, B.; Ahmad, R.N.; Nguyen, T.T.D.; Umaña, A.; Roberts, L.D.M.; Sobol, P.; Jones, S.D.; Munson, J.M.; Slade, D.J.; Verbridge, S.S. Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signalling. Sci. Signal. 2022, 15, eabn4948. [Google Scholar] [CrossRef]
- Lehouritis, P.; Cummins, J.; Stanton, M.; Murphy, C.T.; McCarthy, F.O.; Reid, G.; Urbaniak, C.; Byrne, W.L.; Tangney, M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015, 5, 14554. [Google Scholar] [CrossRef]
- Voorde, J.V.; Sabuncuoǧlu, S.; Noppen, S.; Hofer, A.; Ranjbarian, F.; Fieuws, S.; Balzarini, J.; Liekens, S. Nucleoside-catabolising enzymes in mycoplasma-infected tumour cell cultures compromise the cytostatic activity of the anti-cancer drug gemcitabine. J. Biol. Chem. 2014, 289, 13054–13065. [Google Scholar] [CrossRef]
- Sunakawa, Y.; Arai, H.; Izawa, N.; Mizukami, T.; Horie, Y.; Doi, A.; Hirakawa, M.; Ogura, T.; Tsuda, T.; Nakajima, T.E. Antibiotics may enhance the efficacy of gemcitabine treatment for advanced pancreatic cancer. Ann. Oncol. 2018, 29, viii251–viii252. [Google Scholar] [CrossRef]
- Imai, H.; Saijo, K.; Komine, K.; Otsuki, Y.; Ohuchi, K.; Sato, Y.; Okita, A.; Takahashi, M.; Takahashi, S.; Shirota, H.; et al. Antibiotic therapy augments the efficacy of gemcitabine-containing regimens for advanced cancer: A retrospective study. Cancer Manag. Res. 2019, 11, 7953–7965. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Komatsu, Y.; Kawamoto, Y.; Saito, R.; Ito, K.; Nakatsumi, H.; Yuki, S.; Sakamoto, N. Association between the use of antibiotics and efficacy of gemcitabine plus nab-paclitaxel in advanced pancreatic cancer. Medicine 2020, 99, e22250. [Google Scholar] [CrossRef] [PubMed]
- Fulop, D.J.; Zylberberg, H.M.; Wu, Y.L.; Aronson, A.; Labiner, A.J.; Wisnivesky, J.; Cohen, D.J.; Sigel, K.M.; Lucas, A.L. Association of Antibiotic Receipt With Survival Among Patients With Metastatic Pancreatic Ductal Adenocarcinoma Receiving Chemotherapy. JAMA Netw. Open 2023, 6, e234254. [Google Scholar] [CrossRef]
- Beberok, A.; Rzepka, Z.; Respondek, M.; Rok, J.; Stradowski, M.; Wrześniok, D. Moxifloxacin as an inducer of apoptosis in melanoma cells: A study at the cellular and molecular level. Toxicol. Vitr. 2019, 55, 75–92. [Google Scholar] [CrossRef]
- Song, M.; Wu, H.; Wu, S.; Ge, T.; Wang, G.; Zhou, Y.; Sheng, S.; Jiang, J. The antibiotic drug levofloxacin inhibits proliferation and induces apoptosis of lung cancer cells through inducing mitochondrial dysfunction and oxidative damage. Biomed. Pharmacother. 2016, 55, 75–92. [Google Scholar] [CrossRef]
- Idowu, T.; Schweizer, F. Ubiquitous nature of fluoroquinolones: The oscillation between antibacterial and anti-cancer activities. Antibiotics 2017, 6, 26. [Google Scholar] [CrossRef]
- Yadav, V.; Sultana, S.; Yadav, J.; Saini, N. Gatifloxacin Induces S and G2-Phase Cell Cycle Arrest in Pancreatic Cancer Cells via p21/p27/p53. PLoS ONE 2012, 7, e47796. [Google Scholar] [CrossRef]
- Wei, M.Y.; Shi, S.; Liang, C.; Meng, Q.C.; Hua, J.; Zhang, Y.Y.; Liu, J.; Zhang, B.; Xu, J.; Yu, X.J. The microbiota and microbiome in pancreatic cancer: More influential than expected. Mol. Cancer 2019, 18, 97. [Google Scholar] [CrossRef]
- Selvanesan, B.C.; Chandra, D.; Quispe-Tintaya, W.; Jahangir, A.; Patel, A.; Meena, K.; Da Silva, R.A.A.; Friedman, M.; Gabor, L.; Khouri, O.; et al. Listeria delivers tetanus toxoid protein to pancreatic tumours and induces cancer cell death in mice. Sci. Transl. Med. 2022, 14, eabc1600. [Google Scholar] [CrossRef]
- Tempero, M.A.; Malafa, M.P.; Al-Hawary, M.; Behrman, S.W.; Benson, A.B.; Cardin, D.B.; Chiorean, E.G.; Chung, V.; Czito, B.; Del Chiaro, M.; et al. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 439–457. [Google Scholar] [CrossRef]
- Winograd, R.; Byrne, K.T.; Evans, R.A.; Odorizzi, P.M.; Meyer, A.R.L.; Bajor, D.L.; Clendenin, C.; Stanger, B.Z.; Furth, E.E.; Wherry, E.J.; et al. Induction of T-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol. Res. 2015, 3, 399–411. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Farrow, B.; Rychahou, P.; O’Connor, K.L.; Evers, B.M. Butyrate inhibits pancreatic cancer invasion. J. Gastrointest. Surg. 2003, 7, 864–870. [Google Scholar] [CrossRef]
- Corra, S.; Kazakoff, K.; Mogaki, M.; Cano, M.; Pour, P.M. Modification of antigen expression in human and hamster pancreatic cancer cell lines induced by sodium butyrate. Teratog. Carcinog. Mutagen. 1993, 13, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Stebel, R.; Vojtilová, L.; Husa, P. Fecal microbiota transplantation—Past, present, and future. Gastroenterol. Hepatol. 2020, 74, 541–561. [Google Scholar] [CrossRef]
- Ianiro, G.; Rossi, E.; Thomas, A.M.; Schinzari, G.; Masucci, L.; Quaranta, G.; Settanni, C.R.; Lopetuso, L.R.; Armanini, F.; Blanco-Miguez, A.; et al. Faecal microbiota transplantation for the treatment of diarrhoea induced by tyrosine-kinase inhibitors in patients with metastatic renal cell carcinoma. Nat. Commun. 2020, 11, 4333. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.S.; Chang, H.H.; Hauer, M.; Lagishetty, V.; Katzka, W.; Rozengurt, E.; Jacobs, J.P.; Eibl, G. Metformin alters the duodenal microbiome and decreases the incidence of pancreatic ductal adenocarcinoma promoted by diet-induced obesity. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G763–G772. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, J.; Büchler, M.W.; Friess, H.; Martignoni, M.E. Cachexia in patients with chronic pancreatitis and pancreatic cancer: Impact on survival and outcome. Nutr. Cancer 2013, 65, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Beck, R.; Schakman, O.; Martin, J.C.; de Backer, F.; Sohet, F.M.; Dewulf, E.M.; Pachikian, B.D.; Neyrinck, A.M.; Thissen, J.P.; et al. Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukaemia mouse model. PLoS ONE 2012, 7, e37971. [Google Scholar] [CrossRef]
- Davidson, W.; Ash, S.; Capra, S.; Bauer, J.; Fearon, K.C.H.; von Meyenfeldt, M.F.; Roy, A.; Gouma, D.J.; Giacosa, A.; Van Gossum, A.; et al. Weight stabilisation is associated with improved survival duration and quality of life in unresectable pancreatic cancer. Clin. Nutr. 2004, 23, 239–247. [Google Scholar] [CrossRef]
- Werner, K.; Küllenberg de Gaudry, D.; Taylor, L.A.; Keck, T.; Unger, C.; Hopt, U.T.; Massing, U. Dietary supplementation with n-3-fatty acids in patients with pancreatic cancer and cachexia: Marine phospholipids versus fish oil—A randomized controlled double-blind trial. Lipids Health Dis. 2017, 16, 104. [Google Scholar] [CrossRef]
- Li, D.; Tang, H.; Wei, P.; Zheng, J.; Daniel, C.R.; Hassan, M.M. Vitamin C and Vitamin E Mitigate the Risk of Pancreatic Ductal Adenocarcinoma from Meat-Derived Mutagen Exposure in Adults in a Case-Control Study. J. Nutr. 2019, 149, 1443–1450. [Google Scholar] [CrossRef]
- Michl, P.; Löhr, M.; Neoptolemos, J.P.; Capurso, G.; Rebours, V.; Malats, N.; Ollivier, M.; Ricciardiello, L. UEG position paper on pancreatic cancer. Bringing pancreatic cancer to the 21st century: Prevent, detect, and treat the disease earlier and better. United Eur. Gastroenterol. J. 2021, 9, 860–871. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papa, V.; Schepis, T.; Coppola, G.; Chiappetta, M.F.; Del Vecchio, L.E.; Rozera, T.; Quero, G.; Gasbarrini, A.; Alfieri, S.; Papa, A. The Role of Microbiota in Pancreatic Cancer. Cancers 2023, 15, 3143. https://doi.org/10.3390/cancers15123143
Papa V, Schepis T, Coppola G, Chiappetta MF, Del Vecchio LE, Rozera T, Quero G, Gasbarrini A, Alfieri S, Papa A. The Role of Microbiota in Pancreatic Cancer. Cancers. 2023; 15(12):3143. https://doi.org/10.3390/cancers15123143
Chicago/Turabian StylePapa, Valerio, Tommaso Schepis, Gaetano Coppola, Michele Francesco Chiappetta, Livio Enrico Del Vecchio, Tommaso Rozera, Giuseppe Quero, Antonio Gasbarrini, Sergio Alfieri, and Alfredo Papa. 2023. "The Role of Microbiota in Pancreatic Cancer" Cancers 15, no. 12: 3143. https://doi.org/10.3390/cancers15123143
APA StylePapa, V., Schepis, T., Coppola, G., Chiappetta, M. F., Del Vecchio, L. E., Rozera, T., Quero, G., Gasbarrini, A., Alfieri, S., & Papa, A. (2023). The Role of Microbiota in Pancreatic Cancer. Cancers, 15(12), 3143. https://doi.org/10.3390/cancers15123143






