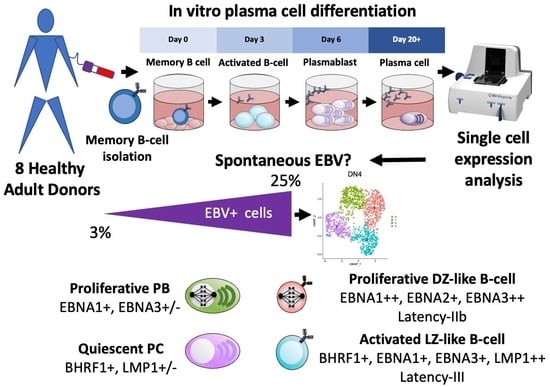
Spontaneous EBV-Reactivation during B Cell Differentiation as a Model for Polymorphic EBV-Driven Lymphoproliferation
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Reagents
2.2. Donors and Cell Isolation
2.3. Cell Cultures
2.4. Flow Cytometric Analysis
2.5. Single-Cell RNAseq
2.6. Data Processing
2.7. Cell Subsets
2.8. Clustering and Differential Gene Expression Analysis
2.9. Gene Signature Data and Enrichment Analysis
2.10. Significance of Cell Groups
2.11. Data Visualisation
2.12. Ethical Approval
3. Results
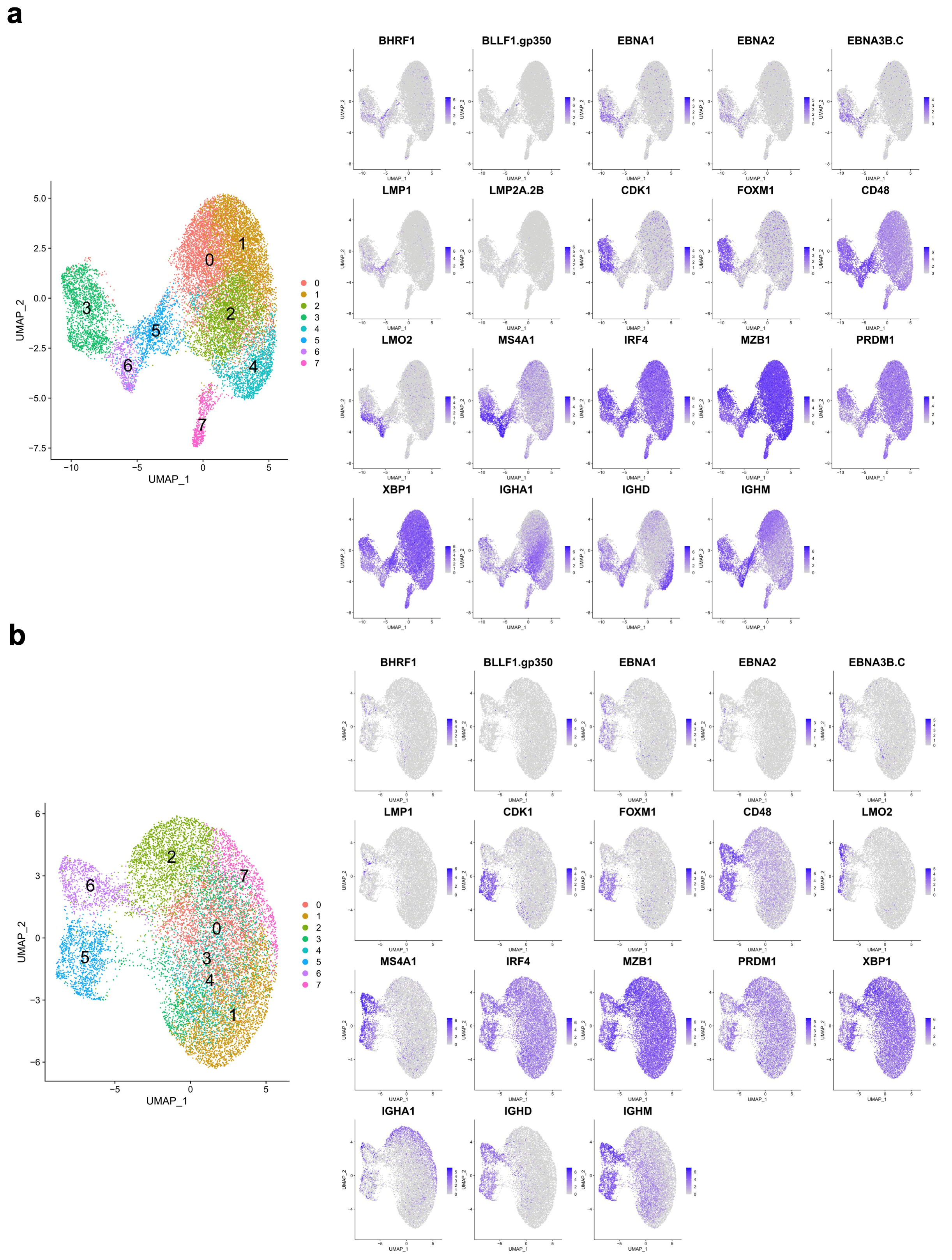
3.1. A Targeted Single-Cell Expression Panel for B Cell and PC Analysis
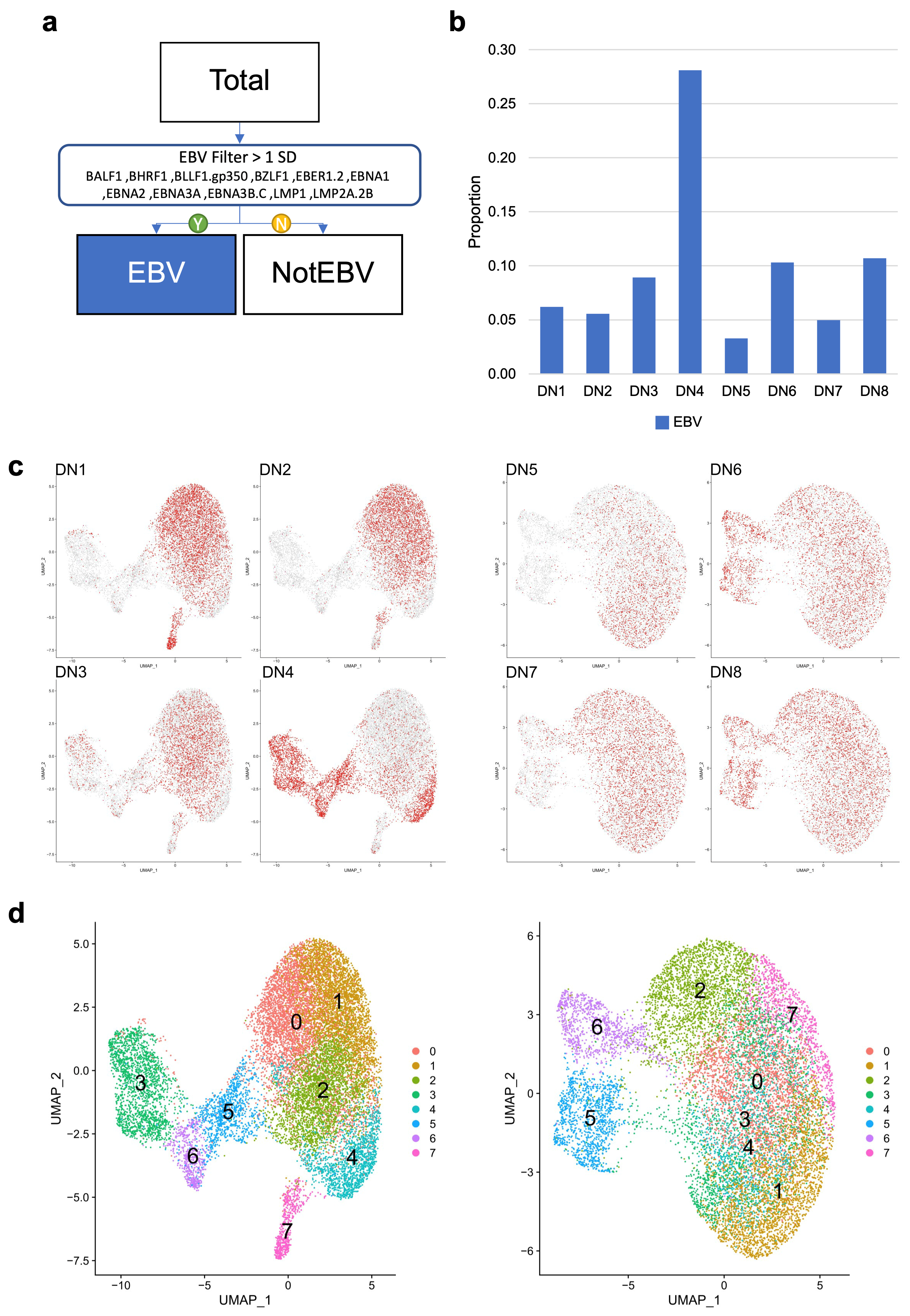
3.2. Assessing Overall EBV Contribution in Late B Cell Differentiation
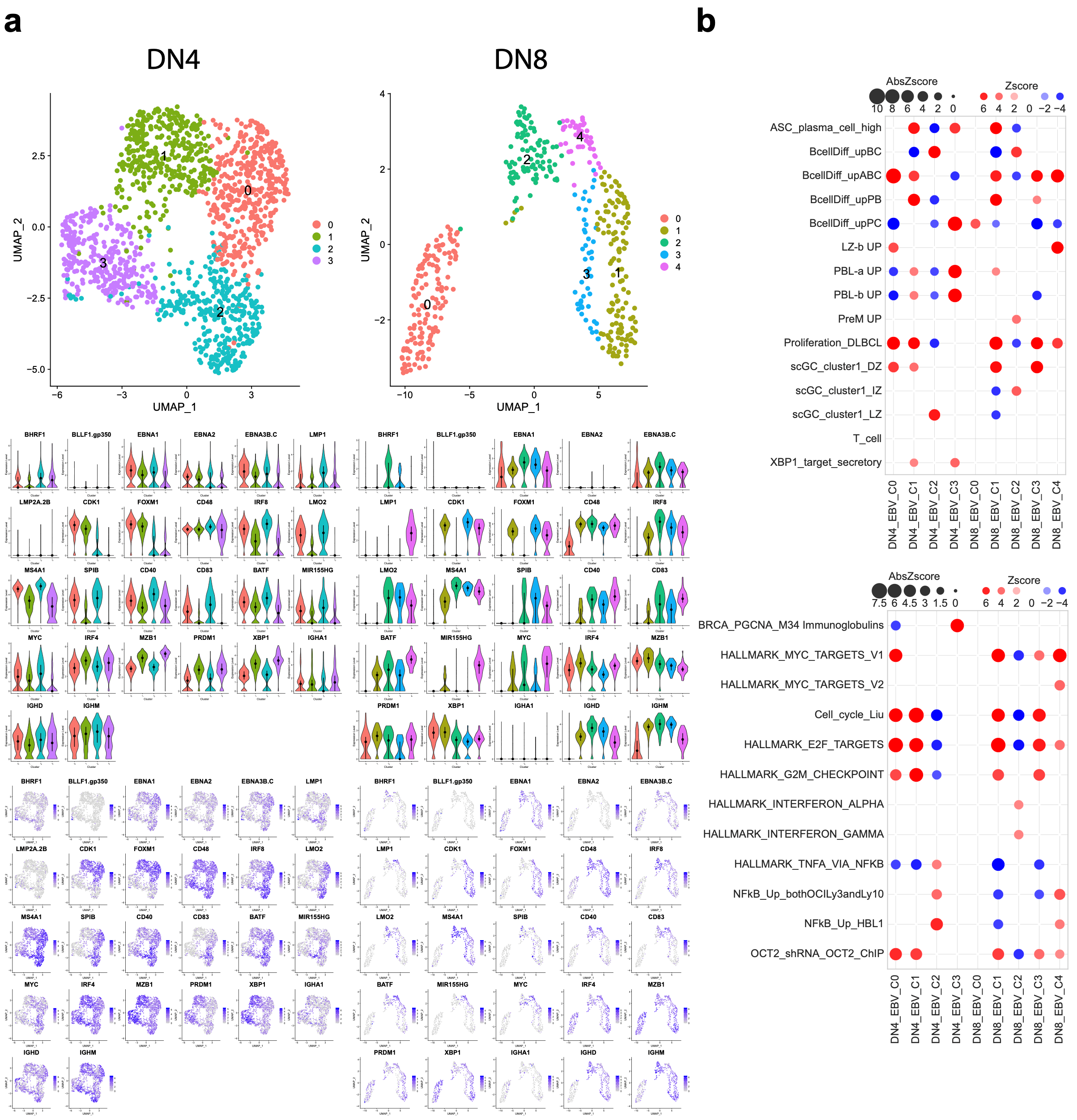
3.3. EBV-Associated Cells Separate Based on Differentiation and Cell-Cycle State
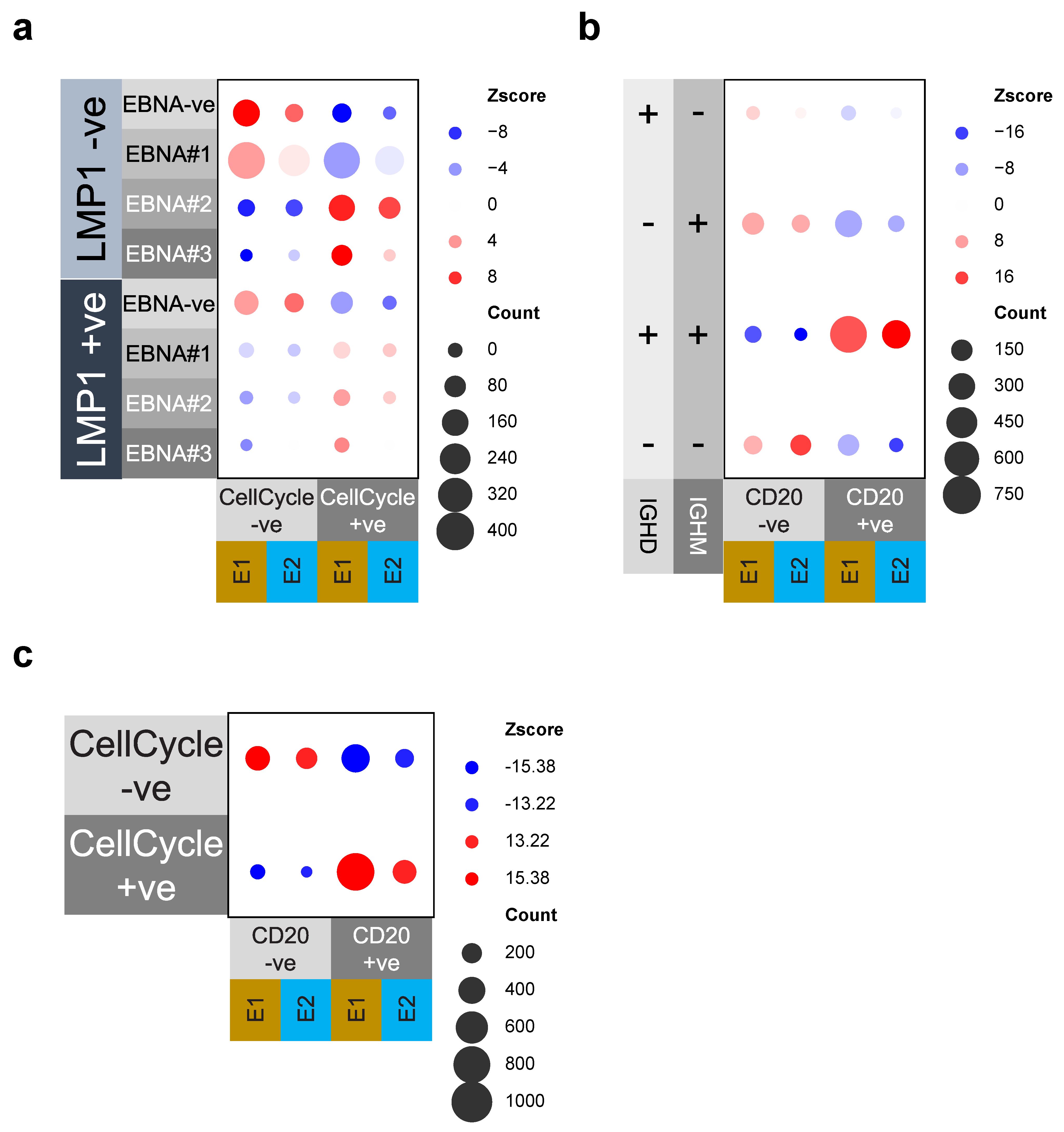
3.4. Cell Cycle and MS4A1/CD20 Expression Are Linked to EBV-Gene and IgM/IgD Expression
3.5. Donor-Specific Patterns of EBV-Associated B Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munz, C. Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 2019, 17, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Munz, C. Epstein Barr virus—A tumor virus that needs cytotoxic lymphocytes to persist asymptomatically. Curr. Opin. Virol. 2016, 20, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; de Oliveira Araujo, I.B.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.A.; Stollar, B.D.; Sullivan, J.L.; Luzuriaga, K.; Thorley-Lawson, D.A. Peripheral B cells latently infected with Epstein-Barr virus display molecular hallmarks of classical antigen-selected memory B cells. Proc. Natl. Acad. Sci. USA 2005, 102, 18093–18098. [Google Scholar] [CrossRef]
- Klein, U.; Rajewsky, K.; Kuppers, R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 1998, 188, 1679–1689. [Google Scholar] [CrossRef]
- Weisel, N.M.; Joachim, S.M.; Smita, S.; Callahan, D.; Elsner, R.A.; Conter, L.J.; Chikina, M.; Farber, D.L.; Weisel, F.J.; Shlomchik, M.J. Surface phenotypes of naive and memory B cells in mouse and human tissues. Nat. Immunol. 2022, 23, 135–145. [Google Scholar] [CrossRef]
- Chaganti, S.; Heath, E.M.; Bergler, W.; Kuo, M.; Buettner, M.; Niedobitek, G.; Rickinson, A.B.; Bell, A.I. Epstein-Barr virus colonization of tonsillar and peripheral blood B-cell subsets in primary infection and persistence. Blood 2009, 113, 6372–6381. [Google Scholar] [CrossRef]
- Joseph, A.M.; Babcock, G.J.; Thorley-Lawson, D.A. EBV persistence involves strict selection of latently infected B cells. J. Immunol. 2000, 165, 2975–2981. [Google Scholar] [CrossRef]
- Chaganti, S.; Ma, C.S.; Bell, A.I.; Croom-Carter, D.; Hislop, A.D.; Tangye, S.G.; Rickinson, A.B. Epstein-Barr virus persistence in the absence of conventional memory B cells: IgM+IgD+CD27+ B cells harbor the virus in X-linked lymphoproliferative disease patients. Blood 2008, 112, 672–679. [Google Scholar] [CrossRef]
- Heath, E.; Begue-Pastor, N.; Chaganti, S.; Croom-Carter, D.; Shannon-Lowe, C.; Kube, D.; Feederle, R.; Delecluse, H.-J.; Rickinson, A.B.; Bell, A.I. Epstein-Barr virus infection of naive B cells in vitro frequently selects clones with mutated immunoglobulin genotypes: Implications for virus biology. PLoS Pathog. 2012, 8, e1002697. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. Epstein-Barr virus: Exploiting the immune system. Nat. Rev. Immunol. 2001, 1, 75–82. [Google Scholar] [CrossRef]
- Tovey, M.G.; Lenoir, G.; Begon-Lours, J. Activation of latent Epstein-Barr virus by antibody to human IgM. Nature 1978, 276, 270–272. [Google Scholar] [CrossRef]
- Laichalk, L.L.; Thorley-Lawson, D.A. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 2005, 79, 1296–1307. [Google Scholar] [CrossRef]
- Price, A.M.; Luftig, M.A. To be or not IIb: A multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015, 11, e1004656. [Google Scholar] [CrossRef]
- Caldwell, R.G.; Wilson, J.B.; Anderson, S.J.; Longnecker, R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 1998, 9, 405–411. [Google Scholar] [CrossRef]
- Uchida, J.; Yasui, T.; Takaoka-Shichijo, Y.; Muraoka, M.; Kulwichit, W.; Raab-Traub, N.; Kikutani, H. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 1999, 286, 300–303. [Google Scholar] [CrossRef]
- Minamitani, T.; Ma, Y.; Zhou, H.; Kida, H.; Tsai, C.-Y.; Obana, M.; Okuzaki, D.; Fujio, Y.; Kumanogoh, A.; Zhao, B.; et al. Mouse model of Epstein-Barr virus LMP1- and LMP2A-driven germinal center B-cell lymphoproliferative disease. Proc. Natl. Acad. Sci. USA 2017, 114, 4751–4756. [Google Scholar] [CrossRef]
- Jiang, S.; Willox, B.; Zhou, H.; Holthaus, A.M.; Wang, A.; Shi, T.T.; Maruo, S.; Kharchenko, P.V.; Johannsen, E.C.; Kieff, E.; et al. Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc. Natl. Acad. Sci. USA 2014, 111, 421–426. [Google Scholar] [CrossRef]
- Schmidt, S.C.S.; Jiang, S.; Zhou, H.; Willox, B.; Holthaus, A.M.; Kharchenko, P.V.; Johannsen, E.C.; Kieff, E.; Zhao, B. Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes. Proc. Natl. Acad. Sci. USA 2015, 112, 554–559. [Google Scholar] [CrossRef]
- Jiang, S.; Zhou, H.; Liang, J.; Gerdt, C.; Wang, C.; Ke, L.; Schmidt, S.C.; Narita, Y.; Ma, Y.; Wang, S.; et al. The Epstein-Barr Virus Regulome in Lymphoblastoid Cells. Cell Host Microbe 2017, 22, 561–573.e4. [Google Scholar] [CrossRef]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Frontzek, F.; Staiger, A.M.; Wullenkord, R.; Grau, M.; Zapukhlyak, M.; Kurz, K.S.; Horn, H.; Erdmann, T.; Fend, F.; Richter, J.; et al. Molecular profiling of EBV associated diffuse large B-cell lymphoma. Leukemia 2023, 37, 670–679. [Google Scholar] [CrossRef] [PubMed]
- SoRelle, E.D.; Dai, J.; Bonglack, E.N.; Heckenberg, E.M.; Zhou, J.Y.; Giamberardino, S.N.; Bailey, J.A.; Gregory, S.G.; Chan, C.; Luftig, M.A. Single-cell RNA-seq reveals transcriptomic heterogeneity mediated by host-pathogen dynamics in lymphoblastoid cell lines. Elife 2021, 10, e62586. [Google Scholar] [CrossRef] [PubMed]
- SoRelle, E.D.; Reinoso-Vizcaino, N.M.; Horn, G.Q.; Luftig, M.A. Epstein-Barr virus perpetuates B cell germinal center dynamics and generation of autoimmune-associated phenotypes in vitro. Front. Immunol. 2022, 13, 1001145. [Google Scholar] [CrossRef] [PubMed]
- Price, A.M.; Tourigny, J.P.; Forte, E.; Salinas, R.E.; Dave, S.S.; Luftig, M.A. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-kappaB activation. J. Virol. 2012, 86, 11096–11106. [Google Scholar] [CrossRef]
- SoRelle, E.D.; Dai, J.; Reinoso-Vizcaino, N.M.; Barry, A.P.; Chan, C.; Luftig, M.A. Time-resolved transcriptomes reveal diverse B cell fate trajectories in the early response to Epstein-Barr virus infection. Cell Rep. 2022, 40, 111286. [Google Scholar] [CrossRef]
- zur Hausen, H.; O’Neill, F.J.; Freese, U.K.; Hecker, E. Persisting oncogenic herpesvirus induced by the tumour promotor TPA. Nature 1978, 272, 373–375. [Google Scholar] [CrossRef]
- Cocco, M.; Stephenson, S.; Care, M.A.; Newton, D.; Barnes, N.A.; Davison, A.; Rawstron, A.; Westhead, D.R.; Doody, G.M.; Tooze, R.M. In vitro generation of long-lived human plasma cells. J. Immunol. 2012, 189, 5773–5785. [Google Scholar] [CrossRef]
- Stephenson, S.; Care, M.A.; Fan, I.; Zougman, A.; Westhead, D.R.; Doody, G.M.; Tooze, R.M. Growth Factor-like Gene Regulation Is Separable from Survival and Maturation in Antibody-Secreting Cells. J. Immunol. 2019, 202, 1287–1300. [Google Scholar] [CrossRef]
- Stephenson, S.; Care, M.A.; Doody, G.M.; Tooze, R.M. APRIL Drives a Coordinated but Diverse Response as a Foundation for Plasma Cell Longevity. J. Immunol. 2022, 209, 926–937. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Kuri, A.; Jacobs, B.M.; Vickaryous, N.; Pakpoor, J.; Middeldorp, J.; Giovannoni, G.; Dobson, R. Epidemiology of Epstein-Barr virus infection and infectious mononucleosis in the United Kingdom. BMC Public Health 2020, 20, 912. [Google Scholar] [CrossRef]
- Milpied, P.; Cervera-Marzal, I.; Mollichella, M.-L.; Tesson, B.; Brisou, G.; Traverse-Glehen, A.; Salles, G.; Spinelli, L.; Nadel, B. Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat. Immunol. 2018, 19, 1013–1024. [Google Scholar] [CrossRef]
- Holmes, A.B.; Corinaldesi, C.; Shen, Q.; Kumar, R.; Compagno, N.; Wang, Z.; Nitzan, M.; Grunstein, E.; Pasqualucci, L.; Dalla-Favera, R.; et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J. Exp. Med. 2020, 217, e20200483. [Google Scholar] [CrossRef]
- Vakiani, E.; Basso, K.; Klein, U.; Mansukhani, M.M.; Narayan, G.; Smith, P.M.; Murty, V.V.; Dalla-Favera, R.; Pasqualucci, L.; Bhagat, G. Genetic and phenotypic analysis of B-cell post-transplant lymphoproliferative disorders provides insights into disease biology. Hematol. Oncol. 2008, 26, 199–211. [Google Scholar] [CrossRef]
- Serafini, B.; Severa, M.; Columba-Cabezas, S.; Rosicarelli, B.; Veroni, C.; Chiappetta, G.; Magliozzi, R.; Reynolds, R.; Coccia, E.M.; Aloisi, F. Epstein-Barr virus latent infection and BAFF expression in B cells in the multiple sclerosis brain: Implications for viral persistence and intrathecal B-cell activation. J. Neuropathol. Exp. Neurol. 2010, 69, 677–693. [Google Scholar] [CrossRef]
- Tosato, G.; Tanner, J.; Jones, K.D.; Revel, M.; E Pike, S. Identification of interleukin-6 as an autocrine growth factor for Epstein-Barr virus-immortalized B cells. J. Virol. 1990, 64, 3033–3041. [Google Scholar] [CrossRef]
- Haddad, E.; Paczesny, S.; Leblond, V.; Seigneurin, J.-M.; Stern, M.; Achkar, A.; Bauwens, M.; Delwail, V.; Debray, M.; Duvoux, C.; et al. Treatment of B-lymphoproliferative disorder with a monoclonal anti-interleukin-6 antibody in 12 patients: A multicenter phase 1-2 clinical trial. Blood 2001, 97, 1590–1597. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Care, M.A.; Stephenson, S.; Owen, R.; Doody, G.M.; Tooze, R.M. Spontaneous EBV-Reactivation during B Cell Differentiation as a Model for Polymorphic EBV-Driven Lymphoproliferation. Cancers 2023, 15, 3083. https://doi.org/10.3390/cancers15123083
Care MA, Stephenson S, Owen R, Doody GM, Tooze RM. Spontaneous EBV-Reactivation during B Cell Differentiation as a Model for Polymorphic EBV-Driven Lymphoproliferation. Cancers. 2023; 15(12):3083. https://doi.org/10.3390/cancers15123083
Chicago/Turabian StyleCare, Matthew A., Sophie Stephenson, Roger Owen, Gina M. Doody, and Reuben M. Tooze. 2023. "Spontaneous EBV-Reactivation during B Cell Differentiation as a Model for Polymorphic EBV-Driven Lymphoproliferation" Cancers 15, no. 12: 3083. https://doi.org/10.3390/cancers15123083
APA StyleCare, M. A., Stephenson, S., Owen, R., Doody, G. M., & Tooze, R. M. (2023). Spontaneous EBV-Reactivation during B Cell Differentiation as a Model for Polymorphic EBV-Driven Lymphoproliferation. Cancers, 15(12), 3083. https://doi.org/10.3390/cancers15123083