Radiogenomics Analysis Linking Multiparametric MRI and Transcriptomics in Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
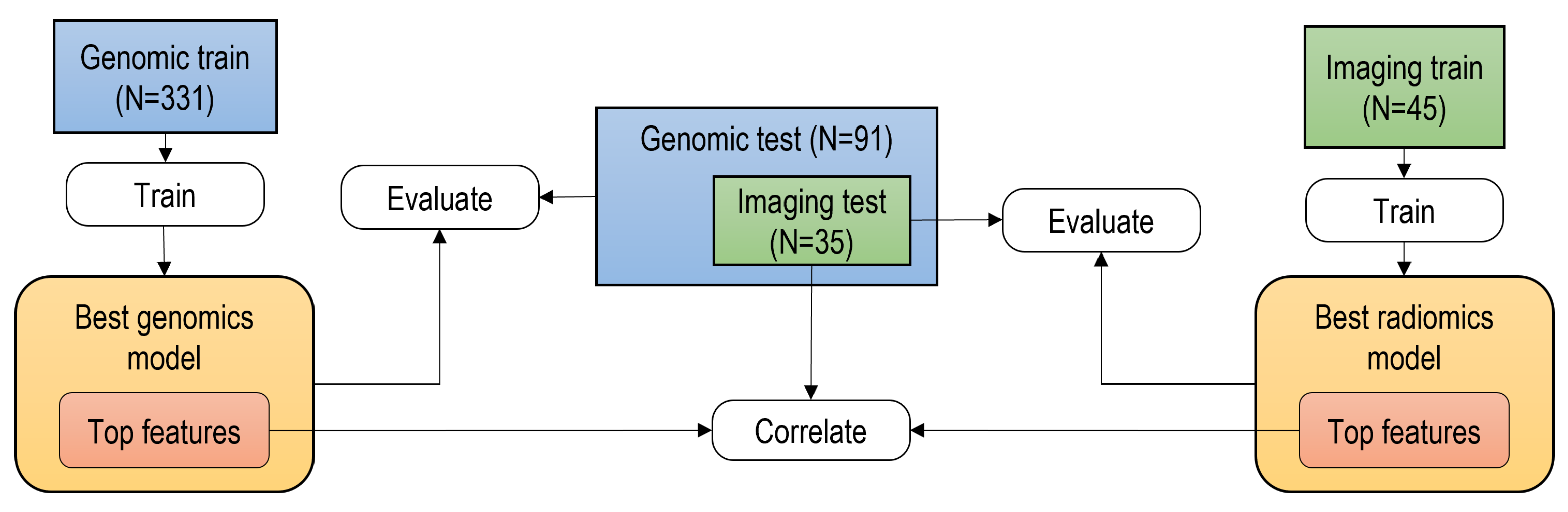
2. Materials and Methods
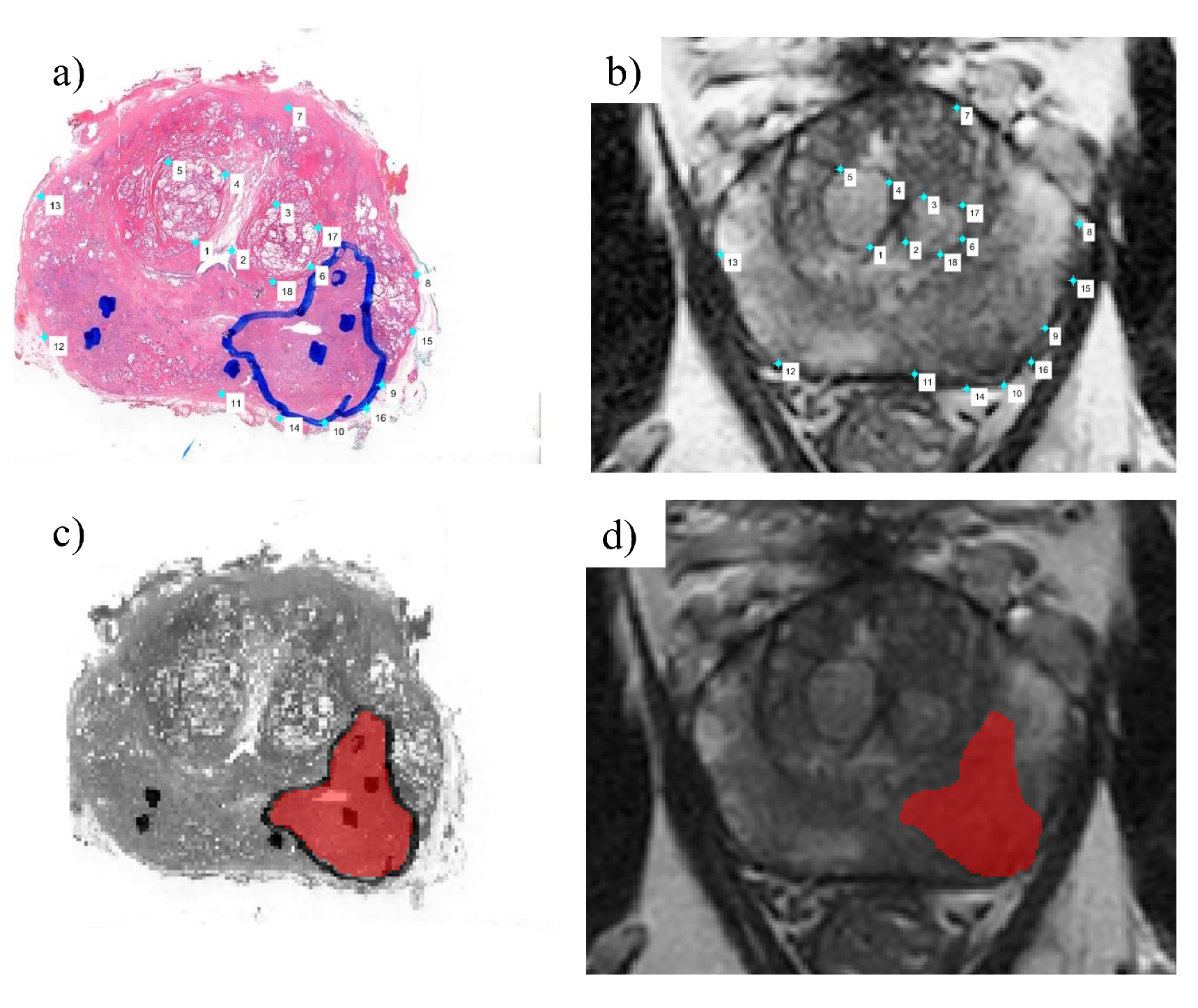
2.1. Data Collection
2.1.1. Imaging Data Pre-Processing
2.2. Transcriptomics Analysis Pipeline
2.2.1. Feature Extraction
2.2.2. Machine Learning for Feature Selection
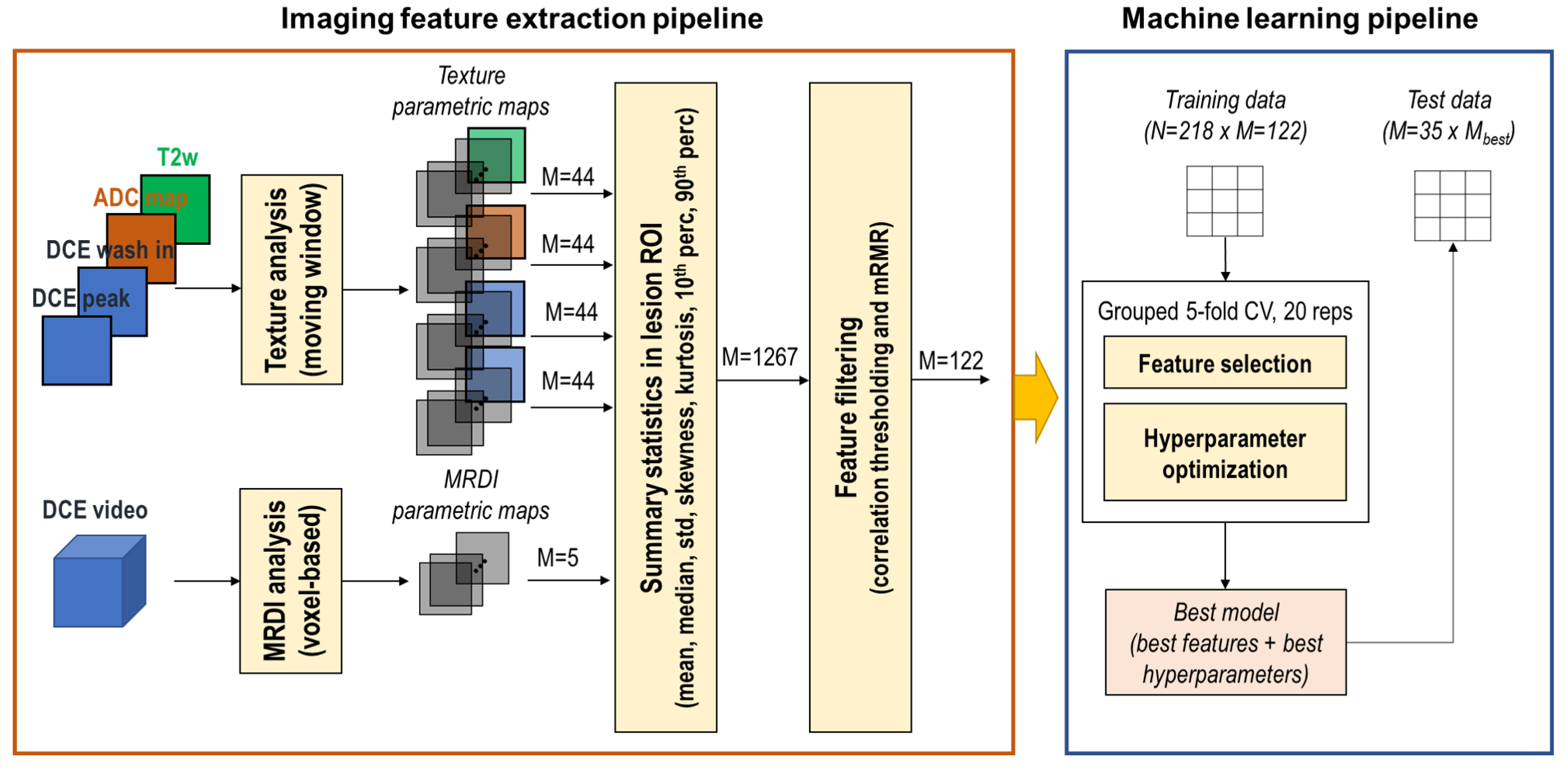
2.3. Image Analysis Pipeline
2.3.1. Feature Extraction
2.3.2. Summary Statistics and Feature Filtering
2.3.3. Machine Learning Pipeline
2.4. Correlation with Transcriptomic Features
3. Results
3.1. Patient Stratification
3.2. Dimensionality Reduction and Feature Selection
3.3. Imaging Classifiers
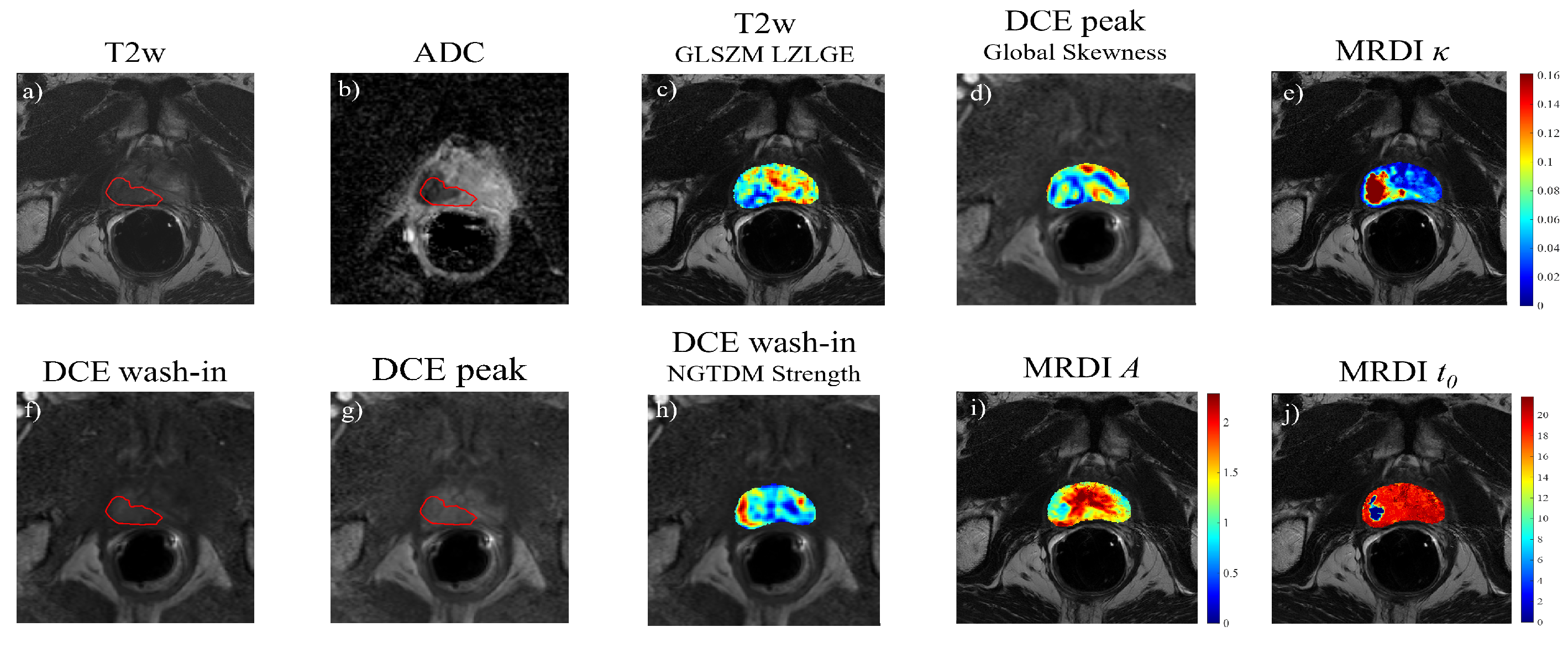
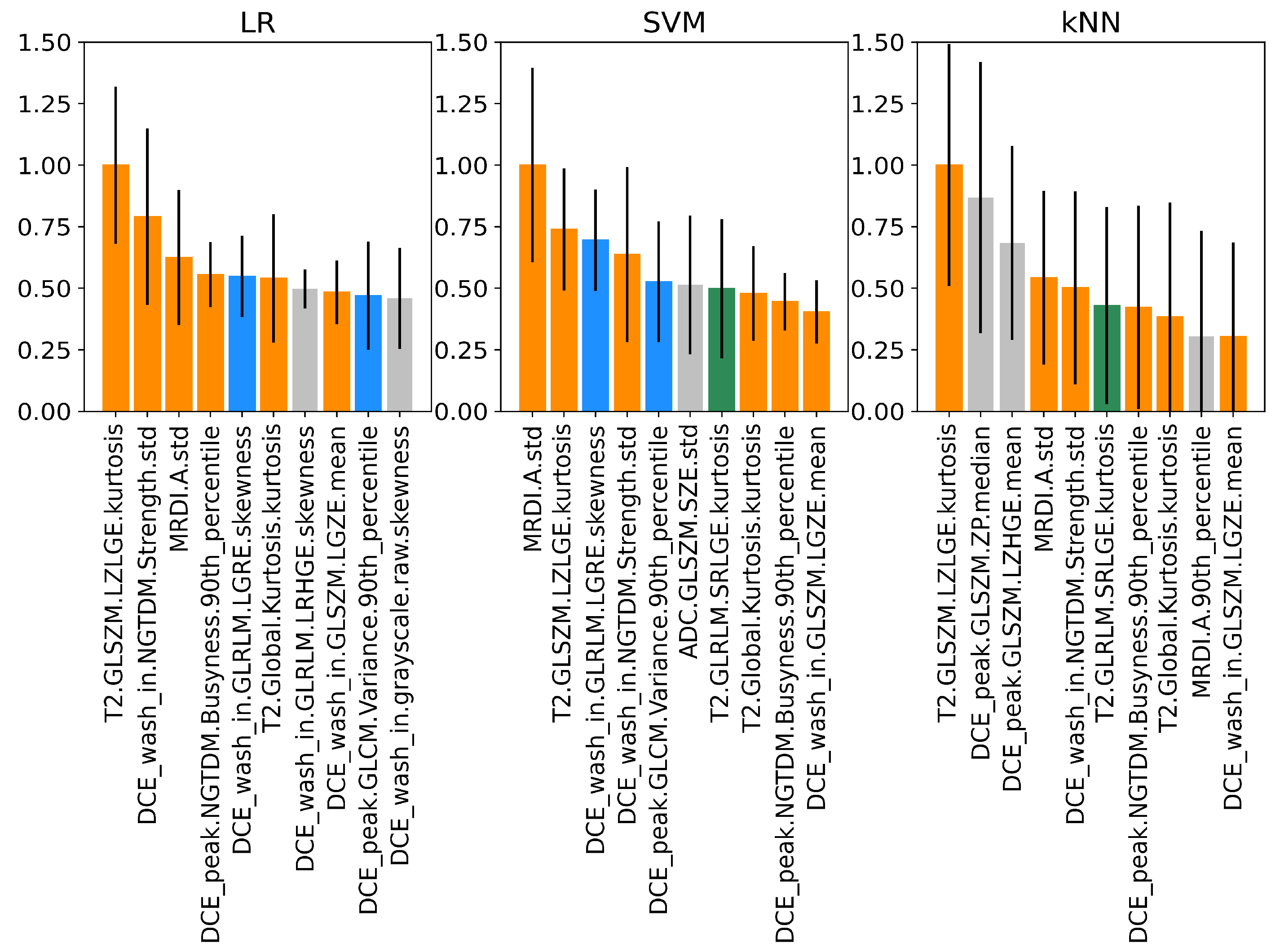
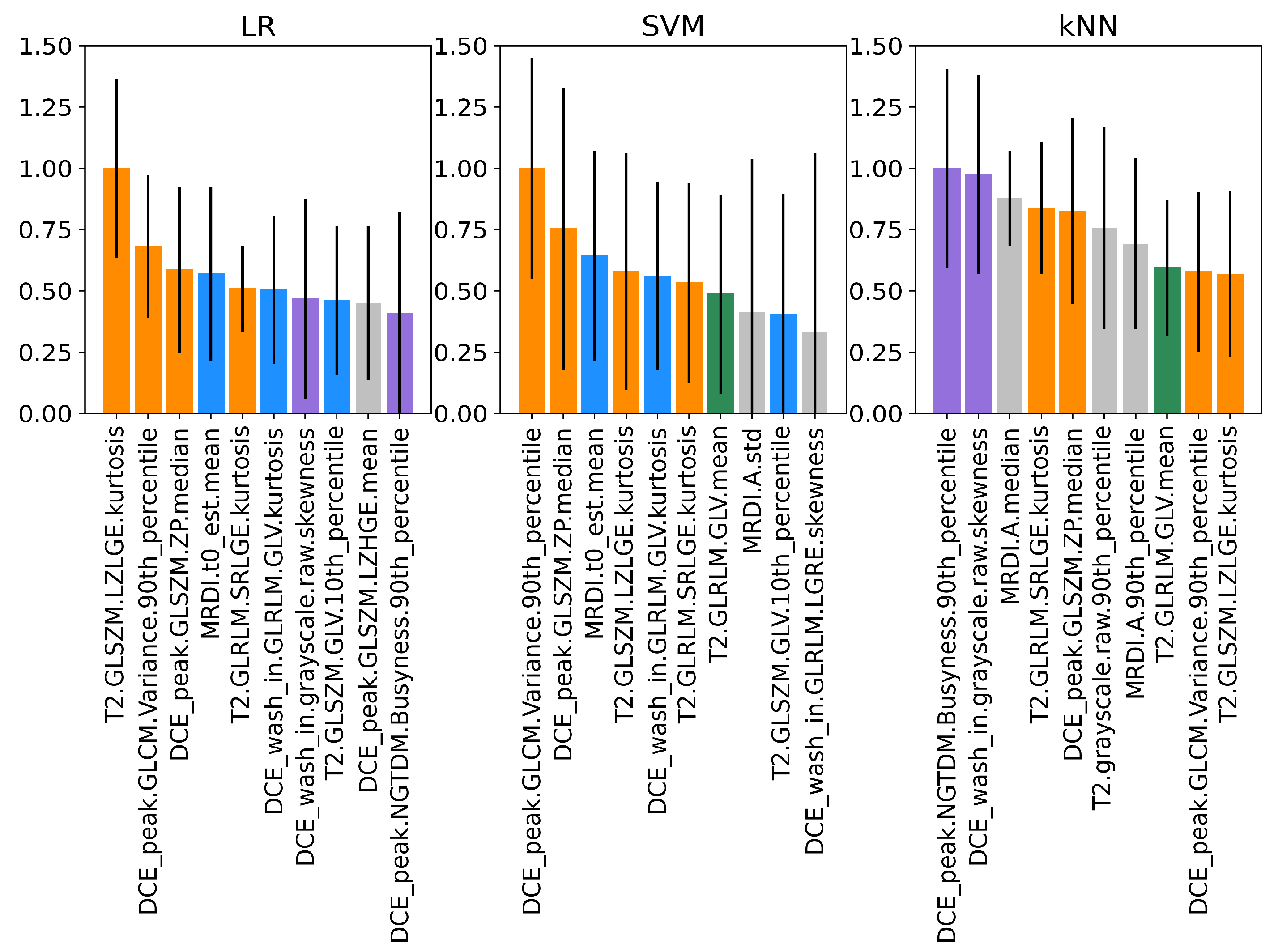
3.4. Permutation Feature Importance of Imaging Features
3.5. Correlation with Transcriptomic Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- EAU Guidelines on Prostate Cancer. Available online: https://uroweb.org/guidelines/prostate-cancer (accessed on 4 April 2023).
- Mottet, N.; van den Bergh, R.C.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer—2020 update. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Stabile, A.; Giganti, F.; Rosenkrantz, A.B.; Taneja, S.S.; Villeirs, G.; Gill, I.S.; Allen, C.; Emberton, M.; Moore, C.M.; Kasivisvanathan, V. Multiparametric MRI for prostate cancer diagnosis: Current status and future directions. Nat. Rev. Urol. 2020, 17, 41–61. [Google Scholar] [CrossRef]
- Van Poppel, H.; Roobol, M.J.; Chapple, C.R.; Catto, J.W.; N’Dow, J.; Sønksen, J.; Stenzl, A.; Wirth, M. Prostate-specific antigen testing as part of a risk-adapted early detection strategy for prostate cancer: European Association of Urology position and recommendations for 2021. Eur. Urol. 2021, 80, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Jairath, N.K.; Dal Pra, A.; Vince Jr, R.; Dess, R.T.; Jackson, W.C.; Tosoian, J.J.; McBride, S.M.; Zhao, S.G.; Berlin, A.; Mahal, B.A.; et al. A systematic review of the evidence for the decipher genomic classifier in prostate cancer. Eur. Urol. 2021, 79, 374–383. [Google Scholar] [CrossRef]
- Fine, N.D.; LaPolla, F.; Epstein, M.; Loeb, S.; Dani, H. Genomic classifiers for treatment selection in newly diagnosed prostate cancer. BJU Int. 2019, 124, 578–586. [Google Scholar] [CrossRef]
- Eastham, J.A.; Auffenberg, G.B.; Barocas, D.A.; Chou, R.; Crispino, T.; Davis, J.W.; Eggener, S.; Horwitz, E.M.; Kane, C.J.; Kirkby, E.; et al. Clinically localized prostate cancer: AUA/ASTRO guideline, part I: Introduction, risk assessment, staging, and risk-based management. J. Urol. 2022, 208, 10–18. [Google Scholar] [CrossRef]
- Merseburger, A.S.; Waldron, N.; Ribal, M.J.; Heidenreich, A.; Perner, S.; Fizazi, K.; Sternberg, C.N.; Mateo, J.; Wirth, M.P.; Castro, E.; et al. Genomic testing in patients with metastatic castration-resistant prostate cancer: A pragmatic guide for clinicians. Eur. Urol. 2021, 79, 519–529. [Google Scholar] [CrossRef]
- Xiang, J.; Yan, H.; Li, J.; Wang, X.; Chen, H.; Zheng, X. Transperineal versus transrectal prostate biopsy in the diagnosis of prostate cancer: A systematic review and meta-analysis. World J. Surg. Oncol. 2019, 17, 1–11. [Google Scholar] [CrossRef]
- Sun, Y.; Reynolds, H.M.; Parameswaran, B.; Wraith, D.; Finnegan, M.E.; Williams, S.; Haworth, A. Multiparametric MRI and radiomics in prostate cancer: A review. Australas. Phys. Eng. Sci. Med. 2019, 42, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Drost, F.J.H.; Osses, D.; Nieboer, D.; Bangma, C.H.; Steyerberg, E.W.; Roobol, M.J.; Schoots, I.G. Prostate magnetic resonance imaging, with or without magnetic resonance imaging-targeted biopsy, and systematic biopsy for detecting prostate cancer: A Cochrane systematic review and meta-analysis. Eur. Urol. 2020, 77, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Eastham, J.A.; Auffenberg, G.B.; Barocas, D.A.; Chou, R.; Crispino, T.; Davis, J.W.; Eggener, S.; Horwitz, E.M.; Kane, C.J.; Kirkby, E.; et al. Clinically localized prostate cancer: AUA/ASTRO guideline, part II: Principles of active surveillance, principles of surgery, and follow-up. J. Urol. 2022, 208, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Rajesh, A.; Rosenkrantz, A.B.; Choyke, P.L.; Turkbey, B. PI-RADS version 2.1: One small step for prostate MRI. Clin. Radiol. 2019, 74, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, C.J.; Bao, M.L.; Zhang, J.; Wang, X.N.; Zhang, Y.D. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur. Radiol. 2017, 27, 4082–4090. [Google Scholar] [CrossRef]
- Hambrock, T.; Vos, P.C.; Hulsbergen-van de Kaa, C.A.; Barentsz, J.O.; Huisman, H.J. Prostate cancer: Computer-aided diagnosis with multiparametric 3-T MR imaging—Effect on observer performance. Radiology 2013, 266, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Wienke, A.; Surov, A. Discrimination between clinical significant and insignificant prostate cancer with apparent diffusion coefficient—A systematic review and meta analysis. BMC Cancer 2020, 20, 482. [Google Scholar] [CrossRef]
- Franiel, T.; Hamm, B.; Hricak, H. Dynamic contrast-enhanced magnetic resonance imaging and pharmacokinetic models in prostate cancer. Eur. Radiol. 2011, 21, 616–626. [Google Scholar] [CrossRef]
- Turco, S.; Lavini, C.; Heijmink, S.; Barentsz, J.; Wijkstra, H.; Mischi, M. Evaluation of dispersion MRI for improved prostate cancer diagnosis in a multicenter study. AJR Am. J. Roentgenol. 2018, 211, 242–251. [Google Scholar] [CrossRef]
- Ferro, M.; de Cobelli, O.; Vartolomei, M.D.; Lucarelli, G.; Crocetto, F.; Barone, B.; Sciarra, A.; Del Giudice, F.; Muto, M.; Maggi, M.; et al. Prostate cancer radiogenomics—From imaging to molecular characterization. Int. J. Mol. Sci. 2021, 22, 9971. [Google Scholar] [CrossRef] [PubMed]
- Ardakani, A.A.; Bureau, N.J.; Ciaccio, E.J.; Acharya, U.R. Interpretation of radiomics features—A pictorial review. Comput. Methods Programs Biomed. 2022, 215, 106609. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging imaging and genomics. Abdom. Radiol. 2019, 44, 1960–1984. [Google Scholar] [CrossRef]
- Perlis, N.; Al-Kasab, T.; Ahmad, A.; Goldberg, E.; Fadak, K.; Sayid, R.; Finelli, A.; Kulkarni, G.; Hamilton, R.; Zlotta, A.; et al. Defining a cohort that may not require repeat prostate biopsy based on PCA3 score and magnetic resonance imaging: The dual negative effect. J. Urol. 2018, 199, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Fenstermaker, M.; Mendhiratta, N.; Bjurlin, M.A.; Meng, X.; Rosenkrantz, A.B.; Huang, R.; Deng, F.M.; Zhou, M.; Huang, W.C.; Lepor, H.; et al. Risk stratification by urinary prostate cancer gene 3 testing before magnetic resonance imaging-ultrasound fusion-targeted prostate biopsy among men with no history of biopsy. Urology 2017, 99, 174–179. [Google Scholar] [CrossRef]
- De Luca, S.; Passera, R.; Cattaneo, G.; Manfredi, M.; Mele, F.; Fiori, C.; Bollito, E.; Cirillo, S.; Porpiglia, F. High prostate cancer gene 3 (PCA 3) scores are associated with elevated Prostate Imaging Reporting and Data System (PI-RADS) grade and biopsy Gleason score, at magnetic resonance imaging/ultrasonography fusion software-based targeted prostate biopsy after a previous negative standard biopsy. BJU Int. 2016, 118, 723–730. [Google Scholar] [PubMed]
- Martin, D.T.; Ghabili, K.; Levi, A.; Humphrey, P.A.; Sprenkle, P.C. Prostate cancer genomic classifier relates more strongly to Gleason grade group than Prostate Imaging Reporting and Data System score in multiparametric prostate magnetic resonance imaging-ultrasound fusion targeted biopsies. Urology 2019, 125, 64–72. [Google Scholar] [CrossRef]
- Leapman, M.S.; Westphalen, A.C.; Ameli, N.; Lawrence, H.J.; Febbo, P.G.; Cooperberg, M.R.; Carroll, P.R. Association between a 17-gene genomic prostate score and multi-parametric prostate MRI in men with low and intermediate risk prostate cancer (PCa). PLoS ONE 2017, 12, e0185535. [Google Scholar] [CrossRef] [PubMed]
- Beksac, A.T.; Cumarasamy, S.; Falagario, U.; Xu, P.; Takhar, M.; Alshalalfa, M.; Gupta, A.; Prasad, S.; Martini, A.; Thulasidass, H.; et al. Multiparametric magnetic resonance imaging features identify aggressive prostate cancer at the phenotypic and transcriptomic level. J. Urol. 2018, 200, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Salmasi, A.; Said, J.; Shindel, A.W.; Khoshnoodi, P.; Felker, E.R.; Sisk, A.E., Jr.; Grogan, T.; McCullough, D.; Bennett, J.; Bailey, H.; et al. A 17-gene genomic prostate score assay provides independent information on adverse pathology in the setting of combined multiparametric magnetic resonance imaging fusion targeted and systematic prostate biopsy. J. Urol. 2018, 200, 564–572. [Google Scholar] [CrossRef] [PubMed]
- McCann, S.M.; Jiang, Y.; Fan, X.; Wang, J.; Antic, T.; Prior, F.; VanderWeele, D.; Oto, A. Quantitative multiparametric MRI features and PTEN expression of peripheral zone prostate cancer: A pilot study. Am. J. Roentgenol. 2016, 206, 559–565. [Google Scholar] [CrossRef]
- Stoyanova, R.; Pollack, A.; Takhar, M.; Lynne, C.; Parra, N.; Lam, L.L.; Alshalalfa, M.; Buerki, C.; Castillo, R.; Jorda, M.; et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget 2016, 7, 53362. [Google Scholar] [CrossRef]
- Hectors, S.J.; Cherny, M.; Yadav, K.K.; Beksaç, A.T.; Thulasidass, H.; Lewis, S.; Davicioni, E.; Wang, P.; Tewari, A.K.; Taouli, B. Radiomics features measured with multiparametric magnetic resonance imaging predict prostate cancer aggressiveness. J. Urol. 2019, 202, 498–505. [Google Scholar] [CrossRef]
- Switlyk, M.D.; Salberg, U.B.; Geier, O.M.; Vlatkovic, L.; Lilleby, W.; Lyng, H.; Seierstad, T. PTEN expression in prostate cancer: Relationship with clinicopathologic features and multiparametric MRI findings. Am. J. Roentgenol. 2019, 212, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Tahoun, M.; Klaan, B.; Thierfelder, K.M.; Weber, M.A.; Krause, B.J.; Hakenberg, O.; Fuellen, G.; Hamed, M. A radiogenomic approach for decoding molecular mechanisms underlying tumor progression in prostate cancer. Cancers 2019, 11, 1293. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Williams, S.; Byrne, D.; Keam, S.; Reynolds, H.M.; Mitchell, C.; Wraith, D.; Murphy, D.; Haworth, A. Association analysis between quantitative MRI features and hypoxia-related genetic profiles in prostate cancer: A pilot study. Br. J. Radiol. 2019, 92, 20190373. [Google Scholar] [CrossRef] [PubMed]
- Mischi, M.; Turco, S.; Lavini, C.; Kompatsiari, K.; de la Rosette, J.J.; Breeuwer, M.; Wijkstra, H. Magnetic resonance dispersion imaging for localization of angiogenesis and cancer growth. Investig. Radiol. 2014, 49, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Abeshouse, A.; Ahn, J.; Akbani, R.; Ally, A.; Amin, S.; Andry, C.D.; Annala, M.; Aprikian, A.; Armenia, J.; Arora, A.; et al. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Srigley, J.R.; Delahunt, B.; Samaratunga, H.; Billis, A.; Cheng, L.; Clouston, D.; Evans, A.; Furusato, B.; Kench, J.; Leite, K.; et al. Controversial issues in Gleason and International Society of Urological Pathology (ISUP) prostate cancer grading: Proposed recommendations for international implementation. Pathology 2019, 51, 463–473. [Google Scholar] [CrossRef]
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Schnall, M.D.; Shtern, F.; Tempany, C.M.; et al. PI-RADS prostate imaging—Reporting and data system: 2015, version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef] [PubMed]
- Dinis Fernandes, C.; Turco, S.; Mischi, M.; Wijkstra, H. Deep learning for prostate and zonal segmentation on a multicenter MRI dataset. In Proceedings of the 9th EAU Section of Urological Imaging 2021, Athens, Greece, 25 November 2021. [Google Scholar]
- Yushkevich, P.A.; Piven, J.; Cody Hazlett, H.; Gimpel Smith, R.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 1–20. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Lapuente-Santana, Ó.; van Genderen, M.; Hilbers, P.A.; Finotello, F.; Eduati, F. Interpretable systems biomarkers predict response to immune-checkpoint inhibitors. Patterns 2021, 2, 100293. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Klinger, B.; Klünemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Blüthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Zhao, S.G.; Chang, S.L.; Spratt, D.E.; Erho, N.; Yu, M.; Ashab, H.A.D.; Alshalalfa, M.; Speers, C.; Tomlins, S.A.; Davicioni, E.; et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: A matched, retrospective analysis. Lancet Oncol. 2016, 17, 1612–1620. [Google Scholar] [CrossRef] [PubMed]
- Erho, N.; Crisan, A.; Vergara, I.A.; Mitra, A.P.; Ghadessi, M.; Buerki, C.; Bergstralh, E.J.; Kollmeyer, T.; Fink, S.; Haddad, Z.; et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE 2013, 8, e66855. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Messina, J.L.; Fenstermacher, D.A.; Eschrich, S.; Qu, X.; Berglund, A.E.; Lloyd, M.C.; Schell, M.J.; Sondak, V.K.; Weber, J.S.; Mulé, J.J. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: Potential for patient selection for immunotherapy? Sci. Rep. 2012, 2, 765. [Google Scholar] [CrossRef] [PubMed]
- Szumilas, M. Explaining odds ratios. J. Can. Acad. Child Adolesc. Psychiatry 2010, 19, 227. [Google Scholar] [PubMed]
- Zhao, Z.; Anand, R.; Wang, M. Maximum relevance and minimum redundancy feature selection methods for a marketing machine learning platform. In Proceedings of the 2019 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Washington, DC, USA, 5–8 October 2019; pp. 442–452. [Google Scholar]
- Verma, S.; Turkbey, B.; Muradyan, N.; Rajesh, A.; Cornud, F.; Haider, M.A.; Choyke, P.L.; Harisinghani, M. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR. Am. J. Roentgenol. 2012, 198, 1277. [Google Scholar] [CrossRef]
- Vallieres, M.; Kay-Rivest, E.; Perrin, L.J.; Liem, X.; Furstoss, C.; Aerts, H.J.; Khaouam, N.; Nguyen-Tan, P.F.; Wang, C.S.; Sultanem, K.; et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 2017, 7, 10117. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; Volume 57, pp. 10–25080. [Google Scholar]
- Roy, S.; Whitehead, T.D.; Quirk, J.D.; Salter, A.; Ademuyiwa, F.O.; Li, S.; An, H.; Shoghi, K.I. Optimal co-clinical radiomics: Sensitivity of radiomic features to tumour volume, image noise and resolution in co-clinical T1-weighted and T2-weighted magnetic resonance imaging. EBioMedicine 2020, 59, 102963. [Google Scholar] [CrossRef] [PubMed]
- Boesen, L.; Chabanova, E.; Løgager, V.; Balslev, I.; Thomsen, H.S. Apparent diffusion coefficient ratio correlates significantly with prostate cancer gleason score at final pathology. J. Magn. Reson. Imaging JMRI 2015, 42, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Rajesh, A.; Morales, H.; Lemen, L.; Bills, G.; Delworth, M.; Gaitonde, K.; Ying, J.; Samartunga, R.; Lamba, M. Assessment of aggressiveness of prostate cancer: Correlation of apparent diffusion coefficient with histologic grade after radical prostatectomy. AJR. Am. J. Roentgenol. 2011, 196, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, J.; Thimansson, E.; Baubeta, E.; Zackrisson, S.; Sundgren, P.C.; Bjartell, A.; Flondell-Sité, D. Correlation between ADC, ADC ratio, and Gleason Grade group in prostate cancer patients undergoing radical prostatectomy: Retrospective multicenter study with different MRI scanners. Front. Oncol. 2023, 13, 1079040. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Roth, C.P.; Wasson, L.M.; Vishwanatha, J.K. Signal transducer and activator of transcription-6 (STAT6) is a constitutively expressed survival factor in human prostate cancer. Prostate 2007, 67, 1550–1564. [Google Scholar] [CrossRef] [PubMed]
- Schorle, H.; Meier, P.; Buchert, M.; Jaenisch, R.; Mitchell, P.J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 1996, 381, 235–238. [Google Scholar] [CrossRef]
- Lipponen, P.; Aaltomaa, S.; Kellokoski, J.; Ala-Opas, M.; Kosma, V.M. Expression of activator protein 2 in prostate cancer is related to tumor differentiation and cell proliferation. Eur. Urol. 2000, 37, 573–578. [Google Scholar] [CrossRef] [PubMed]
- de la Taille, A.; Katz, A.E.; Bagiella, E.; Buttyan, R.; Sharir, S.; Olsson, C.A.; Burchardt, T.; Ennis, R.D.; Rubin, M.A. Microvessel density as a predictor of PSA recurrence after radical prostatectomy: A comparison of CD34 and CD31. Am. J. Clin. Pathol. 2000, 113, 555–562. [Google Scholar] [CrossRef]
- Miller, J.C.; Pien, H.H.; Sahani, D.; Sorensen, A.G.; Thrall, J.H. Imaging angiogenesis: Applications and potential for drug development. J. Natl. Cancer Inst. 2005, 97, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Oto, A.; Yang, C.; Kayhan, A.; Tretiakova, M.; Antic, T.; Schmid-Tannwald, C.; Eggener, S.; Karczmar, G.S.; Stadler, W.M. Diffusion-weighted and dynamic contrast-enhanced MRI of prostate cancer: Correlation of quantitative MR parameters with Gleason score and tumor angiogenesis. Am. J. Roentgenol. 2011, 197, 1382–1390. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K pathway in human disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Wong, H.Y.; Sheng, Q.; Hesterberg, A.B.; Croessmann, S.; Rios, B.L.; Giri, K.; Jackson, J.; Miranda, A.X.; Watkins, E.; Schaffer, K.R.; et al. Single cell analysis of cribriform prostate cancer reveals cell intrinsic and tumor microenvironmental pathways of aggressive disease. Nat. Commun. 2022, 13, 6036. [Google Scholar] [CrossRef]
- Berglund, E.; Maaskola, J.; Schultz, N.; Friedrich, S.; Marklund, M.; Bergenstråhle, J.; Tarish, F.; Tanoglidi, A.; Vickovic, S.; Larsson, L.; et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018, 9, 2419. [Google Scholar] [CrossRef] [PubMed]
- Fournier, L.; Costaridou, L.; Bidaut, L.; Michoux, N.; Lecouvet, F.E.; de Geus-Oei, L.F.; Boellaard, R.; Oprea-Lager, D.E.; Obuchowski, N.A.; Caroli, A.; et al. Incorporating radiomics into clinical trials: Expert consensus endorsed by the European Society of Radiology on considerations for data-driven compared to biologically driven quantitative biomarkers. Eur. Radiol. 2021, 31, 6001–6012. [Google Scholar] [CrossRef] [PubMed]
- Saltybaeva, N.; Tanadini-Lang, S.; Vuong, D.; Burgermeister, S.; Mayinger, M.; Bink, A.; Andratschke, N.; Guckenberger, M.; Bogowicz, M. Robustness of radiomic features in magnetic resonance imaging for patients with glioblastoma: Multi-center study. Phys. Imaging Radiat. Oncol. 2022, 22, 131–136. [Google Scholar] [CrossRef] [PubMed]

| Transcriptomics | Imaging | |||
|---|---|---|---|---|
| Training | Test | Training | Test | |
| ISUP 1 | 64 (19.3%) | 5 (5.5%) | 11 (24.4%) | 1 (2.9%) |
| ISUP 2 | 102 (30.8%) | 29 (31.9%) | 19 (42.2%) | 10 (28.6%) |
| ISUP 3 | 78 (23.6%) | 27 (29.7%) | 9 (20.0%) | 10 (28.6%) |
| ISUP 4 | 44 (13.3%) | 15 (16.5%) | 3 (6.7%) | 8 (22.9%) |
| ISUP 5 | 43 (13.0%) | 15 (16.5%) | 3 (6.7%) | 6 (17.1%) |
| Texture Type | Features | Number of Features |
|---|---|---|
| Global | Variance, skewness, kurtosis | 3 |
| Gray-level co-occurrence matrix (GLCM) | Energy, contrast, correlation, homogeneity, variance, Sum average, entropy, dissimilarity, autocorrelation | 9 |
| Gray-level run-length matrix (GLRLM) | Short-run emphasis (SRE), long-run emphasis (LRE), gray-level non-uniformity (GLN), run-length non-uniformity (RLN), run percentage (RP), low gray-level run emphasis (LGRE), high gray-level run emphasis (HGRE), short-run low gray-level emphasis (SRLGE), short-run high gray-level emphasis (SRHGE), long-run low gray-level emphasis (LRLGE), long-run high gray-level emphasis (LRHGE), gray-level variance (GLV), run-length variance (RLV) | 13 |
| Gray-level size zone matrix (GLSZM) | Small zone emphasis (SZE), large zone emphasis (LZE), gray-level non-uniformity (GLN), zone-size non-uniformity (ZSN), zone percentage (ZP), low gray-level zone emphasis (LGZE), high gray-level zone emphasis (HGZE), small zone low gray-level emphasis (SZLGE), small zone high gray-level emphasis (SZHGE), large zone low gray-level emphasis (LZLGE), large zone high gray-level emphasis (LZHGE), gray-level variance (GLV), zone-size variance (ZSV) | 13 |
| Neighborhood gray-tone difference matrix (NGTDM) | Coarseness, contrast, business, Complexity, strength | 5 |
| Type of Transcriptomic Feature | Acronym | Full Name |
|---|---|---|
| Transcription factors | REST | RE1 silencing transcription factor |
| FOXA2 | Forkhead box protein A2 | |
| TFDP1 | Transcription factor Dp-1 | |
| ERG | Erythroblast transformation specific (ETS) transcription factor ERG | |
| SOX9 | SRY-box transcription factor 9 | |
| TFAP2A | Transcription factor AP-2 alpha | |
| TFAP2C | Transcription factor AP-2 gamma | |
| HNF4A | Hepatocyte nuclear factor 4 alpha | |
| EPAS1 | Endothelial PAS domain-containing protein 1 | |
| E2F1 | E2F transcription factor 1 | |
| E2F2 | E2F transcription factor 2 | |
| E2F3 | E2F transcription factor 3 | |
| E2F4 | E2F transcription factor 4 | |
| BACH1 | BTB and CNC homology 1 | |
| STAT6 | Signal transducer and activator of transcription 6 | |
| AR | Androgen receptor | |
| NR2F2 | Nuclear receptor subfamily 2 group F member 2 | |
| Pathways * | EGFR | Epidermal growth factor receptor—regulates growth, survival, migration, apoptosis, proliferation, and differentiation in mammalian cells. |
| PI3K | Phosphatidylinositol 3-kinase—promotes growth and proliferation. | |
| Androgen | Involved in the growth and development of the male reproductive organs. | |
| Estrogen | Promotes the growth and development of the female reproductive organs. | |
| JAK-STAT | Janus kinases (JAKs), signal transducer and activator of transcription proteins (STATs)—involved in immunity, cell division, cell death, and tumor formation. | |
| MAPK | Mitogen-activated protein kinases (MAPKs)—integrates external signals and promotes cell growth and proliferation. | |
| TGFb | Transforming growth factor beta—involved in the development, homeostasis, and repair of most tissues. | |
| Trail | TNF-related apoptosis-inducing ligand—induces apoptosis. | |
| VEGF | Vascular endothelial growth factor—mediates angiogenesis, vascular permeability, and cell migration. | |
| WNT | Created from the names wingless and Int-1—regulates organ morphogenesis during development and tissue repair. | |
| Signatures | Decipher geometric mean | Decipher signature, derived using the geometric mean |
| Decipher PCA | Decipher signature, derived using PCA | |
| Decipher ssGSEA | Decipher signature, derived using single-sample gene set enrichment analysis | |
| PORTOS geometric mean | PORTOS signature, derived using the geometric mean | |
| PORTOS PCA | PORTOS signature, derived using PCA | |
| PORTOS ssGSEA | PORTOS signature, derived using single-sample gene set enrichment analysis |
| Classifier | Hyperparameter Description | Optimal Value |
|---|---|---|
| LR | Solver | Liblinear |
| Penalty | l2 | |
| C | 0.02 | |
| 31 | ||
| KNN | Weights | Uniform |
| Metric | Minkowski | |
| K | 28 | |
| 30 | ||
| SVM | Kernel | Sigmoid |
| 0.01 | ||
| C | 0.95 | |
| 27 |
| Train | Test | |||||
|---|---|---|---|---|---|---|
| LR | KNN | SVM | LR | KNN | SVM | |
| ACC | 0.72 (0.07) | 0.67 (0.04) | 0.75 (0.08) | 0.71 | 0.34 | 0.69 |
| bACC | 0.72 (0.08) | 0.55 (0.05) | 0.74 (0.08) | 0.74 | 0.52 | 0.72 |
| Sensitivity | 0.72 (0.14) | 0.15 (0.11) | 0.72 (0.16) | 0.67 | 0.04 | 0.63 |
| Specificity | 0.72 (0.09) | 0.96 (0.05) | 0.76 (0.09) | 0.82 | 1.00 | 0.82 |
| AUC | 0.82 (0.08) | 0.72 (0.08) | 0.83 (0.08) | 0.74 | 0.52 | 0.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dinis Fernandes, C.; Schaap, A.; Kant, J.; van Houdt, P.; Wijkstra, H.; Bekers, E.; Linder, S.; Bergman, A.M.; van der Heide, U.; Mischi, M.; et al. Radiogenomics Analysis Linking Multiparametric MRI and Transcriptomics in Prostate Cancer. Cancers 2023, 15, 3074. https://doi.org/10.3390/cancers15123074
Dinis Fernandes C, Schaap A, Kant J, van Houdt P, Wijkstra H, Bekers E, Linder S, Bergman AM, van der Heide U, Mischi M, et al. Radiogenomics Analysis Linking Multiparametric MRI and Transcriptomics in Prostate Cancer. Cancers. 2023; 15(12):3074. https://doi.org/10.3390/cancers15123074
Chicago/Turabian StyleDinis Fernandes, Catarina, Annekoos Schaap, Joan Kant, Petra van Houdt, Hessel Wijkstra, Elise Bekers, Simon Linder, Andries M. Bergman, Uulke van der Heide, Massimo Mischi, and et al. 2023. "Radiogenomics Analysis Linking Multiparametric MRI and Transcriptomics in Prostate Cancer" Cancers 15, no. 12: 3074. https://doi.org/10.3390/cancers15123074
APA StyleDinis Fernandes, C., Schaap, A., Kant, J., van Houdt, P., Wijkstra, H., Bekers, E., Linder, S., Bergman, A. M., van der Heide, U., Mischi, M., Zwart, W., Eduati, F., & Turco, S. (2023). Radiogenomics Analysis Linking Multiparametric MRI and Transcriptomics in Prostate Cancer. Cancers, 15(12), 3074. https://doi.org/10.3390/cancers15123074








