Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Consent
2.2. Standard Diagnostics
2.3. Optical Genome Mapping and Rare Variant Pipeline
2.4. Reverse-Transcription Polymerase Chain Reaction
3. Results
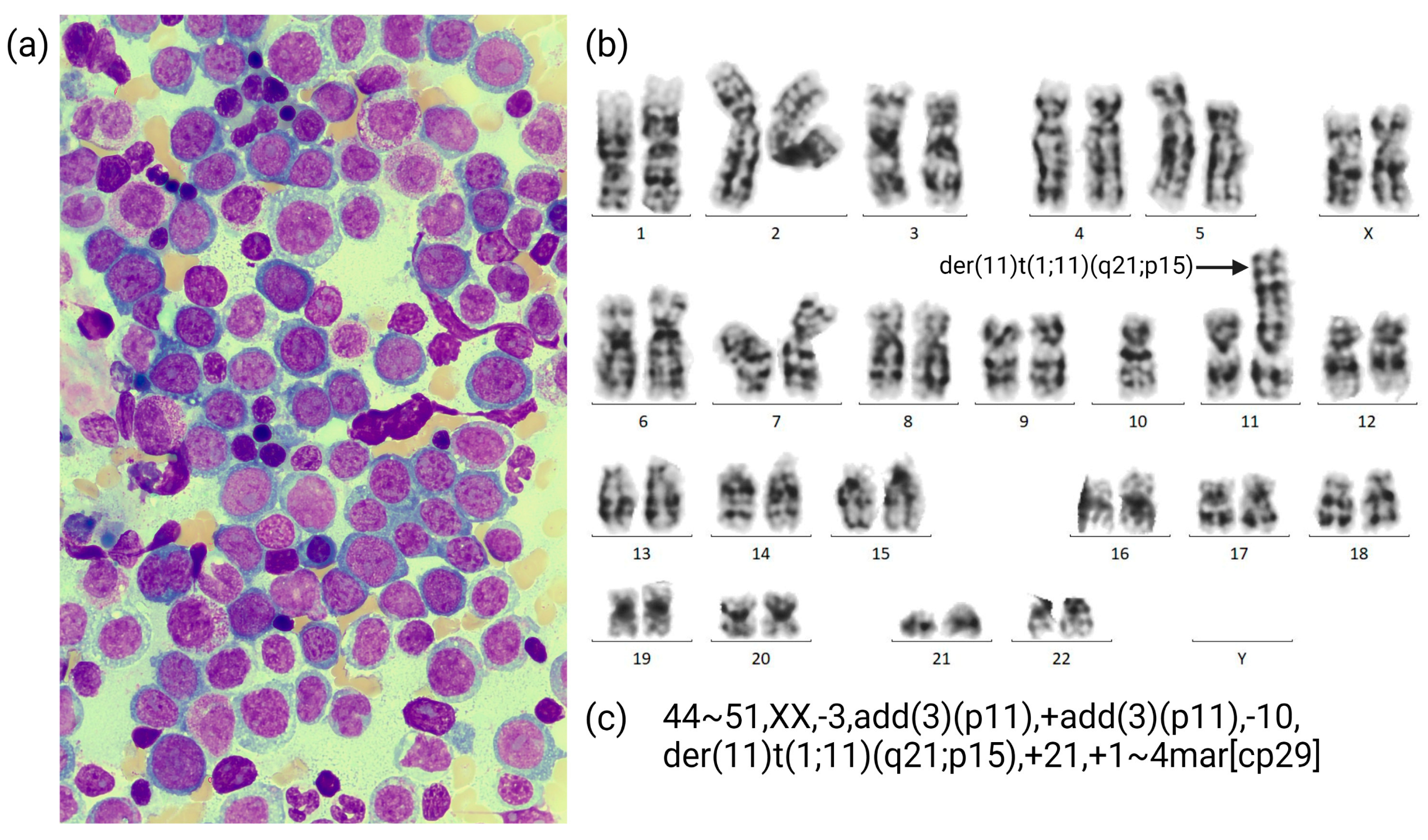
3.1. Case Description
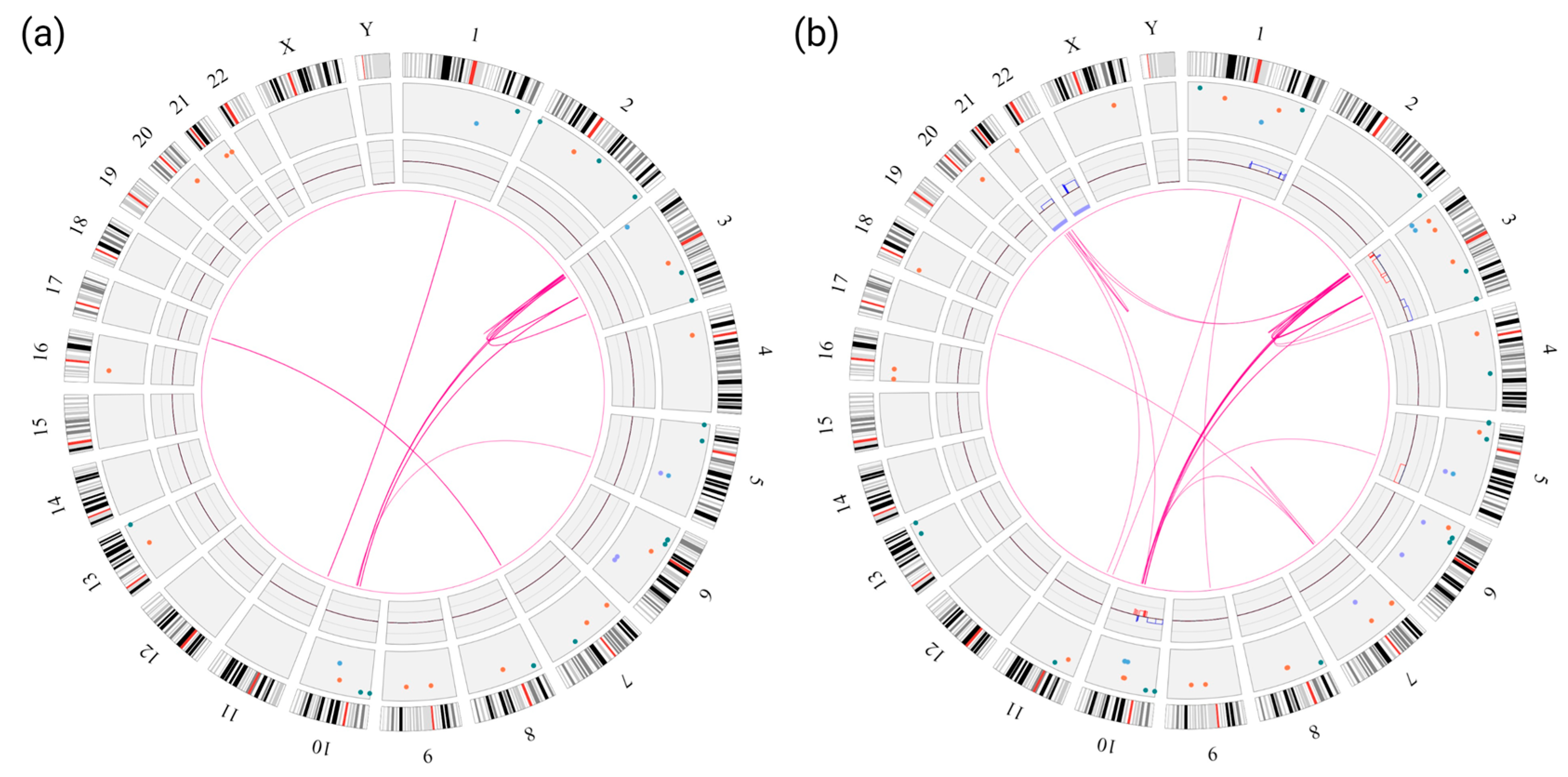
3.2. OGM Findings
3.3. Confirmation of the NUP98::ASH1L Gene Fusion

4. Discussion
4.1. OGM as a Tool for Cytogenetic Diagnostics of AML
4.2. NUP98::ASH1L in Context of NUP98-Rearrangements
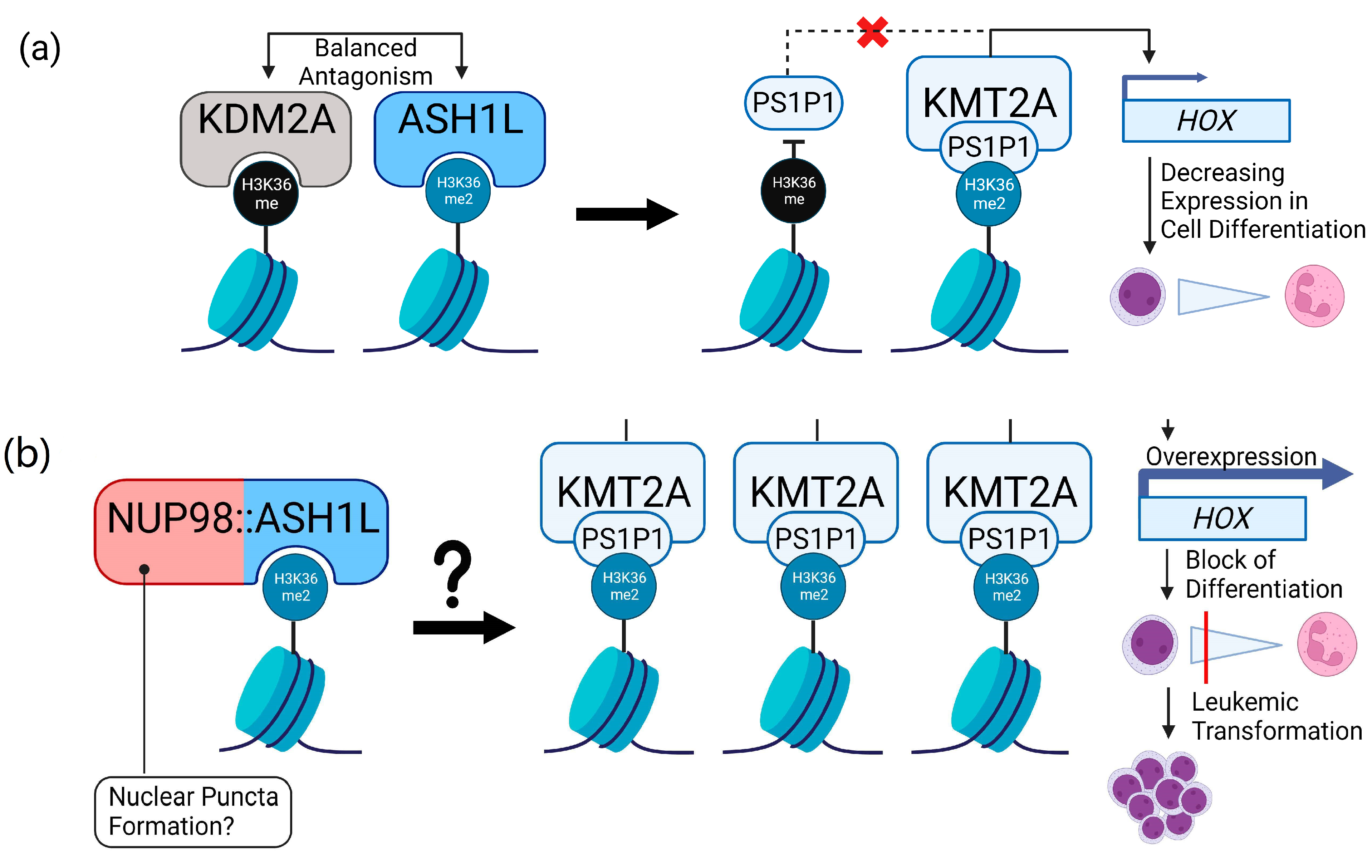
4.3. Role of ASH1L in Leukemia and Putative Implications for NUP98::ASH1L
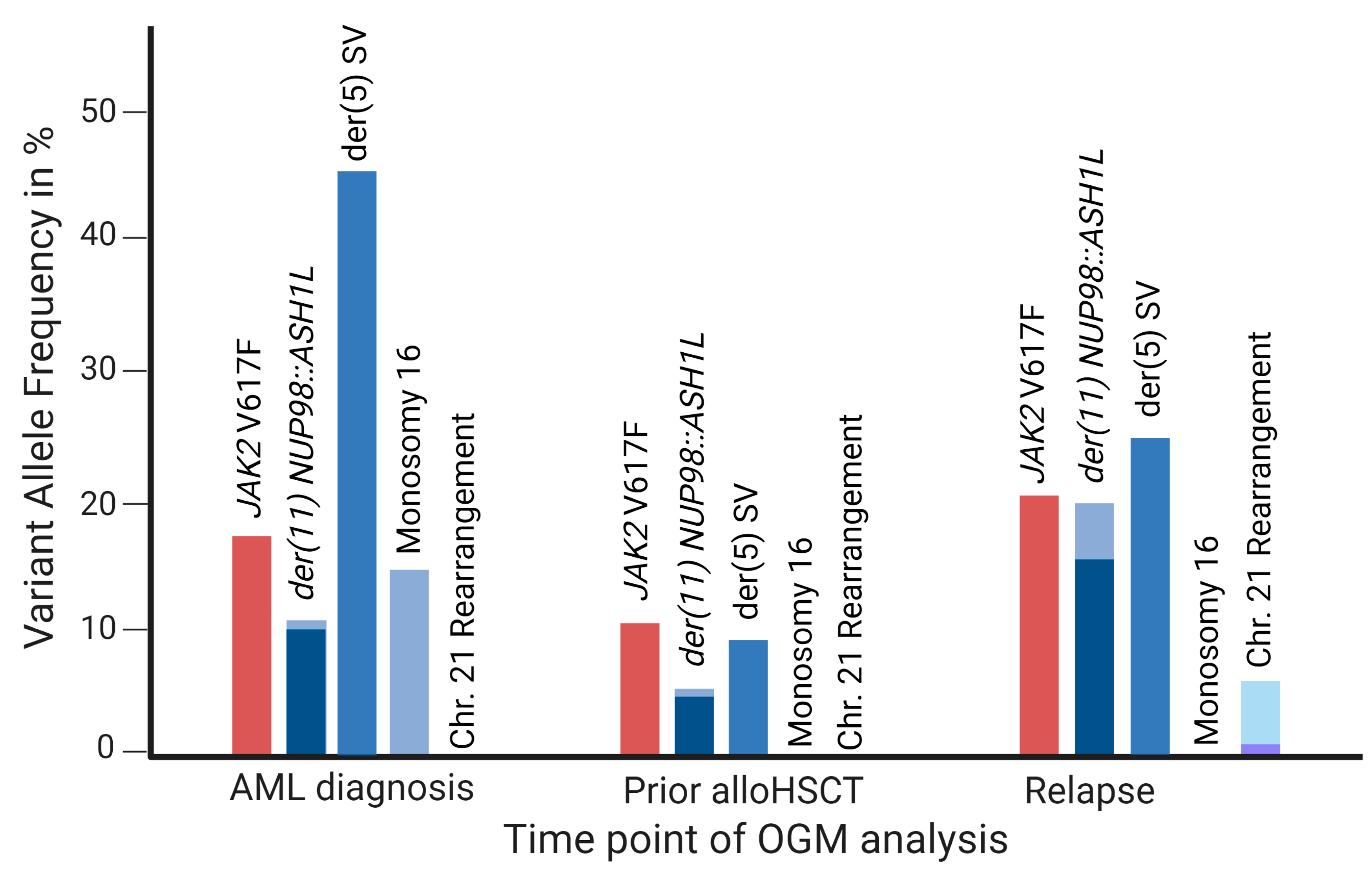
4.4. OGM as a Tool for Disease Monitoring in AML
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1
- Initial AML diagnosis:44~51,XX,-3,add(3)(p11),+add(3)(p11),-10,der(11)t(1;11)(q21;p15),+21,+1~4mar[cp29]
- Post-Induction II:46,XX [25]
- Post-alloHSCT:46~49,XX,add(3)(p11),-10,der(11)t(1;11)(q21;p15),+21,+22,1~2mar,?inc[cp3]/49~50,XX,-3,-10,der(11)t(1:11)(q21;p15),+21,+22,+3~4mar,?inc[cp3]/46,XX [17]
- Relapse:46~49,XX,-3,-10,der(11)t(1;11)(q21;p15),+21,+22,+1~5mar,?inc[cp14]/45~49,XX,add(3)(p11),-10,der(11)t(1;11)(q21;p15),+21,+22,+2~5mar,?inc[cp8]/48~49,XX,add(3)(p11),-10,der(11)t(1;11)(q21;p15),add(11)(p1?4),+21,+22,+2~6mar,?inc[cp3]
Appendix B
References
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 ELN Recommendations from an International Expert Panel. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.G.; Hoadley, K.; Triche, T.J.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Michmerhuizen, N.L.; Klco, J.M.; Mullighan, C.G. Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood 2020, 136, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef] [PubMed]
- Struski, S.; Lagarde, S.; Bories, P.; Puiseux, C.; Prade, N.; Cuccuini, W.; Pages, M.-P.; Bidet, A.; Gervais, C.; Lafage-Pochitaloff, M.; et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia 2017, 31, 565–572. [Google Scholar] [CrossRef]
- Xie, W.; Raess, P.W.; Dunlap, J.; Hoyos, C.M.; Li, H.; Li, P.; Swords, R.; Olson, S.B.; Yang, F.; Anekpuritanang, T.; et al. Adult acute myeloid leukemia patients with NUP98 rearrangement have frequent cryptic translocations and unfavorable outcome. Leuk. Lymphoma 2022, 63, 1907–1916. [Google Scholar] [CrossRef]
- Huber, S.; Baer, C.; Hutter, S.; Dicker, F.; Meggendorfer, M.; Pohlkamp, C.; Kern, W.; Haferlach, T.; Haferlach, C.; Hoermann, G. AML and MDS Classification according to Who 2022 and International Consensus Classification: Do We Invent a Babylonian Confusion of Languages? Blood 2022, 140, 555–556. [Google Scholar] [CrossRef]
- Marceau-Renaut, A.; Duployez, N.; Ducourneau, B.; Labopin, M.; Petit, A.; Rousseau, A.; Geffroy, S.; Bucci, M.; Cuccuini, W.; Fenneteau, O.; et al. Molecular Profiling Defines Distinct Prognostic Subgroups in Childhood AML: A Report from the French ELAM02 Study Group. Hemasphere 2018, 2, e31. [Google Scholar] [CrossRef]
- Shiba, N.; Yoshida, K.; Hara, Y.; Yamato, G.; Shiraishi, Y.; Matsuo, H.; Okuno, Y.; Chiba, K.; Tanaka, H.; Kaburagi, T.; et al. Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood Adv. 2019, 3, 3157–3169. [Google Scholar] [CrossRef]
- Hollink, I.H.I.M.; van den Heuvel-Eibrink, M.M.; Arentsen-Peters, S.T.C.J.M.; Pratcorona, M.; Abbas, S.; Kuipers, J.E.; van Galen, J.F.; Beverloo, H.B.; Sonneveld, E.; Kaspers, G.-J.J.L.; et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 2011, 118, 3645–3656. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Kölking, B.; Hollink, I.H.I.; Damm, F.; van den Heuvel-Eibrink, M.M.; Michel Zwaan, C.; Bug, G.; Ottmann, O.; Wagner, K.; Morgan, M.; et al. Analysis of NUP98/NSD1 translocations in adult AML and MDS patients. Leukemia 2013, 27, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Wang, S.; Qiu, S.; Qi, J.; Mi, Y.; Lin, D.; Zhou, C.; Liu, B.; Li, W.; Wang, Y.; et al. Clinical and laboratory studies of 17 patients with acute myeloid leukemia harboring t(7;11)(p15;p15) translocation. Leuk. Res. 2013, 37, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-C.; Chen, C.-Y.; Hou, H.-A.; Lin, L.-I.; Tang, J.-L.; Yao, M.; Tsay, W.; Ko, B.-S.; Wu, S.-J.; Huang, S.-Y.; et al. Acute myeloid leukemia bearing t(7;11)(p15;p15) is a distinct cytogenetic entity with poor outcome and a distinct mutation profile: Comparative analysis of 493 adult patients. Leukemia 2009, 23, 1303–1310. [Google Scholar] [CrossRef]
- Fasan, A.; Haferlach, C.; Alpermann, T.; Kern, W.; Haferlach, T.; Schnittger, S. A rare but specific subset of adult AML patients can be defined by the cytogenetically cryptic NUP98-NSD1 fusion gene. Leukemia 2013, 27, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Niktoreh, N.; Walter, C.; Zimmermann, M.; von Neuhoff, C.; von Neuhoff, N.; Rasche, M.; Waack, K.; Creutzig, U.; Hanenberg, H.; Reinhardt, D. Mutated WT1, FLT3-ITD, and NUP98-NSD1 Fusion in Various Combinations Define a Poor Prognostic Group in Pediatric Acute Myeloid Leukemia. J. Oncol. 2019, 2019, 1609128. [Google Scholar] [CrossRef]
- Shiba, N.; Ichikawa, H.; Taki, T.; Park, M.-J.; Jo, A.; Mitani, S.; Kobayashi, T.; Shimada, A.; Sotomatsu, M.; Arakawa, H.; et al. NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes Chromosomes Cancer 2013, 52, 683–693. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Ed.; International Agency for Research on Cancer: Lyon, France, 2017; ISBN 9789283244943. [Google Scholar]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Reader, J.C.; Meekins, J.S.; Gojo, I.; Ning, Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia 2007, 21, 842–844. [Google Scholar] [CrossRef]
- Balducci, E.; Kaltenbach, S.; Villarese, P.; Duroyon, E.; Zalmai, L.; Friedrich, C.; Suarez, F.; Marcais, A.; Bouscary, D.; Decroocq, J.; et al. Optical genome mapping refines cytogenetic diagnostics, prognostic stratification and provides new molecular insights in adult MDS/AML patients. Blood Cancer J. 2022, 12, 126. [Google Scholar] [CrossRef] [PubMed]
- Gerding, W.M.; Tembrink, M.; Nilius-Eliliwi, V.; Mika, T.; Dimopoulos, F.; Ladigan-Badura, S.; Eckhardt, M.; Pohl, M.; Wünnenberg, M.; Farshi, P.; et al. Optical genome mapping reveals additional prognostic information compared to conventional cytogenetics in AML/MDS patients. Int. J. Cancer 2022, 150, 1998–2011. [Google Scholar] [CrossRef] [PubMed]
- Neveling, K.; Mantere, T.; Vermeulen, S.; Oorsprong, M.; van Beek, R.; Kater-Baats, E.; Pauper, M.; van der Zande, G.; Smeets, D.; Weghuis, D.O.; et al. Next-generation cytogenetics: Comprehensive assessment of 52 hematological malignancy genomes by optical genome mapping. Am. J. Hum. Genet. 2021, 108, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Mondal, A.K.; Tvrdik, T.; Hauenstein, J.; Shi, H.; Deeb, K.K.; Saxe, D.; Hastie, A.R.; Chaubey, A.; Savage, N.M.; et al. Clinical Validation and Diagnostic Utility of Optical Genome Mapping for Enhanced Cytogenomic Analysis of Hematological Neoplasms. J. Mol. Diagn. 2022, 24, 1279–1291. [Google Scholar] [CrossRef]
- Suttorp, J.; Lühmann, J.L.; Behrens, Y.L.; Göhring, G.; Steinemann, D.; Reinhardt, D.; von Neuhoff, N.; Schneider, M. Optical Genome Mapping as a Diagnostic Tool in Pediatric Acute Myeloid Leukemia. Cancers 2022, 14, 2058. [Google Scholar] [CrossRef]
- Levy, B.; Baughn, L.B.; Akkari, Y.M.N.; Chartrand, S.; LaBarge, B.; Claxton, D.F.; Lennon, P.A.; Cujar, C.; Kolhe, R.; Kroeger, K.; et al. Optical Genome Mapping in Acute Myeloid Leukemia: A Multicenter Evaluation. Blood Adv. 2022, 7, 1297–1307. [Google Scholar] [CrossRef]
- Vangala, D.B.; Nilius-Eliliwi, V.; Gerding, W.M.; Schroers, R.; Nguyen, H.P. Optical Genome Mapping in MDS and AML as tool for structural variant profiling-comment and data update on Yang et al.: “High-resolution structural variant profiling of myelodysplastic syndromes by optical genome mapping uncovers cryptic aberrations of prognostic and therapeutic significance”. Leukemia 2023, 37, 248–249. [Google Scholar] [CrossRef]
- Nilius-Eliliwi, V.; Gerding, W.M.; Schroers, R.; Nguyen, H.P.; Vangala, D.B. Optical Genome Mapping for Cytogenetic Diagnostics in AML. Cancers 2023, 15, 1684. [Google Scholar] [CrossRef]
- Vieler, L.-M.; Nilius-Eliliwi, V.; Schroers, R.; Vangala, D.B.; Nguyen, H.P.; Gerding, W.M. Optical Genome Mapping Reveals and Characterizes Recurrent Aberrations and New Fusion Genes in Adult ALL. Genes 2023, 14, 686. [Google Scholar] [CrossRef]
- Nilius-Eliliwi, V.; Tembrink, M.; Gerding, W.M.; Lubieniecki, K.P.; Lubieniecka, J.M.; Kankel, S.; Liehr, T.; Mika, T.; Dimopoulos, F.; Döhner, K.; et al. Broad genomic workup including optical genome mapping uncovers a DDX3X: MLLT10 gene fusion in acute myeloid leukemia. Front. Oncol. 2022, 12, 959243. [Google Scholar] [CrossRef]
- International Standing Committee on Human Cytogenomic Nomenclature. ISCN 2020: An International System for Human Cytogenomic Nomenclature (2020); Karger: Basel, Switzerland, 2020; ISBN 9783318067064. [Google Scholar]
- Rack, K.A.; van den Berg, E.; Haferlach, C.; Beverloo, H.B.; Costa, D.; Espinet, B.; Foot, N.; Jeffries, S.; Martin, K.; O’Connor, S.; et al. European recommendations and quality assurance for cytogenomic analysis of haematological neoplasms. Leukemia 2019, 33, 1851–1867. [Google Scholar] [CrossRef] [PubMed]
- Bionano Genomics. Saphyr Molecule Quality Report Guidelines Document Revision: C. Available online: https://bionanogenomics.com/wp-content/uploads/2018/04/30223-Saphyr-Molecule-Quality-Report-Guidelines.pdf (accessed on 30 March 2023).
- Bionano Genomics. Bionano Solve Theory of Operation: Structural Variant Calling Document Revision K. Available online: https://bionanogenomics.com/wp-content/uploads/2018/04/30110-Bionano-Solve-Theory-of-Operation-Structural-Variant-Calling.pdf (accessed on 30 March 2023).
- Church, D.M.; Schneider, V.A.; Graves, T.; Auger, K.; Cunningham, F.; Bouk, N.; Chen, H.-C.; Agarwala, R.; McLaren, W.M.; Ritchie, G.R.S.; et al. Modernizing reference genome assemblies. PLoS Biol. 2011, 9, e1001091. [Google Scholar] [CrossRef] [PubMed]
- Bachman, J. Reverse-transcription PCR (RT-PCR). Methods Enzymol. 2013, 530, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; International Agency for Research on Cancer: Lyon, France, 2008. [Google Scholar]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Powers, M.A.; Macaulay, C.; Masiarz, F.R.; Forbes, D.J. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 1995, 128, 721–736. [Google Scholar] [CrossRef]
- Fontoura, B.M.; Blobel, G.; Matunis, M.J. A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 1999, 144, 1097–1112. [Google Scholar] [CrossRef]
- Yung, E.; Sekulovic, S.; Argiropoulos, B.; Lai, C.K.; Leung, M.; Berg, T.; Vollett, S.; Chang, V.C.-D.; Wan, A.; Wong, S.; et al. Delineating domains and functions of NUP98 contributing to the leukemogenic activity of NUP98-HOX fusions. Leuk. Res. 2011, 35, 545–550. [Google Scholar] [CrossRef]
- Saito, S.; Yokokawa, T.; Iizuka, G.; Cigdem, S.; Okuwaki, M.; Nagata, K. Function of Nup98 subtypes and their fusion proteins, Nup98-TopIIβ and Nup98-SETBP1 in nuclear-cytoplasmic transport. Biochem. Biophys. Res. Commun. 2017, 487, 96–102. [Google Scholar] [CrossRef]
- Bertrums, E.J.M.; Smith, J.L.; Harmon, L.; Ries, R.E.; Wang, Y.-C.J.; Alonzo, T.A.; Menssen, A.J.; Chisholm, K.M.; Leonti, A.R.; Tarlock, K.; et al. Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia. Haematologica 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Ostronoff, F.; Ries, R.E.; Gerbing, R.B.; Marra, M.A.; Yussanne, M.; Long, W.; Zong, S.; Mungall, K.; Andrew, A.; Gerhard, D.S.; et al. Rearrangements in Nucleoporin Family of Genes in Childhood Acute Myeloid Leukemia: A Report from Children Oncology Group and NCI/COG Target AML Initiative. Blood 2015, 126, 169. [Google Scholar] [CrossRef]
- Tarlock, K.; Zhong, S.; He, Y.; Ries, R.; Severson, E.; Bailey, M.; Morley, S.; Balasubramanian, S.; Erlich, R.; Lipson, D.; et al. Distinct age-associated molecular profiles in acute myeloid leukemia defined by comprehensive clinical genomic profiling. Oncotarget 2018, 9, 26417–26430. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-Y.; Li, J.-F.; Zhu, Y.-M.; Lin, X.-J.; Wen, L.-J.; Zhang, F.; Zhang, Y.-L.; Zhao, M.; Fang, H.; Wang, S.-Y.; et al. Transcriptome-based molecular subtypes and differentiation hierarchies improve the classification framework of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2022, 119, e2211429119. [Google Scholar] [CrossRef]
- Chandra, B.; Michmerhuizen, N.L.; Shirnekhi, H.K.; Tripathi, S.; Pioso, B.J.; Baggett, D.W.; Mitrea, D.M.; Iacobucci, I.; White, M.R.; Chen, J.; et al. Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic Transformation. Cancer Discov. 2022, 12, 1152–1169. [Google Scholar] [CrossRef] [PubMed]
- Terlecki-Zaniewicz, S.; Humer, T.; Eder, T.; Schmoellerl, J.; Heyes, E.; Manhart, G.; Kuchynka, N.; Parapatics, K.; Liberante, F.G.; Müller, A.C.; et al. Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat. Struct. Mol. Biol. 2021, 28, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, R.A.; Pettengell, R.; Pandha, H.S.; Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013, 27, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Sump, B.; Brickner, J.H. Nup98 regulation of histone methylation promotes normal gene expression and may drive leukemogenesis. Genes Dev. 2017, 31, 2201–2203. [Google Scholar] [CrossRef]
- Xu, H.; Valerio, D.G.; Eisold, M.E.; Sinha, A.; Koche, R.P.; Hu, W.; Chen, C.-W.; Chu, S.H.; Brien, G.L.; Park, C.Y.; et al. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell 2016, 30, 863–878. [Google Scholar] [CrossRef]
- Fisher, J.N.; Thanasopoulou, A.; Juge, S.; Tzankov, A.; Bagger, F.O.; Mendez, M.A.; Peters, A.H.F.M.; Schwaller, J. Transforming activities of the NUP98-KMT2A fusion gene associated with myelodysplasia and acute myeloid leukemia. Haematologica 2020, 105, 1857–1867. [Google Scholar] [CrossRef]
- Fagnan, A.; Mercher, T. NUP98 and KMT2A: Usually the bride rather than the bridesmaid. Haematologica 2020, 105, 1757–1760. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.G.; Cai, L.; Pasillas, M.P.; Kamps, M.P. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 2007, 9, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W.; Slape, C.; Zhang, Z.; Aplan, P.D. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 2005, 106, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Slape, C.; Lin, Y.W.; Hartung, H.; Zhang, Z.; Wolff, L.; Aplan, P.D. NUP98-HOX translocations lead to myelodysplastic syndrome in mice and men. J. Natl. Cancer Inst. Monogr. 2008, 39, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Pineault, N.; Buske, C.; Feuring-Buske, M.; Abramovich, C.; Rosten, P.; Hogge, D.E.; Aplan, P.D.; Humphries, R.K. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood 2003, 101, 4529–4538. [Google Scholar] [CrossRef]
- Kroon, E.; Thorsteinsdottir, U.; Mayotte, N.; Nakamura, T.; Sauvageau, G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001, 20, 350–361. [Google Scholar] [CrossRef]
- Mohanty, S.; Jyotsana, N.; Sharma, A.; Kloos, A.; Gabdoulline, R.; Othman, B.; Lai, C.K.; Schottmann, R.; Mandhania, M.; Schmoellerl, J.; et al. Targeted Inhibition of the NUP98-NSD1 Fusion Oncogene in Acute Myeloid Leukemia. Cancers 2020, 12, 2766. [Google Scholar] [CrossRef]
- Rogawski, D.S.; Deng, J.; Li, H.; Miao, H.; Borkin, D.; Purohit, T.; Song, J.; Chase, J.; Li, S.; Ndoj, J.; et al. Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity. Nat. Commun. 2021, 12, 2792. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Jaju, R.J.; Fidler, C.; Haas, O.A.; Strickson, A.J.; Watkins, F.; Clark, K.; Cross, N.C.; Cheng, J.F.; Aplan, P.D.; Kearney, L.; et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 2001, 98, 1264–1267. [Google Scholar] [CrossRef]
- Rosati, R.; La Starza, R.; Veronese, A.; Aventin, A.; Schwienbacher, C.; Vallespi, T.; Negrini, M.; Martelli, M.F.; Mecucci, C. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 2002, 99, 3857–3860. [Google Scholar] [CrossRef]
- van Zutven, L.J.C.M.; Onen, E.; Velthuizen, S.C.J.M.; van Drunen, E.; von Bergh, A.R.M.; van den Heuvel-Eibrink, M.M.; Veronese, A.; Mecucci, C.; Negrini, M.; de Greef, G.E.; et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer 2006, 45, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, I.; Kerndrup, G.; Carlsen, N.; Strömbeck, B.; Isaksson, M.; Johansson, B. Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12). Br. J. Haematol. 2007, 136, 294–296. [Google Scholar] [CrossRef]
- Kaltenbach, S.; Soler, G.; Barin, C.; Gervais, C.; Bernard, O.A.; Penard-Lacronique, V.; Romana, S.P. NUP98-MLL fusion in human acute myeloblastic leukemia. Blood 2010, 116, 2332–2335. [Google Scholar] [CrossRef] [PubMed]
- Roussy, M.; Bilodeau, M.; Jouan, L.; Tibout, P.; Laramée, L.; Lemyre, E.; Léveillé, F.; Tihy, F.; Cardin, S.; Sauvageau, C.; et al. NUP98-BPTF gene fusion identified in primary refractory acute megakaryoblastic leukemia of infancy. Genes Chromosomes Cancer 2018, 57, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-K.; Chan, H.-Y.; Yung, Y.-L.; Wan, T.S.K.; Leung, A.W.K.; Li, C.-K.; Tian, K.; Chan, N.P.H.; Cheung, J.S.; Ng, M.H.L. A novel NUP98-JADE2 fusion in a patient with acute myeloid leukemia resembling acute promyelocytic leukemia. Blood Adv. 2022, 6, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y. A cryptic translocation leading to NUP98-PHF23 fusion in AML. Best Pract. Res. Clin. Haematol. 2016, 29, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, Y.; Gough, S.M.; Zhang, J.; Vann, K.R.; Li, K.; Cai, L.; Shi, X.; Aplan, P.D.; Wang, G.G.; et al. Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion. Nat. Commun. 2020, 11, 3339. [Google Scholar] [CrossRef]
- Park, E.S.; Chung, Y.J.; Aplan, P.D. PO-020 Discrepancy in efficacy of disulfiram between NUP98-PHF23 fusion acute myelogenous leukaemia cell line and in vivo mouse model: Sharing normal hematopoietic stem cells niche. ESMO Open 2018, 3, A235. [Google Scholar] [CrossRef]
- Gough, S.M.; Lee, F.; Yang, F.; Walker, R.L.; Zhu, Y.J.; Pineda, M.; Onozawa, M.; Chung, Y.J.; Bilke, S.; Wagner, E.K.; et al. NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function. Cancer Discov. 2014, 4, 564–577. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Li, Y.; Yang, W.; Patel, R.M.; Casey, E.B.; Denby, E.; Mendoza-Castrejon, J.; Rodriguez-Lopez, P.; Magee, J.A. FLT3ITD drives context-specific changes in cell identity and variable interferon dependence during AML initiation. Blood 2023, 141, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Zhu, Y.-J.; Gu, B.-W.; Cai, X.; Bai, X.-T.; Yun, H.-Y.; Zhu, J.; Chen, B.; Weng, L.; Chen, Z.; et al. A new fusion gene NUP98-IQCG identified in an acute T-lymphoid/myeloid leukemia with a t(3;11)(q29q13;p15)del(3)(q29) translocation. Oncogene 2008, 27, 3414–3423. [Google Scholar] [CrossRef]
- Such, E.; Cervera, J.; Valencia, A.; Barragán, E.; Ibañez, M.; Luna, I.; Fuster, O.; Perez-Sirvent, M.L.; Senent, L.; Sempere, A.; et al. A novel NUP98/RARG gene fusion in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood 2011, 117, 242–245. [Google Scholar] [CrossRef]
- Soler, G.; Kaltenbach, S.; Dobbelstein, S.; Broccardo, C.; Radford, I.; Mozziconacci, M.-J.; Bernard, O.A.; Penard-Lacronique, V.; Delabesse, E.; Romana, S.P. Identification of GSX2 and AF10 as NUP98 partner genes in myeloid malignancies. Blood Cancer J. 2013, 3, e124. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Katagiri, Z.-I.; Kawahashi, K.; Kioussis, D.; Kitajima, S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 2007, 397, 161–168. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Q.; Wong, S.H.K.; Huang, M.; Klein, B.J.; Shen, J.; Ikenouye, L.; Onishi, M.; Schneidawind, D.; Buechele, C.; et al. ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia. Cancer Discov. 2016, 6, 770–783. [Google Scholar] [CrossRef]
- Rogawski, D. The Function of the ASH1L Histone Methyltransferase in Cancer: A Chemical Biology Approach. Ph.D. Thesis, University of Michigan, Ann Arbor, MI, USA, 2018. [Google Scholar]
- Xu, B.; Qin, T.; Yu, J.; Giordano, T.J.; Sartor, M.A.; Koenig, R.J. Novel role of ASH1L histone methyltransferase in anaplastic thyroid carcinoma. J. Biol. Chem. 2020, 295, 8834–8845. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.-Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget 2015, 6, 2466–2482. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef]
- Collins, C.T.; Hess, J.L. Role of HOXA9 in leukemia: Dysregulation, cofactors and essential targets. Oncogene 2016, 35, 1090–1098. [Google Scholar] [CrossRef]
- Jones, M.; Chase, J.; Brinkmeier, M.; Xu, J.; Weinberg, D.N.; Schira, J.; Friedman, A.; Malek, S.; Grembecka, J.; Cierpicki, T.; et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J. Clin. Investig. 2015, 125, 2007–2020. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kawahashi, K.; Katagiri, Z.-I.; Nakayama, Y.; Mahajan, M.; Kioussis, D. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PLoS ONE 2011, 6, e28171. [Google Scholar] [CrossRef] [PubMed]
- Aljazi, M.B.; Gao, Y.; Wu, Y.; Mias, G.I.; He, J. Histone H3K36me2-Specific Methyltransferase ASH1L Promotes MLL-AF9-Induced Leukemogenesis. Front. Oncol. 2021, 11, 754093. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef]

| Time Point | Days after Diagnosis (Days after alloHSCT) | Cyto- Morphology | Histology | Chromosomal Banding Analysis 1 | JAK2 V617FVAF | Donor Chimerism | NUP98-r VAF |
|---|---|---|---|---|---|---|---|
| AML diagnosis | 0 | 60–70% blasts | n.a. | Complex [29] | 17.5% | - | 10% Trans., 11% Inversion |
| Post-Induction I | +35 | CR | CR | n.a. | n.a. | - | n.a. |
| Post-Induction II | +69 | CR | CR | 46,XX [25] | <0.5% | - | n.a. |
| Post-Consolidation | +136 | CR | 5% blasts | n.a. | n.a. | - | n.a. |
| Prior to alloHSCT | +155 (−7) | CR | 15% blasts | n.a. | 11.6% | - | 5% Trans., 4% Inversion |
| Post alloHSCT | +182 (+20) | CR | CR | Complex [6/23], 46,XX [17/23] | n.a. | 98% | n.a. |
| Monitoring | +204 (+42) | n.a. | n.a. | n.a. | 1.3% 2 | n.a. | n.a. |
| Relapse | +225 (+63) | Left shift, single blasts | 50% blasts | Complex [25] | 21.1% | 63% | 16% Trans., 20% Inversion |
| Death | +269 (+107) |
| ChrA | ChrB | ChrA Reference | ChrB Reference | SV Type | Genes | Time Point | VAF |
|---|---|---|---|---|---|---|---|
| Translocation SV ogm[GRCh37] t(1;11)(q22;p15.4) indicating NUP98::ASH1L | |||||||
| 1 | 11 | 155384865 | 3755020 | Translocation | ASH1L, NUP98 | AML diagnosis Prior alloHSCT Relapse | 10% 5% 16% |
| Inversion SV ogm[GRCh37] inv(1)(q22q22) indicating NUP98::ASH1L | |||||||
| 1 | 1 | 155349797 | 155279255 | Inversion | ASH1L | AML diagnosis Prior to alloHSCT Relapse | 11% 4% 20% |
| Translocation SV indicating der(5) ogm[GRCh37] t(5;10)(q31.1;q22.2) | |||||||
| 5 | 10 | 132131355 | 75647791 | Translocation | - | AML diagnosis Prior to alloHSCT Relapse | 46% 9% 25% |
| Translocation SVs indicating Chr21-Rearrangement (only present at Relapse) | |||||||
| 3 | 21 | 16778711 | 30261569 | Translocation | - | Relapse | 2% |
| 3 | 21 | 18733184 | 42711338 | Translocation | FAM3B | Relapse | 3% |
| 10 | 21 | 72180437 | 34005936 | Translocation | EIF4EBP2, SYNJ1 | Relapse | 6% |
| 11 | 21 | 21948118 | 17221321 | Translocation | USP25 | Relapse | 3% |
| 21 | 21 | 24252871 | 42525873 | Translocation | - | Relapse | 1% |
| 21 | 21 | 24495432 | 29856259 | Translocation | - | Relapse | 3% |
| Chr | Reference Start | Reference End | CNV Type | fractional CN | Time point | VAF | |
| CNV losses indicating Monosomy 16 (only present at AML diagnosis) | |||||||
| 16 | 2263650 | 21369108 | CNV loss | 1.67 | AML diagnosis | 16% | |
| 16 | 22822268 | 32019651 | CNV loss | 1.69 | AML diagnosis | 16% | |
| 16 | 46438848 | 85148570 | CNV loss | 1.68 | AML diagnosis | 16% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tembrink, M.; Gerding, W.M.; Wieczorek, S.; Mika, T.; Schroers, R.; Nguyen, H.P.; Vangala, D.B.; Nilius-Eliliwi, V. Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping. Cancers 2023, 15, 2942. https://doi.org/10.3390/cancers15112942
Tembrink M, Gerding WM, Wieczorek S, Mika T, Schroers R, Nguyen HP, Vangala DB, Nilius-Eliliwi V. Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping. Cancers. 2023; 15(11):2942. https://doi.org/10.3390/cancers15112942
Chicago/Turabian StyleTembrink, Marco, Wanda Maria Gerding, Stefan Wieczorek, Thomas Mika, Roland Schroers, Huu Phuc Nguyen, Deepak Ben Vangala, and Verena Nilius-Eliliwi. 2023. "Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping" Cancers 15, no. 11: 2942. https://doi.org/10.3390/cancers15112942
APA StyleTembrink, M., Gerding, W. M., Wieczorek, S., Mika, T., Schroers, R., Nguyen, H. P., Vangala, D. B., & Nilius-Eliliwi, V. (2023). Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping. Cancers, 15(11), 2942. https://doi.org/10.3390/cancers15112942







