Genetic Variants Associated with Longitudinal Cognitive Performance in Older Breast Cancer Patients and Controls †
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Collection
2.3. Statistical Methods and Analyses
3. Results
3.1. GWAS and Gene Analyses
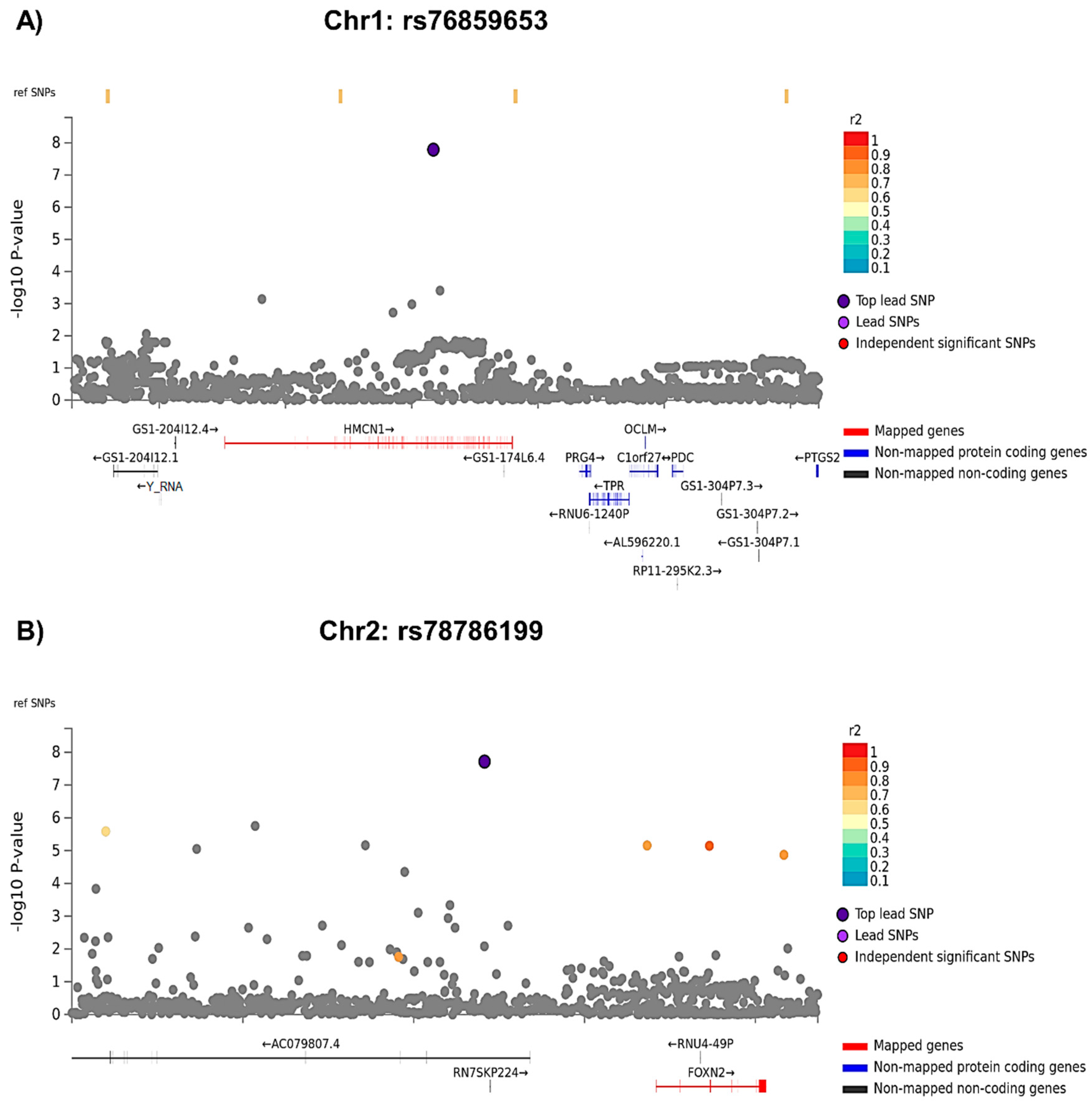
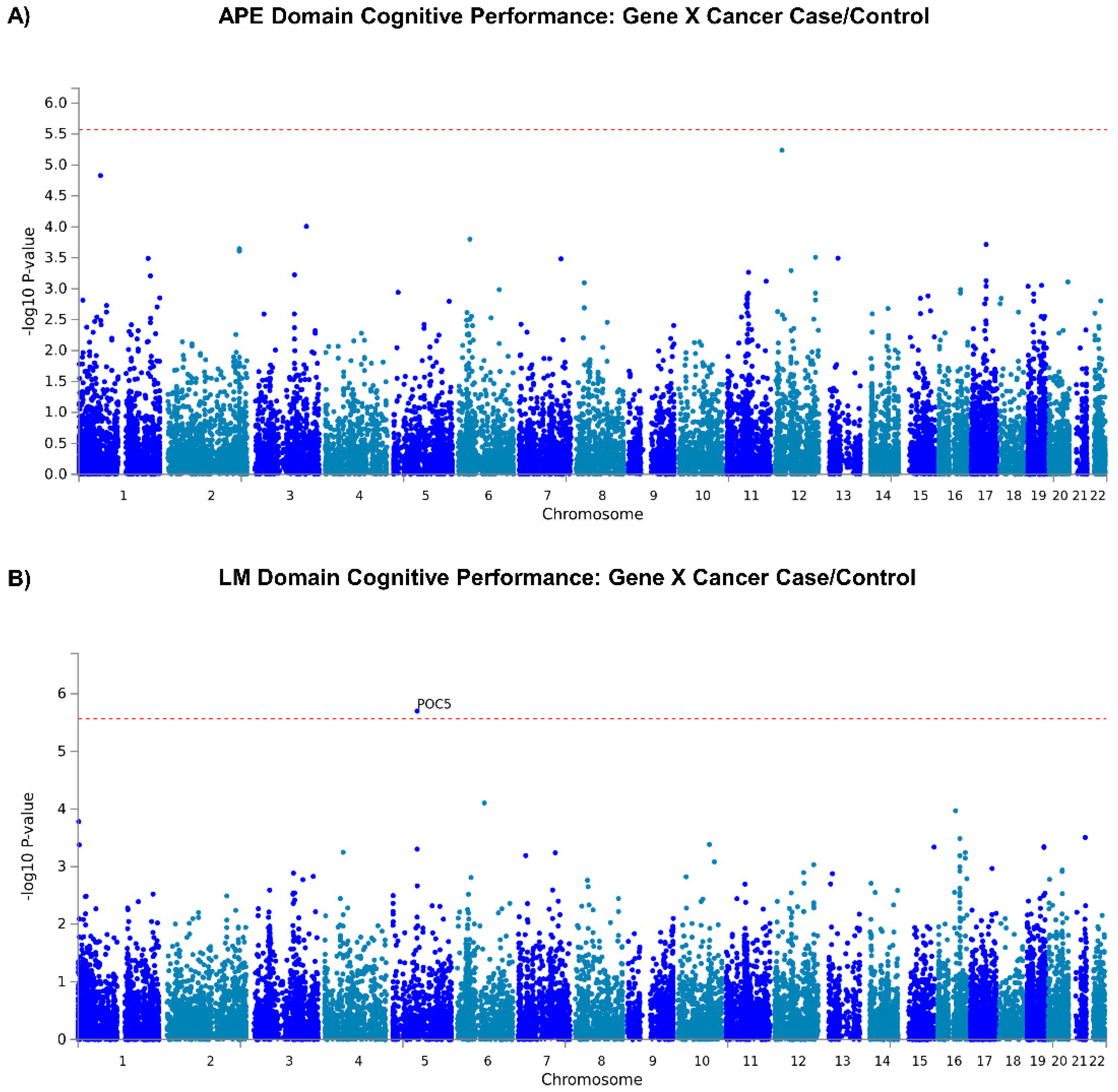
3.1.1. GWAS Analyses
3.1.2. Gene Level Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bluethmann, S.M.; Mariotto, A.B.; Rowland, J.H. Anticipating the “Silver Tsunami”: Prevalence Trajectories and Comorbidity Burden among Older Cancer Survivors in the United States. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Caswell-Jin, J.L.; Plevritis, S.K.; Tian, L.; Cadham, C.J.; Xu, C.; Stout, N.K.; Sledge, G.W.; Mandelblatt, J.S.; Kurian, A.W. Change in Survival in Metastatic Breast Cancer with Treatment Advances: Meta-Analysis and Systematic Review. JNCI Cancer Spectr. 2018, 2, pky062. [Google Scholar] [CrossRef] [PubMed]
- Mandelblatt, J.S.; Small, B.J.; Luta, G.; Hurria, A.; Jim, H.; McDonald, B.C.; Graham, D.; Zhou, X.; Clapp, J.; Zhai, W.; et al. Cancer-Related Cognitive Outcomes Among Older Breast Cancer Survivors in the Thinking and Living with Cancer Study. J. Clin. Oncol. 2018, 36, 3211. [Google Scholar] [CrossRef] [PubMed]
- Mandelblatt, J.S.; Jacobsen, P.B.; Ahles, T. Cognitive effects of cancer systemic therapy: Implications for the care of older patients and survivors. J. Clin. Oncol. 2014, 32, 2617–2626. [Google Scholar] [CrossRef] [PubMed]
- Ahles, T.A.; Li, Y.; McDonald, B.C.; Schwartz, G.N.; Kaufman, P.A.; Tsongalis, G.J.; Moore, J.H.; Saykin, A.J. Longitudinal assessment of cognitive changes associated with adjuvant treatment for breast cancer: The impact of APOE and smoking. Psychooncology 2014, 23, 1382–1390. [Google Scholar] [CrossRef] [PubMed]
- Ahles, T.A.; Saykin, A.J.; Noll, W.W.; Furstenberg, C.T.; Guerin, S.; Cole, B.; Mott, L.A. The relationship of APOE genotype to neuropsychological performance in long-term cancer survivors treated with standard dose chemotherapy. Psychooncology 2003, 12, 612–619. [Google Scholar] [CrossRef]
- Carroll, J.E.; Small, B.J.; Tometich, D.B.; Zhai, W.; Zhou, X.; Luta, G.; Ahles, T.A.; Saykin, A.J.; Nudelman, K.N.H.; Clapp, J.D.; et al. Sleep disturbance and neurocognitive outcomes in older patients with breast cancer: Interaction with genotype. Cancer 2019, 125, 4516–4524. [Google Scholar] [CrossRef]
- Van Dyk, K.; Zhou, X.; Small, B.J.; Ahn, J.; Zhai, W.; Ahles, T.; Graham, D.; Jacobsen, P.B.; Jim, H.; McDonald, B.C.; et al. Protective Effects of APOE epsilon2 Genotype on Cognition in Older Breast Cancer Survivors: The Thinking and Living with Cancer Study. JNCI Cancer Spectr. 2021, 5, pkab013. [Google Scholar] [CrossRef]
- Wilkinson, G.S.; Robertson, G.J. Wide Range Achievement Test (WRAT4); Psychological Assessment Resources: Lutz, FL, USA, 2006. [Google Scholar]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Patterson, J. Controlled Oral Word Association Test. In Encylopedia of Clinical Neuropsychology; Kreutzer, J., DeLuca, J., Caplan, B., Eds.; Springer: New York, NY, USA, 2011; pp. 703–706. [Google Scholar] [CrossRef]
- Reitan, R. Validity of the Trail Making Test as an indicator of organic brain damage. Percept. Mot. Ski. 1958, 8, 271–276. [Google Scholar] [CrossRef]
- Stern, R.; White, T. Neuropsychological Assessment Battery: Administration, Scoring, and Interpretation; Psychological Assessment Resources, Inc.: Lutz, FL, USA, 2003. [Google Scholar]
- Wechsler, D. WAIS-III, Wechsler Adult Intelligence Scale: Administration and Scoring Manual; Psychological Corporation: San Antonio, TX, USA, 1997. [Google Scholar]
- Wechsler, D. Manual for the Wechsler Memory Scale-Revised; Psychological Corporation: San Antonio, TX, USA, 1987. [Google Scholar]
- Clapp, J.D.; Luta, G.; Small, B.J.; Ahles, T.A.; Root, J.C.; Graham, D.; Hurria, A.; Jacobsen, P.B.; Jim, H.; McDonald, B.C.; et al. The Impact of Using Different Reference Populations on Measurement of Breast Cancer-Related Cognitive Impairment Rates. Arch. Clin. Neuropsychol. 2018, 33, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Umicevic Mirkov, M.; de Leeuw, C.A.; van den Heuvel, M.P.; Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 2019, 10, 3222. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Perez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef] [PubMed]
- Vogel, B.E.; Muriel, J.M.; Dong, C.; Xu, X. Hemicentins: What have we learned from worms? Cell. Res. 2006, 16, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Je, E.M.; Yoo, N.J.; Lee, S.H. HMCN1, a cell polarity-related gene, is somatically mutated in gastric and colorectal cancers. Pathol. Oncol. Res. 2015, 21, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.W.; Klein, M.L.; Humpert, A.J.; Luzier, C.W.; Persun, V.; Schain, M.; Mahan, A.; Runckel, C.; Cassera, M.; Vittal, V.; et al. Analysis of the ARMD1 locus: Evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum. Mol. Genet. 2003, 12, 3315–3323. [Google Scholar] [CrossRef] [PubMed]
- McKay, G.J.; Clarke, S.; Hughes, A.; McConnell, V.; Schultz, D.W.; Klein, M.L.; Silvestri, G.; Simpson, D.A. A novel diagnostic test detects a low frequency of the hemicentin Gln5345Arg variant among Northern Irish age related macular degeneration patients. Mol. Vis. 2004, 10, 682–687. [Google Scholar]
- Shang, X.; Zhu, Z.; Huang, Y.; Zhang, X.; Wang, W.; Shi, D.; Jiang, Y.; Yang, X.; He, M. Associations of ophthalmic and systemic conditions with incident dementia in the UK Biobank. Br. J. Ophthalmol. 2021, 107, 275–282. [Google Scholar] [CrossRef]
- Liu, X.H.; Liu, L.P.; Xu, X.M.; Hua, M.; Kang, Q.; Li, A.; Huang, L. FOXN2 suppresses the proliferation and invasion of human hepatocellular carcinoma cells. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 731–737. [Google Scholar] [CrossRef]
- Cheng, C.K.; Chan, N.P.; Wan, T.S.; Lam, L.Y.; Cheung, C.H.; Wong, T.H.; Ip, R.K.; Wong, R.S.; Ng, M.H. Helicase-like transcription factor is a RUNX1 target whose downregulation promotes genomic instability and correlates with complex cytogenetic features in acute myeloid leukemia. Haematologica 2016, 101, 448–457. [Google Scholar] [CrossRef]
- Nudelman, K.N.H.; McDonald, B.C.; Lahiri, D.K.; Saykin, A.J. Biological Hallmarks of Cancer in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 7173–7187. [Google Scholar] [CrossRef]
- Negishi, H.; Taniguchi, T.; Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb. Perspect. Biol. 2018, 10, a028423. [Google Scholar] [CrossRef]
- Xia, X.; Wang, W.; Yin, K.; Wang, S. Interferon regulatory factor 8 governs myeloid cell development. Cytokine Growth Factor. Rev. 2020, 55, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Luo, S.N.; Dong, L.; Liu, T.T.; Shen, X.Z.; Zhang, N.P.; Liang, L. Interferon regulatory factor family influences tumor immunity and prognosis of patients with colorectal cancer. J. Transl. Med. 2021, 19, 379. [Google Scholar] [CrossRef] [PubMed]
- Dornan, D.; Eckert, M.; Wallace, M.; Shimizu, H.; Ramsay, E.; Hupp, T.R.; Ball, K.L. Interferon regulatory factor 1 binding to p300 stimulates DNA-dependent acetylation of p53. Mol. Cell. Biol. 2004, 24, 10083–10098. [Google Scholar] [CrossRef] [PubMed]
- Bowie, M.L.; Ibarra, C.; Seewalt, V.L. IRF-1 promotes apoptosis in p53-damaged basal-type human mammary epithelial cells: A model for early basal-type mammary carcinogenesis. Adv. Exp. Med. Biol. 2008, 617, 367–374. [Google Scholar] [CrossRef]
- Bluyssen, H.A.; Levy, D.E. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 1997, 272, 4600–4605. [Google Scholar] [CrossRef]
- Wei, H.; Ma, W.; Lu, X.; Liu, H.; Lin, K.; Wang, Y.; Ye, Z.; Sun, L.; Huang, Z.; Pan, T.; et al. KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression. Cancer Commun. 2021, 41, 904–920. [Google Scholar] [CrossRef]
- Azimzadeh, J.; Hergert, P.; Delouvee, A.; Euteneuer, U.; Formstecher, E.; Khodjakov, A.; Bornens, M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell. Biol. 2009, 185, 101–114. [Google Scholar] [CrossRef]
- Hassan, A.; Parent, S.; Mathieu, H.; Zaouter, C.; Molidperee, S.; Bagu, E.T.; Barchi, S.; Villemure, I.; Patten, S.A.; Moldovan, F. Adolescent idiopathic scoliosis associated POC5 mutation impairs cell cycle, cilia length and centrosome protein interactions. PLoS ONE 2019, 14, e0213269. [Google Scholar] [CrossRef]
- Weisz Hubshman, M.; Broekman, S.; van Wijk, E.; Cremers, F.; Abu-Diab, A.; Khateb, S.; Tzur, S.; Lagovsky, I.; Smirin-Yosef, P.; Sharon, D.; et al. Whole-exome sequencing reveals POC5 as a novel gene associated with autosomal recessive retinitis pigmentosa. Hum. Mol. Genet. 2018, 27, 614–624. [Google Scholar] [CrossRef]
- Tatour, Y.; Ben-Yosef, T. Syndromic Inherited Retinal Diseases: Genetic, Clinical and Diagnostic Aspects. Diagnostics 2020, 10, 779. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]


| Variable | Case (N = 325) * | Control (N = 340) | p-Value ** |
|---|---|---|---|
| Age, mean years (StDev) | 68.2 (5.7) | 67.9 (6.6) | 0.596 |
| Education, mean years (StDev) | 15.3 (2.1) | 15.7 (2.2) | 0.012 |
| WRAT4 score, mean (StDev) | 111.0 (15.8) | 113.7 (15.4) | 0.028 |
| Chemotherapy treatment, number (%) | 84 (25.8%) | - | - |
| Hormone therapy, number (%) | 257 (79.1%) | - | - |
| APOE e4 carrier, number (%) | 81 (24.9%) | 85 (25.0%) | 1.000 |
| APE baseline score, mean (StDev) | −0.034 (0.634) | 0.094 (0.608) | 0.008 |
| APE one-year score, mean (StDev) | 0.016 (0.643) | 0.173 (0.607) | 0.001 |
| LM baseline score, mean (StDev) | 0.013 (0.792) | 0.056 (0.811) | 0.487 |
| LM one-year score, mean (StDev) | 0.153 (0.839) | 0.238 (0.607) | 0.185 |
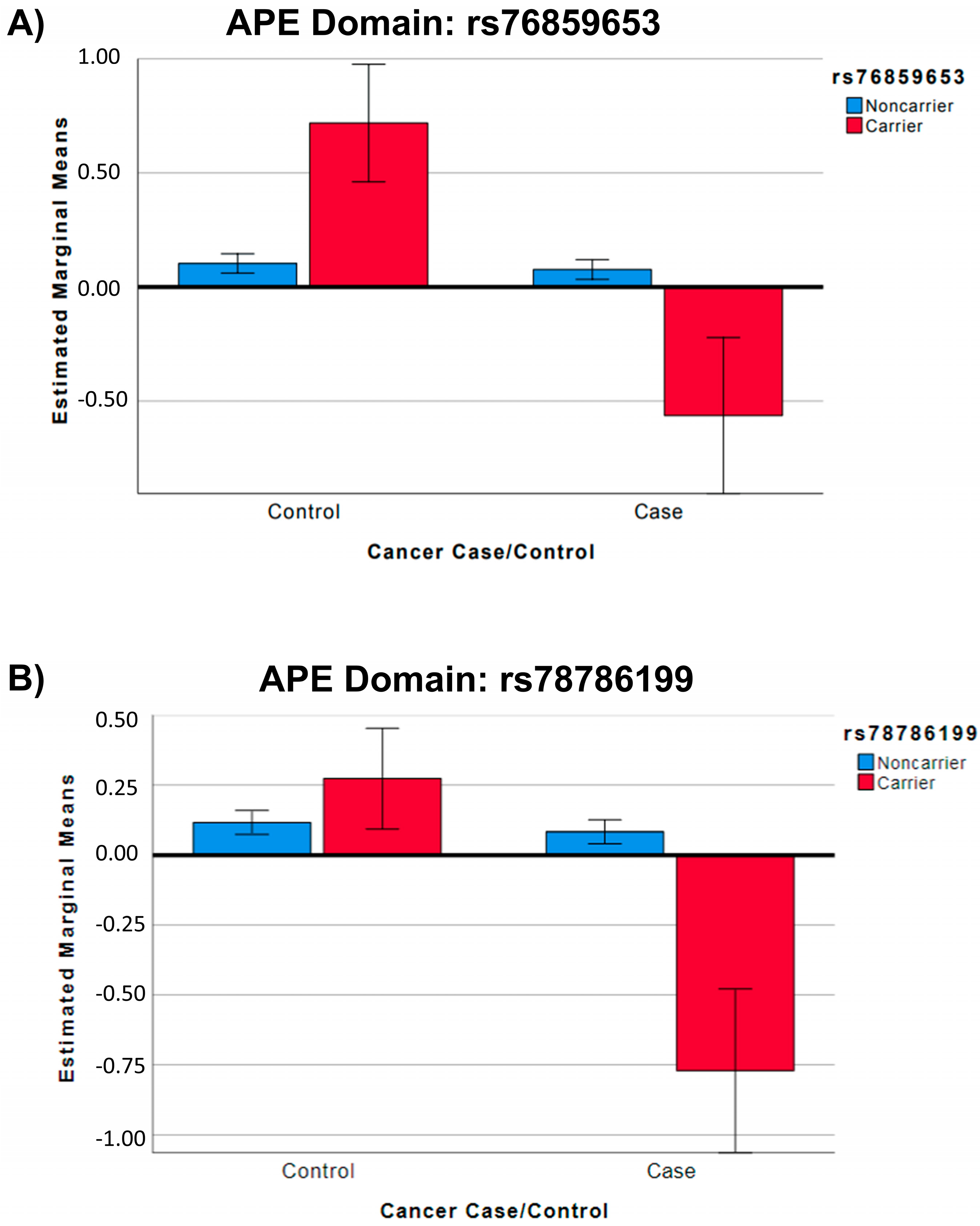
| SNP | Group | SNP MA * | APE Mean ** (StE) | 95% CI Lower Bound | 95% CI Upper Bound | F | p Value ** |
|---|---|---|---|---|---|---|---|
| rs76859653 | Control | 0 (N = 329) | 0.10 (0.02) | 0.06 | 0.15 | 32.68 | <0.001 |
| Control | 1 (N = 9) | 0.71 (0.13) | 0.46 | 0.97 | |||
| Case | 0 (N = 319) | 0.08 (0.02) | 0.03 | 0.12 | |||
| Case | 1 (N = 5) | −0.56 (0.17) | −0.91 | −0.22 | |||
| rs78786199 | Control | 0 (N = 322) | 0.12 (0.02) | 0.07 | 0.16 | 32.27 | <0.001 |
| Control | 1 (N = 18) | 0.27 (0.09) | 0.09 | 0.45 | |||
| Case | 0 (N = 317) | 0.08 (0.02) | 0.04 | 0.13 | |||
| Case | 1 (N = 7) | −0.77 (0.15) | −1.06 | −0.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nudelman, K.; Nho, K.; Zhang, M.; McDonald, B.C.; Zhai, W.; Small, B.J.; Wegel, C.E.; Jacobsen, P.B.; Jim, H.S.L.; Patel, S.K.; et al. Genetic Variants Associated with Longitudinal Cognitive Performance in Older Breast Cancer Patients and Controls. Cancers 2023, 15, 2877. https://doi.org/10.3390/cancers15112877
Nudelman K, Nho K, Zhang M, McDonald BC, Zhai W, Small BJ, Wegel CE, Jacobsen PB, Jim HSL, Patel SK, et al. Genetic Variants Associated with Longitudinal Cognitive Performance in Older Breast Cancer Patients and Controls. Cancers. 2023; 15(11):2877. https://doi.org/10.3390/cancers15112877
Chicago/Turabian StyleNudelman, Kelly, Kwangsik Nho, Michael Zhang, Brenna C. McDonald, Wanting Zhai, Brent J. Small, Claire E. Wegel, Paul B. Jacobsen, Heather S. L. Jim, Sunita K. Patel, and et al. 2023. "Genetic Variants Associated with Longitudinal Cognitive Performance in Older Breast Cancer Patients and Controls" Cancers 15, no. 11: 2877. https://doi.org/10.3390/cancers15112877
APA StyleNudelman, K., Nho, K., Zhang, M., McDonald, B. C., Zhai, W., Small, B. J., Wegel, C. E., Jacobsen, P. B., Jim, H. S. L., Patel, S. K., Graham, D. M. A., Ahles, T. A., Root, J. C., Foroud, T., Breen, E. C., Carroll, J. E., Mandelblatt, J. S., & Saykin, A. J. (2023). Genetic Variants Associated with Longitudinal Cognitive Performance in Older Breast Cancer Patients and Controls. Cancers, 15(11), 2877. https://doi.org/10.3390/cancers15112877









