Efficient Brain Tumor Detection with Lightweight End-to-End Deep Learning Model
Abstract
Simple Summary
Abstract
1. Introduction
- Most of these methods have high computational costs and require large amounts of labeled data for training.
- Potential for overfitting, where the model performs well on the training data but not on new, unseen data.
- Risk of poor performance due to biased training data or incorrect labeling.
- Lack of interpretability of the decision-making process of deep learning models.
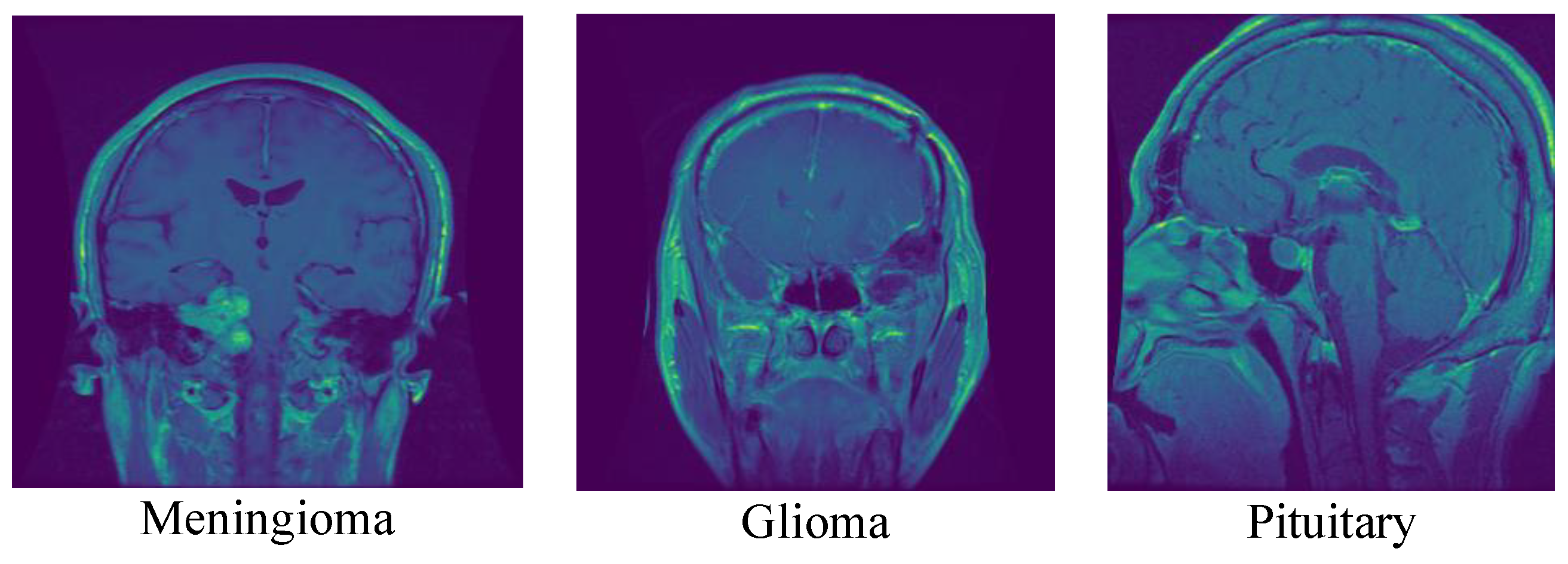
- Propose a new end-to-end structure CNN model for detecting brain tumors. The proposed model can detect the most common intracranial tumors, such as high-grade glioma, low-grade glioma, meningioma, and pituitary adenoma, with acceptable accuracies compared to other previous deep models.
- Propose a new, lightweight deep learning model for brain tumor detection. Our model consists only of eight layers in depth, which makes the system suitable for real-time applications and reduces the time of processing, unlike other previous models in this field that needed deeper layers to obtain good detection accuracy.
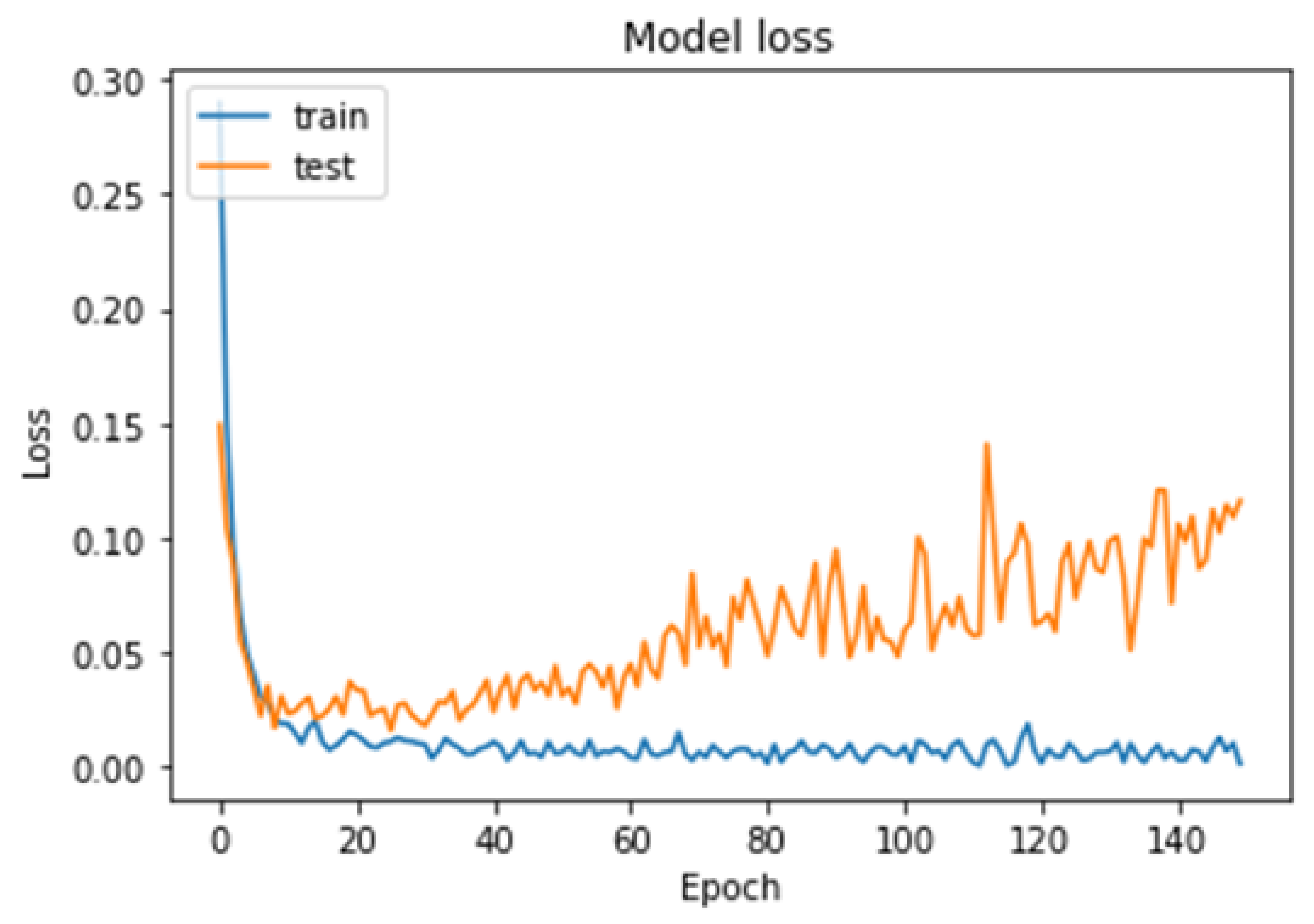
- We tested our model on different datasets for brain tumors using the cross-validation technique, which overcame the overfitting problem and achieved good accuracy for the detection. In addition, we tested our model in different cases, such as binary classification and multi-class classification, which makes the model more robust than other previous models.
2. Related Studies
3. Methodology and Datasets
3.1. The First Dataset
3.2. The Second Dataset
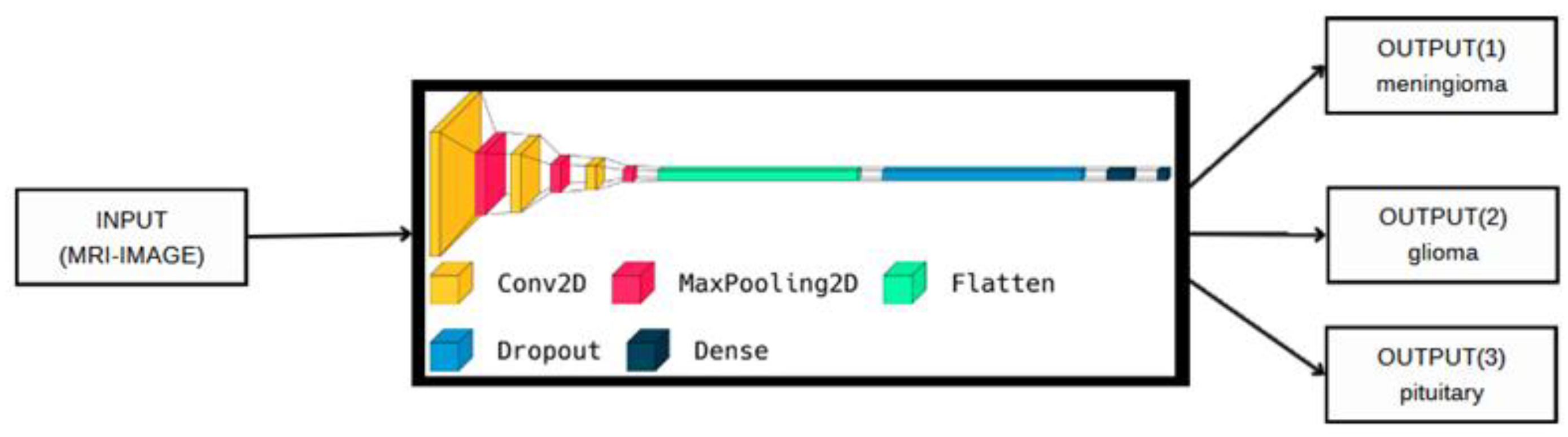
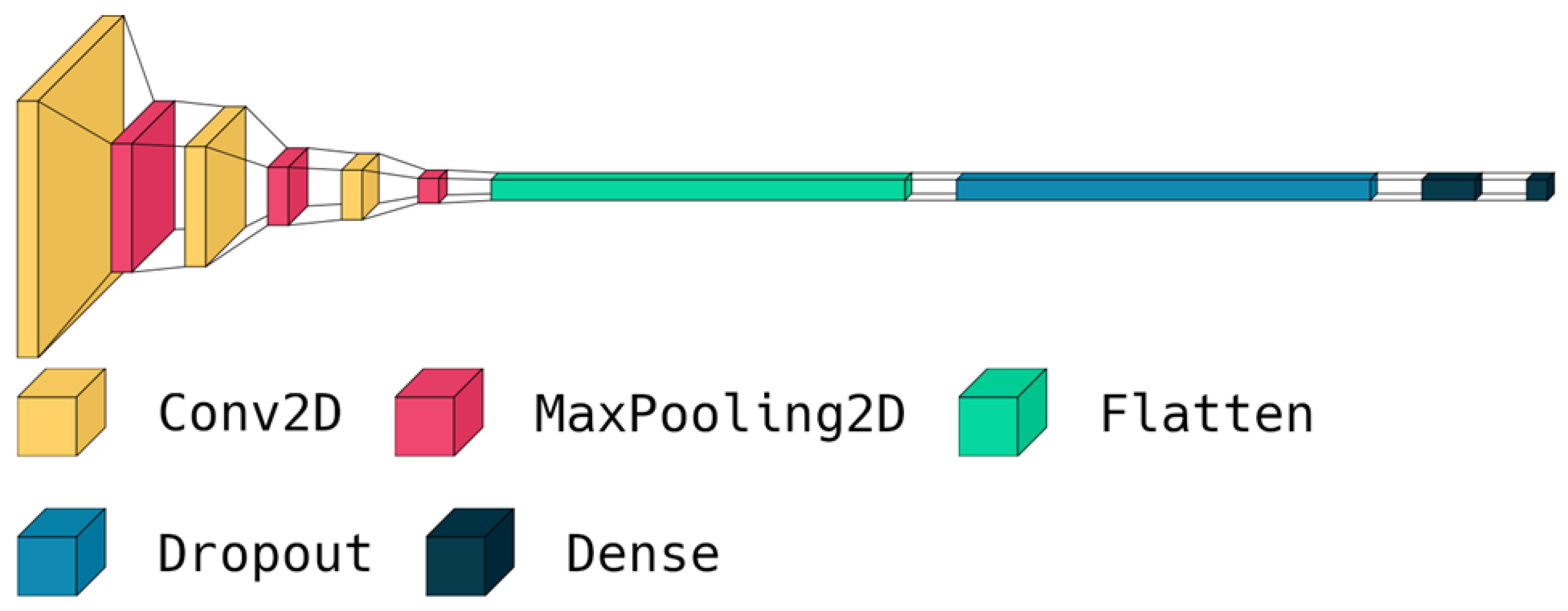
3.3. The Proposed Deep Model
| Algorithm 1: The steps of the proposed deep model |
| 1. Import TensorFlow library as tf 2. Define a sequential model architecture using tf.keras.Sequential() method 3. Add a Conv2D layer to the model with 32 filters of size (3,3) and ReLU activation 4. Add a MaxPooling2D layer to the model 5. Repeat steps 3–4 to increase the depth of the model 6. Add a Flatten layer to the model 7. Add a Dropout layer with a rate of 0.5 to the model 8. Add a Dense layer with 512 neurons and ReLU activation to the model 9. Add a final Dense layer with 3 neurons and SoftMax activation to classify the brain tumors into 3 classes 10. Compile the model using the Adam optimizer, sparse categorical cross-entropy loss function, and accuracy as the evaluation metric 11. Train the model for 150 epochs using a batch size of 32 on the training dataset |
4. Results and Discussion
- GPU: Nvidia Tesla T4
- GPU RAM: 15 GB
- System RAM: 12.72 GB
- Disk Space: 78.2 GB
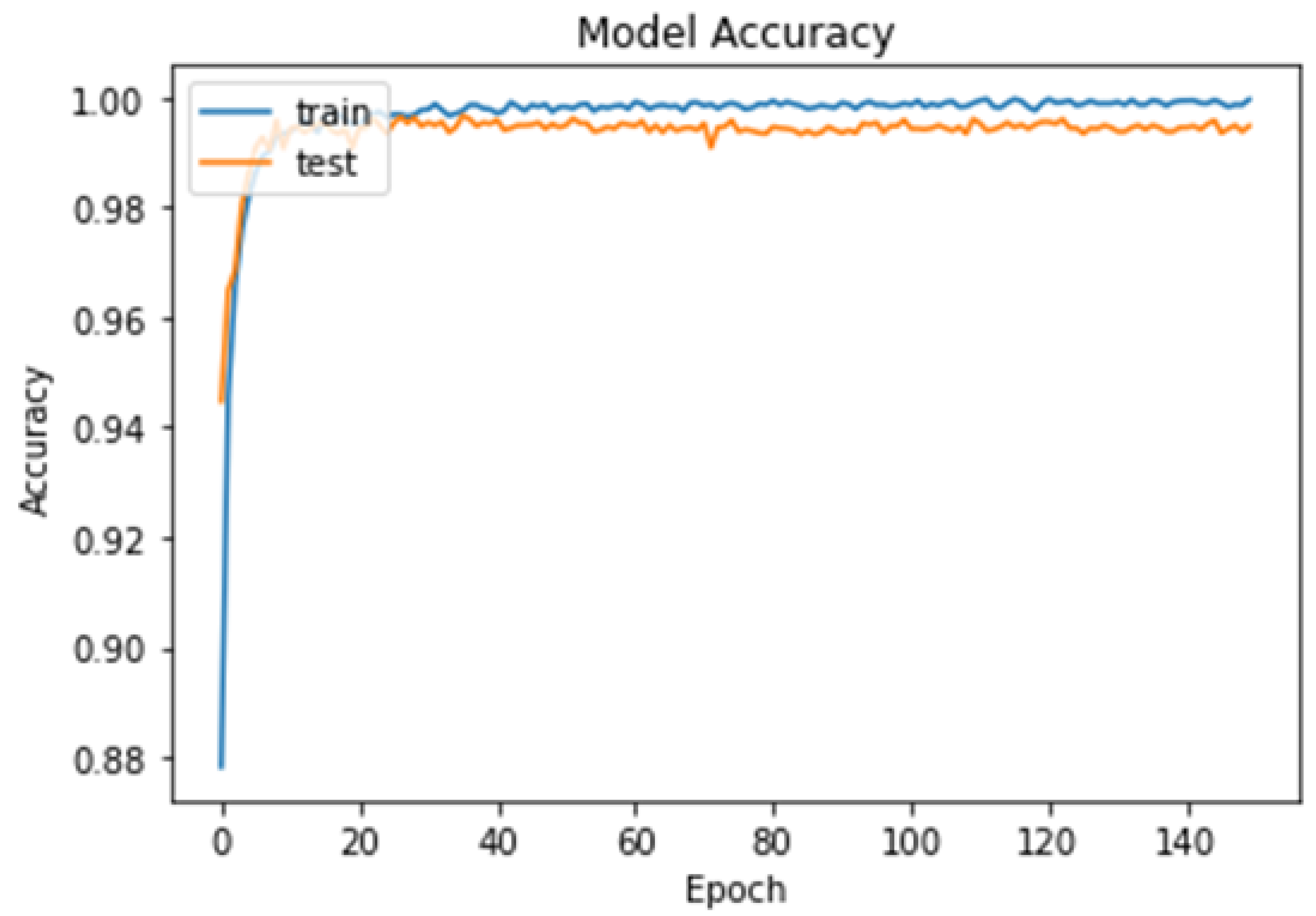
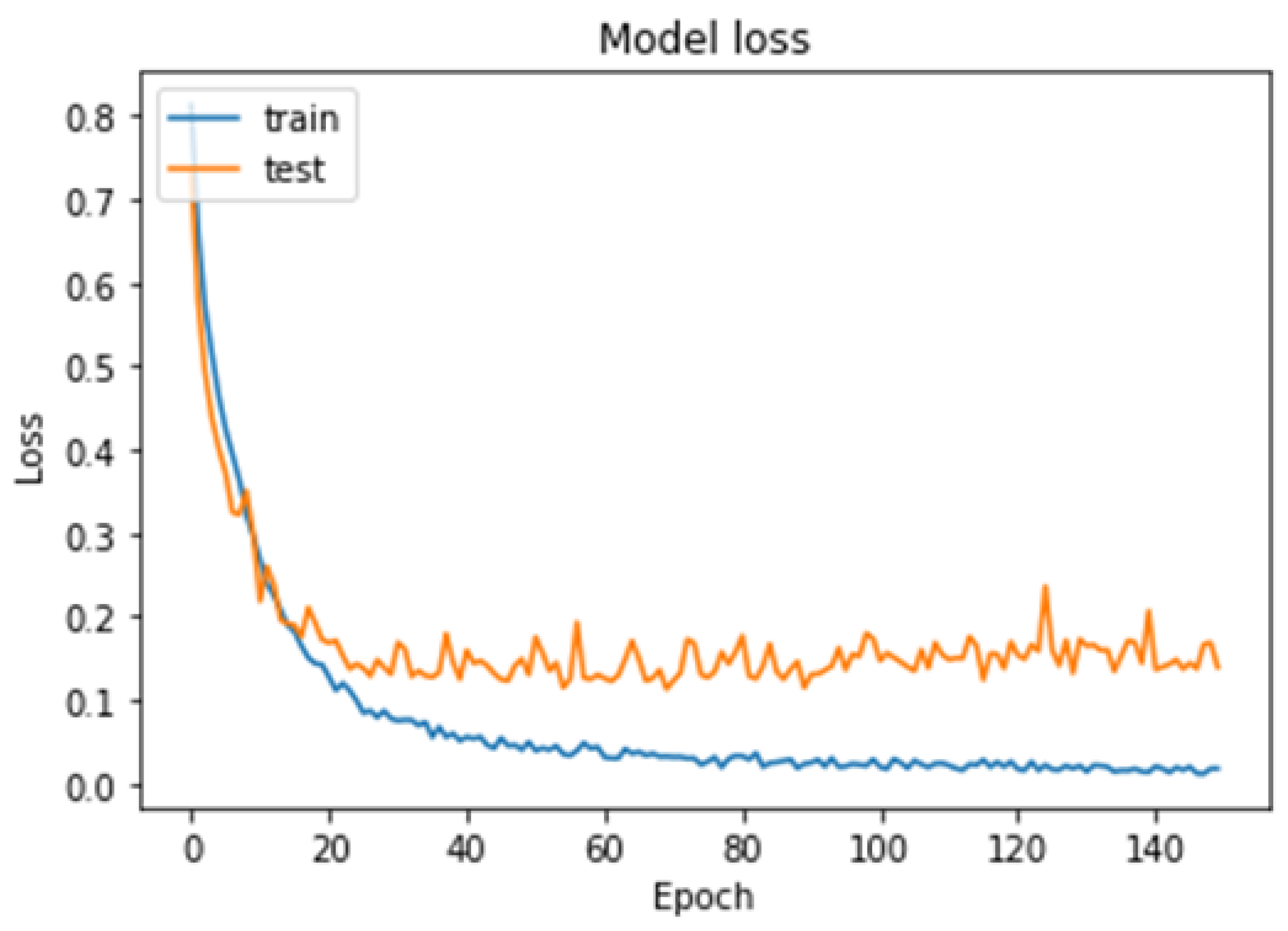
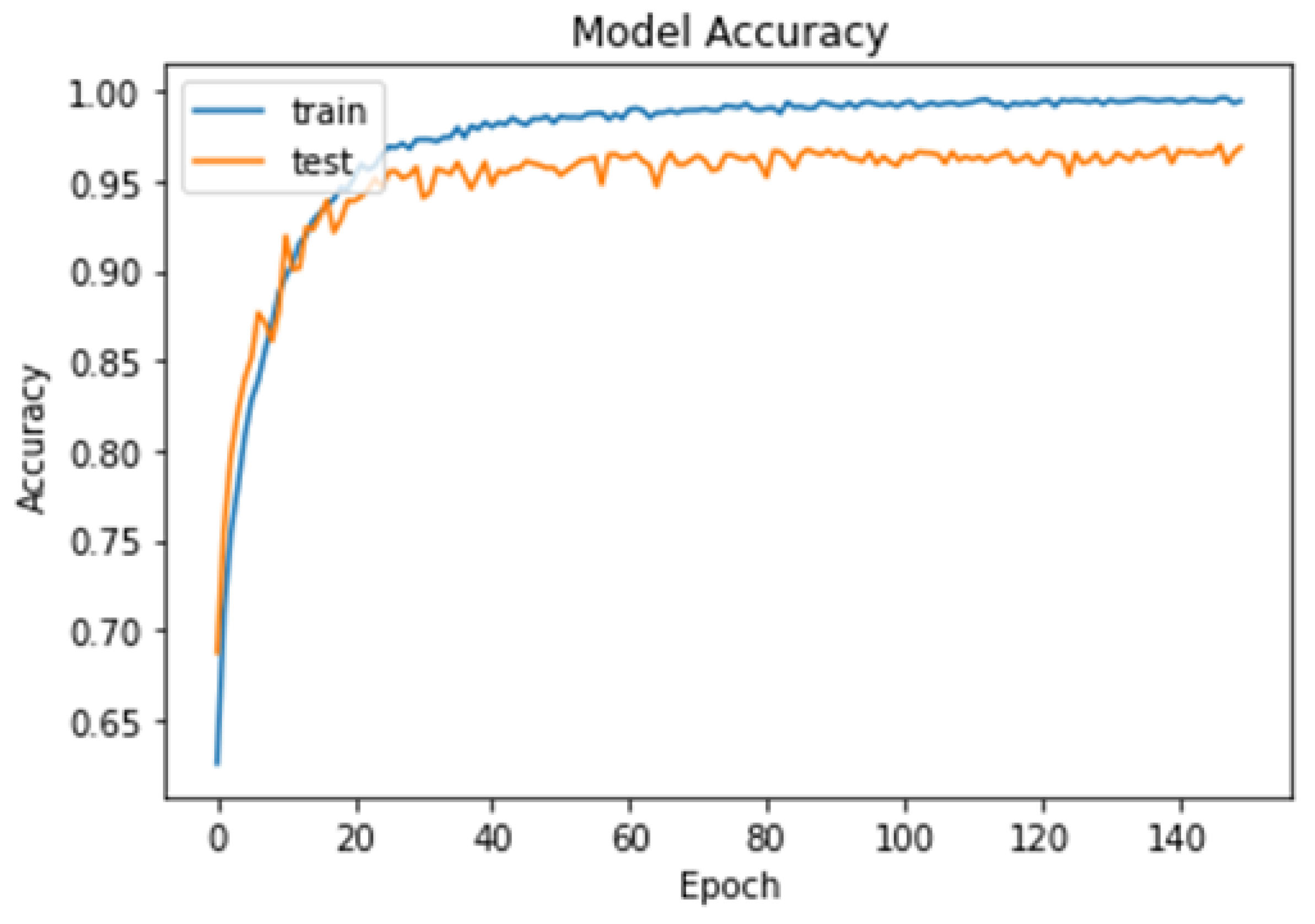
4.1. First Experiment
4.2. Second Experiment
4.3. Third Experiment
- The proposed model achieved high accuracy rates of 99.48% for binary classification and 96.86% for multi-class classification, suggesting that it may be more accurate than previous deep learning models for brain tumor detection.
- The proposed model has a relatively small number of layers (only eight) compared to other deep learning models, which makes it less computationally expensive and more suitable for real-time applications, such as mobile or IoMT devices.
- The proposed model is an end-to-end model, meaning that it can perform feature extraction and classification in a single pipeline, reducing the complexity of the system compared to earlier deep learning models.
- The proposed model was trained and tested using the cross-validation technique, which can help to avoid overfitting and improve the generalizability of the model to new datasets.
- The proposed model can detect multiple types of intracranial tumors, including high-grade glioma, low-grade glioma, meningioma, and pituitary adenoma, making it potentially more versatile than other models that may only detect one type of tumor.
- Despite the advancements in deep learning, brain tumor detection is still a complex problem that requires careful consideration of various factors such as tumor location, shape, size, and enhancement after contrast. The proposed model may not be able to address all these factors and may require further refinement for better accuracy.
- The proposed model has been evaluated on various datasets using cross-validation techniques, but its effectiveness in real-world clinical settings has yet to be fully validated. Additional clinical trials are needed to establish the reliability and accuracy of the model in real-world scenarios.
- The proposed model is designed to detect the most common intracranial tumors such as high-grade glioma, low-grade glioma, meningioma, and pituitary macroadenoma. However, it may not be suitable for detecting other types of brain tumors or diagnosing other neurological conditions.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buckner, J.C.; Brown, P.D.; O’Neill, B.P.; Meyer, F.B.; Wetmore, C.J.; Uhm, J.H. Central nervous system tumors. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2007; Volume 82, pp. 1271–1286. [Google Scholar]
- Saddique, M.; Kazmi, J.H.; Qureshi, K. A hybrid approach of using symmetry technique for brain tumors. Comput. Math. Methods Med. 2014, 2014, 712783. [Google Scholar] [CrossRef] [PubMed]
- Mulhern, R.K.; Merchant, T.E.; Gajjar, A.; Reddick, W.E.; Kun, L.E. Late neurocognitive sequelae in survivors of brain tumours in childhood. Lancet Oncol. 2004, 5, 399–408. [Google Scholar] [CrossRef]
- Omuro, A.M.; Leite, C.C.; Mokhtari, K.; Delattre, J.Y. Pitfalls in the diagnosis of brain tumours. Lancet Neurol. 2006, 5, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Butowski, N.A. Epidemiology and diagnosis of brain tumors. Contin. Lifelong Learn. Neurol. 2015, 21, 301–313. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2018, 43, 98–111. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, Y.; Yang, B.; Hu, S.; Wu, L.; Dhelim, S. Overview of multi-modal brain tumor mr image segmentation. Healthcare 2021, 9, 1051. [Google Scholar] [CrossRef]
- Budati, A.K.; Katta, R.B. An automated brain tumor detection and classification from MRI images using machine learning technique s with IoT. Environ. Dev. Sustain. 2022, 24, 10570–10584. [Google Scholar] [CrossRef]
- Rao, C.S.; Karunakara, K. Efficient detection and classification of brain tumor using kernel based SVM for MRI. Multimed. Tools Appl. 2022, 81, 7393–7417. [Google Scholar] [CrossRef]
- Shinde, A.S.; Mahendra, B.M.; Nejakar, S.; Herur, S.M.; Bhat, N. Performance analysis of machine learning algorithm of detection and classification of brain tumor using computer vision. Adv. Eng. Softw. 2022, 173, 103221. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Marhold, F.; Oberndorfer, S.; Heinz, G.; Buchfelder, M.; Kinfe, T.M.; Meyer-Bäse, A. Radiophysiomics: Brain tumors classification by machine learning and physiological MRI data. Cancers 2022, 14, 2363. [Google Scholar] [CrossRef]
- Jena, B.; Nayak, G.K.; Saxena, S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Mach. Vis. Appl. 2022, 33, 6. [Google Scholar] [CrossRef]
- Sundarasekar, R.; Appathurai, A. Automatic Brain Tumor Detection and Classification Based on IoT and Machine Learning Techniques. Fluct. Noise Lett. 2022, 21, 2250030. [Google Scholar] [CrossRef]
- Vankdothu, R.; Hameed, M.A.; Fatima, H. A brain tumor identification and classification using deep learning based on CNN-LSTM method. Comput. Electr. Eng. 2022, 101, 107960. [Google Scholar] [CrossRef]
- Aamir, M.; Rahman, Z.; Dayo, Z.A.; Abro, W.A.; Uddin, M.I.; Khan, I.; Imran, A.S.; Ali, Z.; Ishfaq, M.; Guan, Y.; et al. A deep learning approach for brain tumor classification using MRI images. Comput. Electr. Eng. 2022, 101, 108105. [Google Scholar] [CrossRef]
- Hashemzehi, R.; Mahdavi, S.J.S.; Kheirabadi, M.; Kamel, S.R. Detection of brain tumors from MRI images base on deep learning using hybrid model CNN and NADE. Biocybern. Biomed. Eng. 2020, 40, 1225–1232. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Maitra, M. MRI-based brain tumor image detection using CNN based deep learning method. Neurosci. Inform. 2022, 100060. [Google Scholar] [CrossRef]
- Younis, A.; Qiang, L.; Nyatega, C.O.; Adamu, M.J.; Kawuwa, H.B. Brain tumor analysis using deep learning and VGG-16 ensembling learning approaches. Appl. Sci. 2022, 12, 7282. [Google Scholar] [CrossRef]
- Rasool, M.; Ismail, N.A.; Boulila, W.; Ammar, A.; Samma, H.; Yafooz, W.M.; Emara, A.H.M. A Hybrid Deep Learning Model for Brain Tumour Classification. Entropy 2022, 24, 799. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A deep learning-based framework for automatic brain tumors classification using transfer learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]
- Muhammad, K.; Khan, S.; Del Ser, J.; De Albuquerque, V.H.C. Deep learning for multigrade brain tumor classification in smart healthcare systems: A prospective survey. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 507–522. [Google Scholar] [CrossRef]
- Figshare Brain Tumor Dataset. 2021. Ashwani Rathee. Available online: https://www.kaggle.com/datasets/ashkhagan/figshare-brain-tumor-dataset (accessed on 25 February 2023).
- Alves, A.F.F.; Miranda, J.R.A.; Reis, F.; de Souza, S.A.S.; Alves, L.L.R.; Feitoza, L.M.; de Castro, J.T.S.; de Pina, D.R. Inflammatory lesions and brain tumors: Is it possible to differentiate them based on texture features in magnetic resonance imaging? J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20200011. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, G.R.; Brusini, L.; Bajrami, A.; Pizzini, F.B.; Calabrese, M.; Reis, F.; Appenzeller, S.; Menegaz, G.; Rittner, L. Diffusion MRI and silver standard masks to improve CNN-based thalamus segmentation. In Medical Imaging 2021: Image Processing; SPIE: Bellingham, WA, USA, 2021; Volume 11596, pp. 692–698. [Google Scholar]
- Amin, J.; Sharif, M.; Raza, M.; Saba, T.; Sial, R.; Shad, S.A. Brain tumor detection: A long short-term memory (LSTM)-based learning model. Neural Comput. Appl. 2020, 32, 15965–15973. [Google Scholar] [CrossRef]
- Albraikan, A.A.; Nemri, N.; Alkhonaini, M.A.; Hilal, A.M.; Yaseen, I.; Motwakel, A. Automated Deep Learning Based Melanoma Detection and Classification Using Biomedical Dermoscopic Images. Comput. Mater. Contin. 2023, 74, 2443–2459. [Google Scholar] [CrossRef]
- Almustafa, K.M.; Sharma, A.K.; Bhardwaj, S. STARC: Deep learning Algorithms’ modelling for STructured analysis of retina classification. Biomed. Signal Process. Control 2023, 80, 104357. [Google Scholar] [CrossRef]
- Brain Tumor Dataset. 2022. Vishwa Patel. Available online: https://www.kaggle.com/datasets/vishwapatel10/brain-tumor-dataset (accessed on 25 February 2023).
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, W.; Huang, M.; Huang, W.; Jiang, J.; Zhou, Y.; Yang, R.; Zhao, J.; Feng, Y.; Feng, Q.; et al. Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation. PLoS ONE 2016, 11, e0157112. [Google Scholar] [CrossRef]
- Parvat, A.; Chavan, J.; Kadam, S.; Dev, S.; Pathak, V. A survey of deep-learning frameworks. In Proceedings of the 2017 International Conference on Inventive Systems and Control (ICISC), Coimbatore, India, 19–20 January 2017; IEEE: Piscataway, NJ, USA; pp. 1–7. [Google Scholar]
- Zhang, Z. Improved adam optimizer for deep neural networks. In Proceedings of the 2018 IEEE/ACM 26th International Symposium on Quality of Service (IWQoS), Banff, AB, Canada, 4–6 June 2018; IEEE: Piscataway, NJ, USA; pp. 1–2. [Google Scholar]
- Ho, Y.; Wookey, S. The real-world-weight cross-entropy loss function: Modeling the costs of mislabeling. IEEE Access 2019, 8, 4806–4813. [Google Scholar] [CrossRef]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Seo, H.; Badiei Khuzani, M.; Vasudevan, V.; Huang, C.; Ren, H.; Xiao, R.; Jia, X.; Xing, L. Machine learning techniques for biomedical image segmentation: An overview of technical aspects and introduction to state-of-art applications. Med. Phys. 2020, 47, e148–e167. [Google Scholar] [CrossRef]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep learning for brain MRI segmentation: State of the art and future directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; AlRuwaili, R.; Alruwaili, M.; Ali, N.H.; Albarrak, A.M. Brain tumor/mass classification framework using magnetic-resonance-imaging-based isolated and developed transfer deep-learning model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef] [PubMed]
- Masood, M.; Maham, R.; Javed, A.; Tariq, U.; Khan, M.A.; Kadry, S. Brain MRI analysis using deep neural network for medical of internet things applications. Comput. Electr. Eng. 2022, 103, 108386. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Khan, M.S.; Khan, W.; Hassan, I.; Obayya, M.; Negm, N.; Salama, A.S. An Effective Approach to Detect and Identify Brain Tumors Using Transfer Learning. Appl. Sci. 2022, 12, 5645. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Raza, M.; Saba, T.; Anjum, M.A. Brain tumor detection using statistical and machine learning method. Comput. Methods Programs Biomed. 2019, 177, 69–79. [Google Scholar] [CrossRef]




| Class | Normal | Brain Tumor |
|---|---|---|
| Normal | 3736 | 14 |
| Brain tumor | 27 | 4119 |
| Metrics | Precision | Recall | F1-Score |
|---|---|---|---|
| Normal | 0.99 | 1.00 | 0.99 |
| Brain tumor | 1.00 | 0.99 | 1.00 |
| Accuracy | 0.99 | ||
| Macro avg | 0.99 | 0.99 | 0.99 |
| Weighted avg | 0.99 | 0.99 | 0.99 |
| Accuracy for testing | 99.48% | ||
| Class | Meningioma | Glioma | Pituitary |
|---|---|---|---|
| Meningioma | 971 | 71 | 20 |
| Glioma | 39 | 2092 | 8 |
| Pituitary | 1 | 5 | 1389 |
| Metrics | Precision | Recall | F1-Score |
|---|---|---|---|
| Meningioma | 0.96 | 0.91 | 0.94 |
| Glioma | 0.96 | 0.98 | 0.97 |
| Pituitary | 0.98 | 1.00 | 0.99 |
| Accuracy | 0.97 | ||
| Macro avg | 0.97 | 0.96 | 0.97 |
| Weighted avg | 0.97 | 0.97 | 0.97 |
| Accuracy for testing | 96.86% | ||
| Study | Methodology | Performance | Limitations | How Our Model Overcomes Limitations |
|---|---|---|---|---|
| Vankdothu et al. [14] | CNN+LSTM | 92% overall accuracy for multi-class classification of normal, glioma, meningioma, and pituitary adenoma | Traditional computer vision techniques used for pre-processing and feature extraction | Our model uses end-to-end deep learning without the need for traditional techniques |
| Aamir et al. [15] | Hybrid deep learning model | 98.83% overall classification accuracy for three classes of brain tumor | Requires a separate algorithm for generating locations and ROI alignment | Our model uses a single end-to-end deep learning model for classification and localization without the need for a separate algorithm |
| Hashemzehi et al. [16] | CNN+NADE | 95% overall accuracy for three-class brain tumor detection | Relatively low accuracy compared to other models | Our model uses an attention-based mechanism to improve accuracy |
| Chattopadhyay and Maitra [17] | CNN+SVM | 99.74% overall accuracy for binary classification of normal and abnormal brain images | Only binary classification performed | Our model performs multi-class classification |
| Younis et al. [18] | Ensemble learning with CNN+VGG-16 | 98.41% accuracy for distinguishing between tumor and healthy brain images | No clear explanation of image processing techniques used | Our model uses a combination of attention and data augmentation techniques to improve performance |
| Rasool et al. [19] | Deep learning (Google-Net)+SVM | 94.12% overall accuracy for multi-class classification of brain tumors | Relatively low accuracy compared to other models | Our model uses a combination of attention and data augmentation techniques to improve accuracy |
| Our model | End-to-end learning using CNN | 96.86% overall accuracy for multi-class classification | Requires further refinement for better accuracy to classify other types | Our model uses end-to-end learning and eliminates the need for traditional pre-processing, feature extraction, and separate classification |
| 99.48% overall accuracy for binary classification |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammad, M.; ElAffendi, M.; Ateya, A.A.; Abd El-Latif, A.A. Efficient Brain Tumor Detection with Lightweight End-to-End Deep Learning Model. Cancers 2023, 15, 2837. https://doi.org/10.3390/cancers15102837
Hammad M, ElAffendi M, Ateya AA, Abd El-Latif AA. Efficient Brain Tumor Detection with Lightweight End-to-End Deep Learning Model. Cancers. 2023; 15(10):2837. https://doi.org/10.3390/cancers15102837
Chicago/Turabian StyleHammad, Mohamed, Mohammed ElAffendi, Abdelhamied A. Ateya, and Ahmed A. Abd El-Latif. 2023. "Efficient Brain Tumor Detection with Lightweight End-to-End Deep Learning Model" Cancers 15, no. 10: 2837. https://doi.org/10.3390/cancers15102837
APA StyleHammad, M., ElAffendi, M., Ateya, A. A., & Abd El-Latif, A. A. (2023). Efficient Brain Tumor Detection with Lightweight End-to-End Deep Learning Model. Cancers, 15(10), 2837. https://doi.org/10.3390/cancers15102837









